Abstract
Aspergillus fumigatus is a model fungal pathogen and a common cause of severe infections and diseases. CD8+ T cells are present in the human and murine T-cell repertoire to the fungus. However, CD8+ T-cell function in infection and the molecular mechanisms that control their priming and differentiation into effector and memory cells in vivo remain elusive. In the present study, we report that both CD4+ and CD8+ T cells mediate protective memory responses to the fungus contingent on the nature of the fungal vaccine. Mechanistically, class I MHC-restricted, CD8+ memory T cells were activated through TLR3 sensing of fungal RNA by cross-presenting dendritic cells. Genetic deficiency of TLR3 was associated with susceptibility to aspergillosis and concomitant failure to activate memory-protective CD8+ T cells both in mice and in patients receiving stem-cell transplantations. Therefore, TLR3 essentially promotes antifungal memory CD8+ T-cell responses and its deficiency is a novel susceptibility factor for aspergillosis in high-risk patients.
Introduction
TLR3 plays a key role in modulating inflammation and innate immunity in the airway. Although best known for recognition of viral double-stranded RNA (dsRNA) and its synthetic analog polyinosinic:polycytidylic acid [poly(I:C)],1 TLR3 also recognizes endogenous ligands,2 including heterologous RNA released from or associated with necrotic cells or generated by in vitro transcription.3 Therefore, TLR3, together with other intracellular signaling proteins,4 induces or otherwise modulates innate immune responses and inflammation in settings that are not associated with viral dsRNA. TLR3 signaling may also modulate adaptive immune responses by providing cross-priming of cytotoxic T lymphocytes through signaling in dendritic cells (DCs)5,6 via type I IFNs7 and in the absence of CD4+ T-cell help.8 Therefore, it is not surprising that people with mutations in key TLR3 signaling components have a selective immunodeficiency manifested by recurrent episodes of herpes simplex virus 1 encephalitis9,10 or enteroviral myocarditis/cardiomyopathy.11
Aspergillus fumigatus is a model fungal pathogen and a common cause of severe infections and diseases. Humans inhale hundreds of conidia every day without adverse consequences,12 except for a minority of persons in whom defense systems fail and a life-threatening form of disease can develop. CD4+ and CD8+ T cells are present in the human T-cell repertoire to the fungus13-15 and adoptive transfer of A fumigatus–specific CD4+ T cells conferred protection against invasive fungal infection.15,16 Recent studies indicate a role for TLR3 in murine aspergillosis. By functioning as an endogenous sensor of fungal RNA,17 TLR3 mediates expression of the enzyme indoleamine 2,3-dioxygenase (IDO) on both epithelial cells18 and DCs,19 contributing to the local regulation of innate and adaptive inflammation to the fungus. However, the findings that protective memory CD8+ T cells are induced against fungi20-23 suggest a possible role for TLR3 in activating memory CD8+ T cell–mediated immunity to the fungus.
We evaluated the contribution of TLR3 to the activation of CD8+ T cells to A fumigatus in preclinical models of aspergillosis and in patients receiving hematopoietic stem cell transplantations (HSCTs) with a single nucleotide polymorphism (SNP) in TLR3. We found that TLR3 on murine and human DCs sorts fungal RNA for activation of class I–restricted protective memory CD8+ T-cell responses to the fungus. TLR3 deficiency was associated with severe infection in mice, and a TLR3 SNP resulting in a loss-of-function phenotype of DCs was associated with increased susceptibility to aspergillosis and concomitant failure to activate antifungal CD8+ T cells in HSCT patients.
Methods
Mice
Female C57BL/6 mice (Charles River Laboratories) and homozygous Tlr3−/− mice from the C57BL/6 background were bred under pathogen-free conditions in the animal facility of the University of Perugia (Perugia, Italy), and experiments were performed according to the Italian Approved Animal Welfare Assurance Act A-3143-01.
Fungal and bacterial strains, infections, and treatments
Viable resting and swollen conidia from the A fumigatus Af293 strain were obtained as described previously.19 A green fluorescent protein (GFP)–expressing strain of A fumigatus (provided by M. M. Moore, Department of Biologic Sciences, Simon Fraser University, Burnaby, BC) was used to track viable fungi. Infection and bronchoalveolar lavage morphometry (at least 200 cells per cytospin preparation were counted) were performed as described previously.19 Mice were monitored for fungal growth (CFUs/organ, mean ± SE), histopathology (periodic acid-Schiff and Gomori's methenamine silver staining of lung tissue sections) and lung immunofluorescence. Histology sections and cytospin preparations were observed using a BX51 microscope (Olympus) and images were captured using a high-resolution DP71 camera (Olympus). Poly(I:C) (Sigma-Aldrich) was given (50 μg) IP twice the day of and the day after infection. Control mice received PBS. Mice were treated with 300 μg of anti-CD4 (GK 1.5) or anti-CD8 (YTS 169) mAbs (provided by L. Boon, Bioceros BV. The Netherlands) the day before and 1 and 3 days after infection. Control mice received isotype control rat IgG2a mAb (eBR2a; eBioscience). Depletion of the corresponding T-cell subsets with this regimen was monitored in each experiment and was consistently between 95% and 98%, lasting for at least 3 days after treatment (data not shown). Mice were infected intranasally with the Pseudomonas aeruginosa PAO1 (ATCC BAA-47) strain. Bacterial CFUs/organ (mean ± SE) were quantified by plating lung homogenates on trypticase soy agar plates.
Vaccination models
Two vaccination models were used, one using live A fumigatus conidia or purified fungal Ags given with murine CpG oligodeoxynucleotide 1862, and the other using fungus-pulsed DCs.24 In the first model, mice were injected intranasally with 2 × 107 conidia/20 μL of saline 14 days before the infection or with 5 μg of the A fumigatus cell wall glucanase Crf1p, which is known to induce MHC class II–restricted memory antifungal CD4+ Th1 responses13,25 together with 10nM CpG in 20 μL of saline administered 14, 7, and 3 days before the infection. Mice were immunosuppressed IP with 150 mg/kg of cyclophosphamide 1 day before the infection. In the adoptive transfer model, purified lung CD11c+ DCs were pulsed with viable resting conidia or fungal RNA together with the cationic lipid N-[1-(2,3-dioleoyloxypropyl]-N,N,N,-trimethylammonium methylsulfate (DOTAP; Boehringer Mannheim) in serum Opti-MEM medium (Life Technologies) for 2 hours, as described previously.26 For total RNA isolation, mixing with DOTAP and detection inside the cells, see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). DCs were adoptively transferred by IP injection (5 × 105 cells/each injection) twice, a week apart, before the intranasal infection. For vaccination with fungal RNA, 10 μg fungal RNA + 20 μg DOTAP were administered intranasally 3 times as above before the infection. A total of 300 μg of anti-CD4, anti-CD8, anti–MHC class I-A (34-5-8S), anti–MHC class II (M5/114) mAbs (provided by L. Boon) or control rat IgG2a mAb was administered along with the vaccines.
Fluorescence microscopy and morphometric analysis
Mice were perfused with 2 mL of cold PBS and 1:1 OCT (Cryomount Histolab). The lung was snap-frozen in OCT and 5-μm sections were cut with a semiautomatic cryostat (MC4000; Histo-Line Laboratories). Rehydrated sections were fixed in 4% paraformaldehyde, blocked with 5% BSA, and stained with a combination of Alexa Fluor 488 anti–mouse CD11c (N418; BioLegend) and anti–mouse CD3 (17A2; BioLegend) mAbs at room temperature for 1 hour. 4′-6-Diamino-2-phenylindole (Molecular Probes) was used to counterstain tissues and to detect nuclei. Images were acquired using a fluorescence microscope (BX51; Olympus) with a 40× objective and the Cell∧P software Version 3.3 build 2108 (Olympus).
Generation of BM chimeras
BM chimeras were generated as described previously18 and as detailed in supplemental Methods.
Cell preparation and cultures
Murine lung cells were isolated as described previously24 and as detailed in supplemental Methods. CD4+ or CD8+ T cells were isolated after incubation with FITC-labeled anti-CD4 or anti-CD8, followed by anti-FITC MicroBeads (Miltenyi Biotec) and DCs were isolated with MicroBeads (Miltenyi Biotec) conjugated to hamster anti–mouse CD11c mAbs (N418) before magnetic cell sorting.24 After written informed consent, human PBMCs were isolated using Histopaque-1077 (Sigma-Aldrich) according to the manufacturer's instructions. Human CD141+ (BDCA-3)+ DCs were isolated from total PBMCs using the CD141 (BDCA-3) MicroBead Kit (Miltenyi Biotec) according to the manufacturer's instructions (see supplemental Methods for purity and phenotypic analysis). Murine or human cells (106 cells/mL) were pulsed with poly(I:C) (50 μg/mL), viable conidia of A fumigatus (5:1 cell:fungus ratio), fungal RNA (10 μg), and DOTAP (20 μg) or DOTAP alone for 18 hours before RNA extraction and flow cytometry. Amphotericin B (Sigma-Aldrich; 2.5 μg/mL) was added after 2 hours of pulsing to prevent fungal overgrowth.
Flow cytometry
Directly conjugated Abs were purchased from BD Pharmingen (see the supplemental Methods for details). Cells were analyzed with a FACScan flow cytofluorometer (BD Biosciences) equipped with CellQuest Version 5.1 software (BD Biosciences).
Uptake tracking of GFP-expressing A fumigatus in vivo
Single-cell suspensions were prepared from lungs or draining lymph nodes depleted of T and B cells. Cells were immunostained with the CD11c markers and analyzed by flow cytometry. For a description of the phagocytosis assay, see supplemental Methods.
Ag presentation in vitro and in cytotoxic and conidiocidal assays
A panel of Ag-presentation inhibitors (all from Sigma-Aldrich) at their optimum concentrations was added to DCs for 120 minutes before the 2-hour pulsing with A fumigatus conidia or RNA + DOTAP. We used 5 μg/mL of chloroquine, 100nM bafilomycin A, 5 μg/mL of brefeldin A, and 50μM lactacystin. After pulsing, DCs were fixed in 0.4% paraformaldehyde (Sigma-Aldrich) and extensively washed. DCs (1 × 105) were incubated with CD4+ or CD8+ T cells (5 × 105) purified from lungs 1 week after infection for lymphoproliferation (after 72 hours of incubation), gene expression, supernatant collection, and cytolytic activity (after 24 hours of incubation). DNA synthesis was measured by 3H-thymidine labeling (Amersham Biosciences) for 6 hours. Cytolytic activity was tested using a standard 51Cr-release assay15 (see supplemental Methods for details). Live or heat-inactivated (85°C for 30 minutes) conidia of A fumigatus were labeled with FUN-1 (5μM; Molecular Probes). Because lymphocytes take up FUN-1, cells could not be added directly in this assay. FUN-1–stained live conidia were incubated overnight in culture medium alone or with medium from naive CD8+ T cells or cells from infected mice either unexposed or exposed to DCs pulsed with conidia or fungal RNA, and were examined under fluorescence microscopy. Metabolically active conidia accumulate orange fluorescence in vacuoles and dormant and dead conidia stain green.
Proliferative activity of human T-lymphocyte clones
Limiting dilution and proliferation assays were performed as described previously15 and as detailed in supplemental Methods.
Western blotting
Blots of cell lysates were incubated with rabbit polyclonal Abs recognizing phospho-IRF3 (Ser396, 4D4G) followed by HRP-conjugated anti–mouse or anti–rabbit IgG, as per the manufacturer's instructions (Cell Signaling Technology). Immunoblotting for IDO was performed with rabbit polyclonal IDO-specific Ab on total lung cells.18 Scanning densitometry was done on a Scion Image apparatus. The pixel density of bands was normalized against total protein or β-tubulin.
ELISA and real-time PCR
The level of cytokines in culture supernatants of DCs was determined with an ELISA kit (R&D Systems) at 24 hours. The detection limits of the assays were < 10 pg/mL for IL-12p70, < 3 pg/mL for IL-10, < 30 pg/mL for IL-23, < 10 pg/mL for IL-17A, and < 10 pg/mL for IFN-γ. Cell lysis, RNA extraction, and real-time PCR were performed as described previously.18,19 Amplification efficiencies were validated and normalized against Gapdh or ACTB. Each data point was examined for integrity by analysis of the amplification plot. The mRNA-normalized data were expressed as relative mRNA expression in treated cells compared with that of unstimulated cells. For details, see supplemental Methods.
Patients
The genetic study included 223 patients undergoing allogeneic HSCT at the University of Perugia (Perugia, Italy) between 2003 and 2010 and their respective donors. Patient characteristics are summarized in supplemental Table 1. Grafts consisted of immunoselected CD34+ peripheral blood cells in all patient, and transplantation procedures, antifungal prophylaxis, and surveillance for fungal infection were performed as described previously27 and as detailed in supplemental Methods. Probable/proven fungal infection was defined according to the revised standard criteria from the European Organization for Research and Treatment of Cancer/Mycology Study Group. Study approval was provided by the local ethics committee and informed written consent was obtained from all participants in accordance with the Declaration of Helsinki.
SNP genotyping
SNPs were selected from a literature review and public databases based on 3 selection criteria: (1) published evidence of association with human diseases; (2) localization to the promoter, untranslated, or coding regions; and (3) minor allele frequencies higher than 5% in the white population. Two TLR3 SNPs complied with the selection criteria: +1234C/T (L412F, rs3775291) and +95C/A (rs3775296). Genotyping was performed as described previously27 (for details and primer sequences, see supplemental Methods).
Statistical analysis
The Student t test or ANOVA with Bonferroni adjustment was used to determine statistical significance (P < .05). Data reported are either from 1 of 3-5 independent experiments (Western blotting and RT-PCR) or are pooled from 3-5 experiments. The in vivo groups consisted of 6-8 mice/group. Data were analyzed with Prism Version 4.03 software (GraphPad). For details on the statistical analysis of genetic associations, see supplemental Methods.
Results
Tlr3−/− mice are highly susceptible to pulmonary aspergillosis
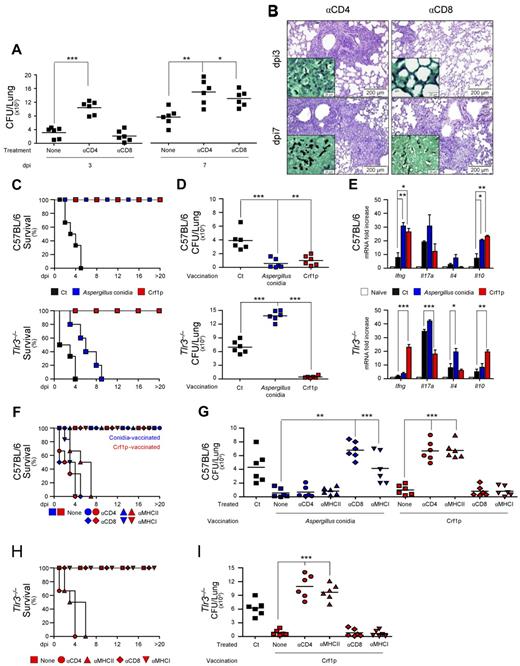
We evaluated resistance to primary pulmonary aspergillosis and reinfection in Tlr3−/− and C57BL/6 mice. In the primary infection, fungal growth was higher in the lungs of Tlr3−/− than control mice throughout the course of the infection (Figure 1A) and was associated with higher and persistent neutrophil recruitment and infiltration in the lung parenchyma and bronchoalveolar lavage fluids (Figure 1B inset shows numerous hyphae) and the presence of peribronchiolar lymphocyte infiltrations (Figure 1B arrows and immunofluorescence staining). Fungal dissemination was also observed in the brains of Tlr3−/− mice (Figure 1A), more than 85% of which survived the infection (data not shown) if not immunosuppressed (Figure 2C). Stimulating TLR3 with the poly(I:C) agonist significantly reduced both the fungal growth and inflammation in the lungs of C57BL/6 mice (supplemental Figure 1A-B), a finding further supporting that TLR3 provides protection in infection. Gene-expression analysis of the lungs confirmed the higher and more persistent inflammatory response in Tlr3−/− mice than control mice, as revealed by the higher mRNA expression of Cxcl1, Cxcl2, and Mpo genes and of genes for inflammatory cytokines such as Il1b (Figure 1C). The levels of Ifna1 and Ifnb1 were lower in Tlr3−/− mice (Figure 1C). In addition, the conidiocidal activity of lung cells was half reduced in Tlr3−/− mice compared with the C57BL/6 control mice (data not shown). Therefore, TLR3 contributes to the effector activity of lung cells and to protection from unintended inflammation.
Tlr3−/− mice are highly susceptible to pulmonary aspergillosis. C57BL/6 or Tlr3−/− mice were infected intranasally with live A fumigatus conidia (6-8 mice/group). (A) Fungal growth (mean CFUs ± SE) in the lungs and brains of infected mice was assessed at different days postinfection (dpi). (B) Lung histology (periodic acid-Schiff staining), bronchoalveolar lavage morphometry, expressed as a percentage (mean ± SD) of mononuclear cells (MNCs) or polymorphonuclear (PMN) cells, and immunofluorescence of infected mice at different dpi. Note the sustained inflammatory cell recruitment in the lungs and bronchoalveolar lavage (May-Grünwald Giemsa staining in the inset using a 100×/1.3 oil objective) of Tlr3−/− mice and the presence of peribronchiolar lymphocyte infiltrates (at 3 dpi; arrows in periodic acid-Schiff staining and immunofluorescence images). Cell-surface markers used for DC and T-cell visualization were CD11c (green, Alexa Fluor 488) and CD3 epsilon followed by PE-secondary Ab (Red-647), respectively. Cell nuclei were stained blue with 4′,6-diamidino-2-phenylindole and then mounted with Vectashield mounting medium (Vector Laboratories). Representative images of 2 independent experiments were visualized with a 40×/0.75 objective. All images were captured on the BX51 upright microscope (Olympus) using a high-resolution Microscopy Olympus DP71 camera (Olympus) and digitally acquired using the Cell∧P software (Version 3.2 build 2108, Olympus). Bars in histology sections indicate magnifications. (C) Lung total RNA from infected mice was extracted from naive (0) or at 1, 3, and 7 dpi, and the relative expression of Cxcl1, Cxcl2, Mpo, Il1b, Ifna1, and Ifnb1 was assessed by RT-PCR. (D) Total RNA was extracted from purified CD4+ T cells from the draining lymph nodes of naive or infected mice at 7 dpi, and the relative expression of Tbet, Rorc, and Foxp3 was assessed by RT-PCR. (E) Total lung homogenates at 3 and 7 dpi were assessed for levels of IFN-γ, IL-17A and IL-10 by specific ELISA (mean values ± SD). Data are pooled from 3 independent experiments. *P < .05; **P < .01; and ***P < .001 for infected versus naive mice (panel D) or Tlr3−/− vs C57BL/6 mice panel (panels A, C, and E).
Tlr3−/− mice are highly susceptible to pulmonary aspergillosis. C57BL/6 or Tlr3−/− mice were infected intranasally with live A fumigatus conidia (6-8 mice/group). (A) Fungal growth (mean CFUs ± SE) in the lungs and brains of infected mice was assessed at different days postinfection (dpi). (B) Lung histology (periodic acid-Schiff staining), bronchoalveolar lavage morphometry, expressed as a percentage (mean ± SD) of mononuclear cells (MNCs) or polymorphonuclear (PMN) cells, and immunofluorescence of infected mice at different dpi. Note the sustained inflammatory cell recruitment in the lungs and bronchoalveolar lavage (May-Grünwald Giemsa staining in the inset using a 100×/1.3 oil objective) of Tlr3−/− mice and the presence of peribronchiolar lymphocyte infiltrates (at 3 dpi; arrows in periodic acid-Schiff staining and immunofluorescence images). Cell-surface markers used for DC and T-cell visualization were CD11c (green, Alexa Fluor 488) and CD3 epsilon followed by PE-secondary Ab (Red-647), respectively. Cell nuclei were stained blue with 4′,6-diamidino-2-phenylindole and then mounted with Vectashield mounting medium (Vector Laboratories). Representative images of 2 independent experiments were visualized with a 40×/0.75 objective. All images were captured on the BX51 upright microscope (Olympus) using a high-resolution Microscopy Olympus DP71 camera (Olympus) and digitally acquired using the Cell∧P software (Version 3.2 build 2108, Olympus). Bars in histology sections indicate magnifications. (C) Lung total RNA from infected mice was extracted from naive (0) or at 1, 3, and 7 dpi, and the relative expression of Cxcl1, Cxcl2, Mpo, Il1b, Ifna1, and Ifnb1 was assessed by RT-PCR. (D) Total RNA was extracted from purified CD4+ T cells from the draining lymph nodes of naive or infected mice at 7 dpi, and the relative expression of Tbet, Rorc, and Foxp3 was assessed by RT-PCR. (E) Total lung homogenates at 3 and 7 dpi were assessed for levels of IFN-γ, IL-17A and IL-10 by specific ELISA (mean values ± SD). Data are pooled from 3 independent experiments. *P < .05; **P < .01; and ***P < .001 for infected versus naive mice (panel D) or Tlr3−/− vs C57BL/6 mice panel (panels A, C, and E).
Because A fumigatus is sensed by TLR3 on both epithelial cells17 and DCs,19 we determined the relative contribution of both types of cells to TLR3-mediated protection by infecting chimeric mice with TLR3-deficient hematopoietic or nonhematopoietic cells. TLR3 deficiency in myeloid, more so than epithelial cells, greatly impaired resistance to the fungus, as revealed by the inability to control fungal growth and restrict inflammation (supplemental Figure 2). Accordingly, DC cytokines were altered in Tlr3−/− mice: levels of IL-12p70 were lower and those of IL-23 were higher than in C57BL/6 mice (supplemental Figure 3). Therefore, the myeloid-DC compartment is dysfunctional in Tlr3−/− mice, and this would predict altered T-cell priming in infection.
CD8+ T cells do not expand in Tlr3−/− mice after infection
We evaluated the priming of CD4+ or CD8+ T cells in Tlr3−/− or C57BL/6 mice by assessing the expansion of either T-cell subset in infection. Both subsets expanded in the lung and the draining lymph nodes of infected C57BL/6 mice, whereas CD4+ but not CD8+ T cells expanded more in the lung and less in the lymph nodes of Tlr3−/− mice (supplemental Figure 4). Not only were CD8+ T cells defective, but CD4+ Th-cell subsets were also abnormally activated in Tlr3−/− mice, as judged by the decreased expression of Tbet/Foxp3–specific transcripts in cells from draining lymph nodes (Figure 1D), the production of IFN-γ/IL-10 in the lungs (Figure 1E), and the increased expression of the Rorc transcript (Figure 1D) and the corresponding cytokine IL-17A (Figure 1E). Therefore, in addition to being required for the expansion of CD8+ T cells, TLR3 is also required for the proper CD4+ Th balance in infection. Activation of IDO via the TLR3/TRIF pathway confers tolerogenic and Th1-activating potential to lung DCs in murine aspergillosis.18,19 Consistently, the enzyme was not induced in infection in the lung of Tlr3−/− mice despite being expressed at the basal level (supplemental Figure 5). Therefore, through IDO, TLR3 likely contributes to the promotion of protective tolerance to the fungus in infection and to the down-regulation of Th17 responses, as shown previously with other lung infections.28
Tlr3−/− mice fail to develop MHC class I–restricted CD8+ T-cell responses after vaccination
To define the contribution of CD8+ T cells in infection, we evaluated fungal growth and histopathology of infected C57BL/6 mice in conditions of CD4+ or CD8+ T-cell depletion. Both CD4+ and CD8+ T-cell depletion impaired antifungal resistance (Figure 2A-B), but with interesting differences. Unlike CD4+ T-cell depletion, CD8+ T-cell depletion did not adversely affect antifungal resistance early in infection as it did during late infection, as revealed by the increased fungal burden and concomitant inflammatory pathology in the lung. This finding suggests that CD8+ T cells, somehow dispensable in the early phase of the infection, are instead needed for optimal clearance of the fungus late in infection, as demonstrated previously in other fungal infections,22 and points to a role for CD8+ T cells in adaptive antifungal immunity. To assess this role, we resorted to well-established experimental models of vaccine-induced resistance.24 Mice were treated with conidia or the protective recombinant fungal Ag Crf1p,24 along with CpG as an adjuvant, and determined resistance to subsequent infection in terms of survival, fungal growth, and pattern of cytokine gene expression. In contrast to control mice, Tlr3−/− mice failed to develop vaccine-induced resistance in response to conidia, as revealed by their inability to survive infection (Figure 2C), restrict fungal growth (Figure 2D), and produce protective IFN-γ and IL-10 (Figure 2E). Surprisingly, however, these mice developed full resistance after Crf1p vaccination (Figure 2C-E).
Tlr3−/− mice fail to develop MHC class I–restricted CD8+ T-cell responses after vaccination. (A) C57BL/6 mice were treated with a total of 300 μg of anti-CD4 or anti-CD8 Ab the day before or 1 or 3 days after the intranasal infection with live A fumigatus conidia (6-8 mice/group). “None” indicates mice receiving isotype control Ab. Depletion of the corresponding T-cell subsets with this regimen was between 95% and 98% at 3 and 7 days postinfection (dpi). (B) Periodic acid-Schiff staining of the lungs from mice treated in panel A. Note the presence of fungi (Gomori methenamine silver staining in the inset) and the sustained inflammation in the lungs of CD4-depleted mice throughout the infection, as opposed to the increased inflammation and fungal growth only observed at 7 dpi in CD8-depleted mice. Images were visualized with the 40×/0.75 objective on the BX51 upright microscope (Olympus) and captured using a high-resolution Microscopy Olympus DP71 camera (Olympus) and digitally acquired using the Cell∧P software (Version 3.3 build 2108, Olympus). Bars indicate magnifications. Lung histology of untreated mice was similar to those shown in Figure 1. (C) Survival (%), (D) fungal growth (CFUs, mean ± SE), and (E) relative lung expression of Ifng, Il17a, Il4, and Il10 by RT-PCR in C57BL/6 or Tlr3−/− mice vaccinated with A fumigatus conidia 14 days before (blue color) or with the recombinant fungal Ag Crf1p + CpG 14 7 and 3 days (red color) before subsequent intranasal infection with live conidia (6-8 mice/group). Mice were given cyclophosphamide 1 day before the infection. Fungal growth and gene expression were assessed at 3 dpi. CpG alone failed to induce vaccine-induced resistance, as reported previously.24 Survival (F and H) and fungal growth (at 3 dpi; G and I) in mice treated with anti-CD4, anti-CD8, anti–MHC class I, or anti–MHC class II mAbs in concomitance with A fumigatus conidia vaccination (blue color) or Crf1p + CpG vaccination (red color). Treatment with anti-CD4 alone, but not anti-CD8, anti–MHC class I-A, or anti–MHC class II mAbs alone, further increased the susceptibility to the infection in cyclophosphamide-treated mice compared with control mice. Ct indicates infected unvaccinated mice; none, vaccinated mice treated with an isotype control Ab. Bars indicate magnifications. Data are representative of at least 3 independent experiments. *P < .05; **P < .01; and ***P < .001 for vaccinated versus unvaccinated mice (panels D-E) or treated versus untreated mice (panels A, G, and I).
Tlr3−/− mice fail to develop MHC class I–restricted CD8+ T-cell responses after vaccination. (A) C57BL/6 mice were treated with a total of 300 μg of anti-CD4 or anti-CD8 Ab the day before or 1 or 3 days after the intranasal infection with live A fumigatus conidia (6-8 mice/group). “None” indicates mice receiving isotype control Ab. Depletion of the corresponding T-cell subsets with this regimen was between 95% and 98% at 3 and 7 days postinfection (dpi). (B) Periodic acid-Schiff staining of the lungs from mice treated in panel A. Note the presence of fungi (Gomori methenamine silver staining in the inset) and the sustained inflammation in the lungs of CD4-depleted mice throughout the infection, as opposed to the increased inflammation and fungal growth only observed at 7 dpi in CD8-depleted mice. Images were visualized with the 40×/0.75 objective on the BX51 upright microscope (Olympus) and captured using a high-resolution Microscopy Olympus DP71 camera (Olympus) and digitally acquired using the Cell∧P software (Version 3.3 build 2108, Olympus). Bars indicate magnifications. Lung histology of untreated mice was similar to those shown in Figure 1. (C) Survival (%), (D) fungal growth (CFUs, mean ± SE), and (E) relative lung expression of Ifng, Il17a, Il4, and Il10 by RT-PCR in C57BL/6 or Tlr3−/− mice vaccinated with A fumigatus conidia 14 days before (blue color) or with the recombinant fungal Ag Crf1p + CpG 14 7 and 3 days (red color) before subsequent intranasal infection with live conidia (6-8 mice/group). Mice were given cyclophosphamide 1 day before the infection. Fungal growth and gene expression were assessed at 3 dpi. CpG alone failed to induce vaccine-induced resistance, as reported previously.24 Survival (F and H) and fungal growth (at 3 dpi; G and I) in mice treated with anti-CD4, anti-CD8, anti–MHC class I, or anti–MHC class II mAbs in concomitance with A fumigatus conidia vaccination (blue color) or Crf1p + CpG vaccination (red color). Treatment with anti-CD4 alone, but not anti-CD8, anti–MHC class I-A, or anti–MHC class II mAbs alone, further increased the susceptibility to the infection in cyclophosphamide-treated mice compared with control mice. Ct indicates infected unvaccinated mice; none, vaccinated mice treated with an isotype control Ab. Bars indicate magnifications. Data are representative of at least 3 independent experiments. *P < .05; **P < .01; and ***P < .001 for vaccinated versus unvaccinated mice (panels D-E) or treated versus untreated mice (panels A, G, and I).
The ability of TLR3 signaling to provide cross-priming of CD8+ T cells5 in the relative absence of CD4+ T-cell help8 prompted us to evaluate whether, similar to what has been described for other fungi,20,21 exogenous fungal Ags activate memory CD8+ T cells in a MHC class I–restricted manner and in the relative absence of Th cells. For this purpose, we evaluated vaccine-induced resistance in conditions of CD4, CD8, and MHC class I or class II ablation by specific neutralizing Abs, thus preventing residual Th activity of genetically deficient mice. We found that blocking CD4 or class II Ags abrogated the resistance induced by vaccination with Crf1p in both C57BL/6 (Figure 2F-G) and Tlr3−/− (Figure 2H-I) mice, whereas blocking CD8 or class I Ags abrogated the resistance induced by vaccination with conidia in C57BL/6 mice, as revealed by the survival (Figure 2F), fungal growth (Figure 2G), and lung histopathology (supplemental Figure 6). These results indicate that both CD4+ and CD8+ T cells mediate memory responses to the fungus contingent on the nature of the fungal vaccine, and that CD8+-dependent, but not CD4+-dependent, memory responses to the fungus are defective in the absence of TLR3.
Migratory CCR7+ DCs are defective in Tlr3−/− mice
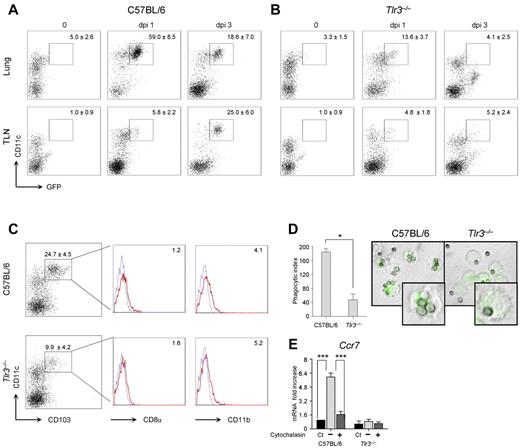
Lung DCs transport A fumigatus conidia or hyphae to the draining lymph nodes, where T-cell priming occurs.29 To assess whether the defective CD8+ T-cell expansion in Tlr3−/−–infected mice could result from defective priming in the draining lymph nodes, we infected Tlr3−/− and C57BL/6 mice with a GFP-expressing strain of A fumigatus and investigated migration of GFP+ DCs from the lung to the draining lymph nodes. We found that a population of CD11chigh DCs expressed GFP early in the lungs of C57BL/6 mice (Figure 3A) and, to a lesser extent, in Tlr3−/− mice in which a population of CD11clow cells (also CD11bhigh, data not shown) expressed GFP (Figure 3B). While still present in the lung, GFP+ DCs also accumulated in the draining lymph nodes of C57BL/6 mice (Figure 3A) but not Tlr3−/− mice (Figure 3B). Therefore, DCs responsible for T-cell priming in the draining lymph nodes are defective in Tlr3−/− mice. This was confirmed by flow cytometry analysis of lung DCs showing a decreased number of CD103+ cells that poorly express both CD8α and CD11b (Figure 3C), as reported previously.30
Migratory CCR7+ DCs are defective in Tlr3−/− mice. C57BL/6 mice (A) or Tlr3−/− mice (B) were infected intranasally with GFP-expressing A fumigatus conidia, and the numbers of GFP+ CD11c+ cells were assessed by flow cytometry on T- and B-cell–depleted lungs or draining lymph nodes at different days postinfection (dpi). Numbers (mean values ± SD, n = 3) refer to the percentage of positive cells on gated CD11c+ cells. Shown is one representative experiment. (C) Phenotype of CD11c+ DCs from naive C57BL/6 or Tlr3−/− mice. Flow cytometry was performed on gated CD11c+ DCs enriched from lungs. Numbers (mean values ± SD, n = 3) refer to the percentage of CD103+ cells. Histograms refer to cells expressing CD8α or CD11b on gated CD103+ cells from one representative experiment. Isotype-matched rat IgG2a (CD8α) or IgG2b (CD11b) control staining is shown in the blue histograms. Phagocytic index (n = 3; D) and relative expression of Ccr7 (RT-PCR, mean values ± SD of at least 3 independent experiments assessed in triplicates; E) by purified lung DCs. Monolayers of DCs were added to live GFP-expressing conidia for 2 hours at 37°C. Cytochalasin D was added for 1 hour at 37°C before phagocytosis. After quenching the fluorescence of bound, uningested conidia with Trypan blue, phagocytosis was quantified via phase contrast and fluorescence microscopy using the 100× objective and the DCs were fixed in 1% paraformaldehyde (shown are representative microscopic images of 2 independent experiments visualized with the 100×/1.3 oil objective on the BX51 upright microscope (Olympus). All images were captured using a high-resolution Microscopy Olympus DP71 camera (Olympus) and digitally acquired using the Cell∧P software (Version 3.3 build 2108, Olympus)). Ct indicates control, unexposed cells.
Migratory CCR7+ DCs are defective in Tlr3−/− mice. C57BL/6 mice (A) or Tlr3−/− mice (B) were infected intranasally with GFP-expressing A fumigatus conidia, and the numbers of GFP+ CD11c+ cells were assessed by flow cytometry on T- and B-cell–depleted lungs or draining lymph nodes at different days postinfection (dpi). Numbers (mean values ± SD, n = 3) refer to the percentage of positive cells on gated CD11c+ cells. Shown is one representative experiment. (C) Phenotype of CD11c+ DCs from naive C57BL/6 or Tlr3−/− mice. Flow cytometry was performed on gated CD11c+ DCs enriched from lungs. Numbers (mean values ± SD, n = 3) refer to the percentage of CD103+ cells. Histograms refer to cells expressing CD8α or CD11b on gated CD103+ cells from one representative experiment. Isotype-matched rat IgG2a (CD8α) or IgG2b (CD11b) control staining is shown in the blue histograms. Phagocytic index (n = 3; D) and relative expression of Ccr7 (RT-PCR, mean values ± SD of at least 3 independent experiments assessed in triplicates; E) by purified lung DCs. Monolayers of DCs were added to live GFP-expressing conidia for 2 hours at 37°C. Cytochalasin D was added for 1 hour at 37°C before phagocytosis. After quenching the fluorescence of bound, uningested conidia with Trypan blue, phagocytosis was quantified via phase contrast and fluorescence microscopy using the 100× objective and the DCs were fixed in 1% paraformaldehyde (shown are representative microscopic images of 2 independent experiments visualized with the 100×/1.3 oil objective on the BX51 upright microscope (Olympus). All images were captured using a high-resolution Microscopy Olympus DP71 camera (Olympus) and digitally acquired using the Cell∧P software (Version 3.3 build 2108, Olympus)). Ct indicates control, unexposed cells.
Because CCR7, which is known to be crucially required for CD8+ T-cell priming in the lymph nodes,31 is up-regulated in DCs after conidia internalization,32 we assessed phagocytosis and Ccr7 expression in Tlr3−/− DCs. We found that, whereas the phagocytosis percentage was not different (data not shown), the ability to internalize more than one conidia was greatly impaired in Tlr3−/− DCs, as revealed by the decreased phagocytic index compared with DCs from C57BL/6 mice (Figure 3D). Tlr3−/− DCs also failed to up-regulate Ccr7 expression after phagocytosis in a manner similar to control DCs after blocking phagocytosis (Figure 3E). Therefore, TLR3 contributes to Ccr7 expression of lung DCs after conidia phagocytosis, likely conditioning their migratory capacity and priming efficiency.
Recognition of fungal RNA is defective in Tlr3−/− DCs
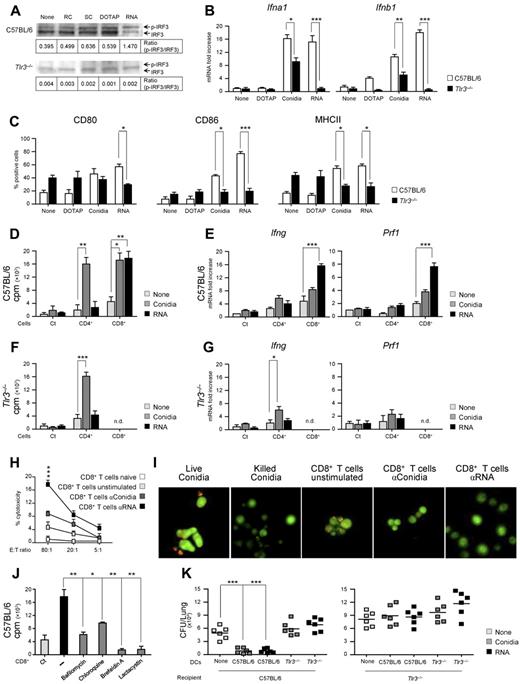
TLR3 is present within endocytic vesicles of cross-presenting DC subsets,6 where it is activated by microbial dsRNA1,33 or endogenous mRNA.2 Fungal RNA is known to activate DCs for vaccine-induced resistance26 and is recognized by TLR3.17 DCs transfected with fungal RNA expressed immunogenic fungal Ags,26 a finding indicating that newly synthesized fungal Ags originate from RNA translation. This was confirmed in experiments in which vaccination of mice with fungal RNA and the cationic lipid DOTAP, which is known to target dsRNA to endosomal TLR3 and to direct Ags into the class I MHC Ag-presentation pathway,34 resulted in the induction of fungus-specific CD8+ T cell–mediated resistance (supplemental Figure 7). Based on these assumptions, we comparatively assessed purified DCs from the lungs of Tlr3−/− and C57BL/6 mice for functional responses to A fumigatus conidia or fungal RNA in terms of signaling events downstream TLR3. Such events included phosphorylation of IFN-regulatory factor 3 (IRF3), cytokine gene expression (type I IFNs), maturation (CD80, CD86, and MHC class II expression), ability to prime Ag-specific CD4+ or CD8+ T cells in vitro, and ability to confer resistance after adoptive transfer in vivo. Fungal RNA potently induced IRF3 phosphorylation in C57BL/6 DCs, an activity also observed with swollen conidia, but not with resting conidia or DOTAP and not in Tlr3−/− DCs (Figure 4A). Fungal RNA also potently activated gene expression of type I IFNs (Figure 4B) and induced DC maturation (Figure 4C) in a TLR3-dependent manner. Although conidia similarly activated DCs, the residual activity observed in Tlr3−/− DCs suggests the participation of other TLRs and/or intracellular RNA sensors in response to conidia. This was confirmed by the ability of conidia-pulsed DCs to prime CD4+ T cells in both C57BL/6 (Figure 4D-E) and Tlr3−/− mice (Figure 4F-G) and, similar to RNA-pulsed DCs, to activate CD8+ T cells. Indeed, conidia-pulsed DCs primed both T-cell subsets from C57BL/6 mice for proliferation (Figure 4D) and Ifng expression (Figure 4E), and primed CD8+ T cells for Prf1 gene expression (Figure 4E). In contrast, RNA-pulsed DCs primed only CD8+ T cells for proliferation (Figure 4D), Ifng and Prf1 gene expression (Figure 4E), and cytolytic activity against fungus-pulsed DCs (Figure 4H) or against the fungus itself (Figure 4I). CD8+ T-cell priming by RNA-pulsed DCs was abrogated in the presence of known inhibitors of the TLR3-dependent Ag-presentation pathway11 (bafilomycin A1 and chloroquine) and of the cytosolic, class I–dependent Ag-presentation pathway35 (brefeldin A and lactacystin; Figure 4J). These findings indicate that both the endosomal/lysosomal and proteasomal degradation pathways are involved in Ag processing after RNA transfection. Finally, fungus- or RNA-pulsed DCs isolated from Tlr3−/− mice failed to confer protection after adoptive transfer in vivo in C57BL/6 mice. However, fungus- or RNA-pulsed DCs from C57BL/6 mice also failed to confer protection after transfer in Tlr3−/− mice (Figure 4K), a finding suggesting that host DCs cross-present fungal Ags from migratory donor DCs, as proposed previously.36
Recognition of fungal RNA is defective in Tlr3−/− DCs. C57BL/6 or Tlr3−/− lung DCs from naive mice were left unpulsed (None) or were pulsed with resting (RC) or swollen (SC) A fumigatus conidia, DOTAP alone, or fungal RNA + DOTAP for 2 hours. (A) Phosphorylation of IRF3 (p-IRF3) in DCs pulsed as above. Shown is a representative Western blot of 3 independent experiments. The IRF3 bands were quantified and the ratios of phosphorylated to total IRF3 are shown. (B) Relative expression of Ifna1 and Ifnb1 (RT-PCR) and (C) expression of CD80, CD86, and MHC class II (flow cytometry) on pulsed DCs (mean values ± SD of at least 3 independent experiments assessed in triplicate). Flow cytometry was performed 6 hours after pulsing and results are expressed as the percentage of positive cells. (D, F) T-cell proliferation induced by conidia- or RNA-pulsed DCs in CD4+ or CD8+ T cells purified from lungs of C57BL/6 mice (D) or Tlr3−/− mice (F) 1 week after the intranasal infection and assessed for lymphoproliferation after 72 hours coculture with pulsed DCs from naive mice. DNA synthesis was measured by 3H-thymidine uptake (cpm, counts per minute). Ct indicates pulsed DCs alone. (E, G) Relative expression of Ifng and Prf1 by RT-PCR in CD4+ and CD8+ T cells exposed to conidia- or fungal RNA–pulsed DCs for 24 hours from C57BL/6 mice (E) or Tlr3−/− mice (G). (H) Cytolytic activity of CD8+ T cells from C57BL/6 mice, obtained as in panel D, against conidia-pulsed DCs at different effector cell:target cell (E:T) ratios. Shown is the percentage of specific cytotoxic activity determined by a standard 4-hour 51Cr-release assay. (I) Conidiocidal activity of culture supernatants from CD8+ T cells exposed as in panel H. Live conidia were labeled with the fluorescent molecular stain FUN-1 and incubated overnight with culture medium from cultured CD8+ T cells before examination by fluorescence microscopy. Metabolically active conidia accumulate orange fluorescence in vacuoles, whereas dormant and dead conidia stain green. Shown are representative fluorescence microscopy images of 3 independent experiments visualized with the 100×/1.3 oil objective on the BX51 upright microscope (Olympus). All images were captured using a high-resolution Microscopy Olympus DP71 camera (Olympus) and digitally acquired using the Cell∧P software (Version 3.3 build 2108, Olympus). (J) CD8+ T-cell proliferation induced by RNA-pulsed C57BL/6 DCs in the presence or not (−) of inhibitors targeting class II (bafilomycin A1 and chloroquine) or class I (brefeldin A and lactacystin) Ag-presentation pathways. Ct, CD8+ T cells alone. (K) Adoptive transfer of C57BL/6 or Tlr3−/− DCs pulsed with conidia or fungal RNA into C57BL/6 or Tlr3−/− mice. DCs were adoptively transferred by IP injection twice a week before the infection. Fungal growth in the lungs (mean CFUs ± SE, representative of at least 3 independent experiments) was assessed 3 days after the intranasal infection. In vivo data are from 4 independent experiments assessed in triplicate. *P < .05; **P < .01; and ***P < .001 for Tlr3−/− versus C57BL/6 mice (panels B-C), Ag-pulsed DC-primed versus unpulsed DC-primed (None) T cells (panels D-G), stimulated versus unstimulated CD8+ T cells (panel H), inhibitors versus no inhibitors (−; panel J), and DC-treated versus untreated (None) mice (panel K).
Recognition of fungal RNA is defective in Tlr3−/− DCs. C57BL/6 or Tlr3−/− lung DCs from naive mice were left unpulsed (None) or were pulsed with resting (RC) or swollen (SC) A fumigatus conidia, DOTAP alone, or fungal RNA + DOTAP for 2 hours. (A) Phosphorylation of IRF3 (p-IRF3) in DCs pulsed as above. Shown is a representative Western blot of 3 independent experiments. The IRF3 bands were quantified and the ratios of phosphorylated to total IRF3 are shown. (B) Relative expression of Ifna1 and Ifnb1 (RT-PCR) and (C) expression of CD80, CD86, and MHC class II (flow cytometry) on pulsed DCs (mean values ± SD of at least 3 independent experiments assessed in triplicate). Flow cytometry was performed 6 hours after pulsing and results are expressed as the percentage of positive cells. (D, F) T-cell proliferation induced by conidia- or RNA-pulsed DCs in CD4+ or CD8+ T cells purified from lungs of C57BL/6 mice (D) or Tlr3−/− mice (F) 1 week after the intranasal infection and assessed for lymphoproliferation after 72 hours coculture with pulsed DCs from naive mice. DNA synthesis was measured by 3H-thymidine uptake (cpm, counts per minute). Ct indicates pulsed DCs alone. (E, G) Relative expression of Ifng and Prf1 by RT-PCR in CD4+ and CD8+ T cells exposed to conidia- or fungal RNA–pulsed DCs for 24 hours from C57BL/6 mice (E) or Tlr3−/− mice (G). (H) Cytolytic activity of CD8+ T cells from C57BL/6 mice, obtained as in panel D, against conidia-pulsed DCs at different effector cell:target cell (E:T) ratios. Shown is the percentage of specific cytotoxic activity determined by a standard 4-hour 51Cr-release assay. (I) Conidiocidal activity of culture supernatants from CD8+ T cells exposed as in panel H. Live conidia were labeled with the fluorescent molecular stain FUN-1 and incubated overnight with culture medium from cultured CD8+ T cells before examination by fluorescence microscopy. Metabolically active conidia accumulate orange fluorescence in vacuoles, whereas dormant and dead conidia stain green. Shown are representative fluorescence microscopy images of 3 independent experiments visualized with the 100×/1.3 oil objective on the BX51 upright microscope (Olympus). All images were captured using a high-resolution Microscopy Olympus DP71 camera (Olympus) and digitally acquired using the Cell∧P software (Version 3.3 build 2108, Olympus). (J) CD8+ T-cell proliferation induced by RNA-pulsed C57BL/6 DCs in the presence or not (−) of inhibitors targeting class II (bafilomycin A1 and chloroquine) or class I (brefeldin A and lactacystin) Ag-presentation pathways. Ct, CD8+ T cells alone. (K) Adoptive transfer of C57BL/6 or Tlr3−/− DCs pulsed with conidia or fungal RNA into C57BL/6 or Tlr3−/− mice. DCs were adoptively transferred by IP injection twice a week before the infection. Fungal growth in the lungs (mean CFUs ± SE, representative of at least 3 independent experiments) was assessed 3 days after the intranasal infection. In vivo data are from 4 independent experiments assessed in triplicate. *P < .05; **P < .01; and ***P < .001 for Tlr3−/− versus C57BL/6 mice (panels B-C), Ag-pulsed DC-primed versus unpulsed DC-primed (None) T cells (panels D-G), stimulated versus unstimulated CD8+ T cells (panel H), inhibitors versus no inhibitors (−; panel J), and DC-treated versus untreated (None) mice (panel K).
TLR3 SNP associates with increased risk of invasive aspergillosis in HSCT recipients
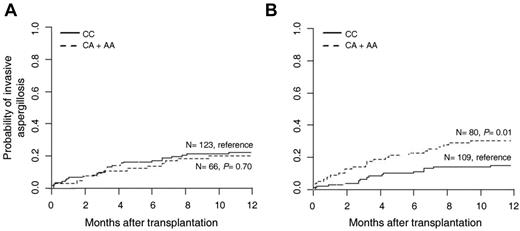
Given results obtained thus far, we also investigated whether genetic variants affecting the function of TLR3 might influence susceptibility to aspergillosis in patients with predisposing conditions. To this purpose, we performed a genetic analysis of selected SNPs in TLR3 in a cohort of 223 consecutive HSCT recipients and their respective donors (supplemental Table 1). To estimate the risk of infection according to patient or donor TLR3 genotypes, we determined cumulative incidences of aspergillosis among transplantation recipients at 12 months after transplantation. The +1234C/T (L412F) SNP did not influence susceptibility to infection when present in either patients (22.1% for CC and 20.3% for CT + TT; P = .73) or donors (24.5% for CC, 15.9% for CT + TT; P = .19). In contrast, the +95C/A SNP was found to significantly increase risk of aspergillosis when present in donors (14.8% for CC and 30.2% for CA + AA; P = .01; Figure 5B), but not in patients (22.2% for CC and 19.9% for CA + AA; P = .89; Figure 5A). Therefore, recapitulating the murine findings, the specific contribution of the TLR3 +95C/A SNP to the infection appears to rely on its presence on myeloid cells. Moreover, the genetic association remained significant after correction for clinical covariables, namely transplantation matching, GVHD, and antifungal prophylaxis. In the multivariate model, donor TLR3 +95C/A SNP (CA + AA genotype) independently increased the risk of IA (adjusted hazard ratio, 2.41; 95% confidence interval, 1.27-4.58; P = .007). No statistically significant influence of the tested SNPs was observed on patient survival (data not shown).
A TLR3 SNP is associated with increased risk of invasive aspergillosis in HSCT recipients. The TLR3 +95C/A genotypes of 223 consecutive hematologic patients undergoing allogeneic HSCT and their respective donors were determined by the bidirectional PCR amplification of specific alleles (Bi-PASA) method and correlated with the incidence of invasive aspergillosis. (A) Cumulative incidence of invasive aspergillosis at 12 months according to patient +95C/A genotype: CC (22.2%, n = 123) and CA + AA (19.9%, n = 66); P = .70 (B) Cumulative incidence of invasive aspergillosis at 12 months according to donor +95C/A genotype: CC (14.8%, n = 109) and CA + AA (30.2%, n = 80); P = .01.
A TLR3 SNP is associated with increased risk of invasive aspergillosis in HSCT recipients. The TLR3 +95C/A genotypes of 223 consecutive hematologic patients undergoing allogeneic HSCT and their respective donors were determined by the bidirectional PCR amplification of specific alleles (Bi-PASA) method and correlated with the incidence of invasive aspergillosis. (A) Cumulative incidence of invasive aspergillosis at 12 months according to patient +95C/A genotype: CC (22.2%, n = 123) and CA + AA (19.9%, n = 66); P = .70 (B) Cumulative incidence of invasive aspergillosis at 12 months according to donor +95C/A genotype: CC (14.8%, n = 109) and CA + AA (30.2%, n = 80); P = .01.
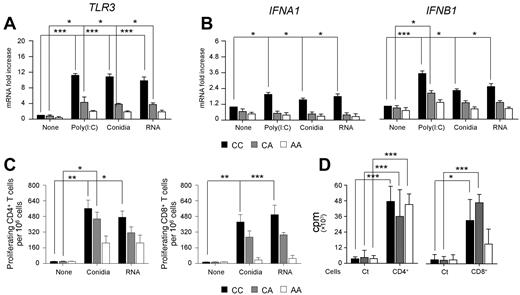
The TLR3 +95C/A SNP leads to defective CD8+ T-cell activation by DCs
Human CD141+ (BDCA-3)+ DCs are known to represent the equivalent of mouse DCs that cross-present Ags to CD8+ T cells after treatment with poly(I:C).37 We purified BDCA-3+ DCs from individuals bearing distinct TLR3 +95C/A genotypes and assessed TLR3 expression after stimulation with poly(I:C), A fumigatus conidia, or fungal RNA. Phenotypic analysis revealed that BDCA-3+ DCs were CD11c+, CD11b−, CD1a−, CD86+, and CD80− (supplemental Figure 8), as described previously.38 We found that TLR3 mRNA expression in WT BDCA-3+ cells was strongly induced after pulsing with each stimulus (Figure 6A). In contrast, BDCA-3+ cells bearing the +95C/A SNP displayed instead a marked decrease in TLR3 expression and responsiveness, a defect found to be genotype dose dependent. We also found that expression of both the IFNA1 and IFNB1 genes was triggered by all of the stimuli in wild-type BDCA-3+ DCs, but not in TLR3 polymorphic cells (Figure 6B). Finally, after assessing the ability of BDCA-3+ DCs to prime CD4+ or CD8+ T cells for proliferation, we found that Ag-specific (Figure 6C) and, to a lesser extent, polyclonal (Figure 6D) CD8+ T-cell proliferation were mostly affected by the TLR3 +95C/A SNP compared with CD4+ T cells. Given that TLR3 is expressed on CD8+ T cells (Edwards et al39 and supplemental Figure 9), we cannot rule out the contribution of direct TLR3 signaling to the defective proliferative activity of CD8+ T cells. Our results point to an essential role for TLR3 in the activation of memory-protective CD8+ T-cell responses to A fumigatus in mice and humans.
The +95C/A SNP in TLR3 associates with defective CD8+ T cells by peripheral DCs. Human CD141+ (BDCA-3+) DCs were isolated from peripheral blood of healthy volunteers bearing distinct TLR3 +95C/A genotypes (CC, CA, and AA) and were either left untreated or pulsed with poly(I:C), A fumigatus conidia, or fungal RNA. Relative expression of TLR3 (A) and type I IFNs (IFNA1 and IFNB1; B) was assessed by RT-PCR. (C) Frequencies of Ag-specific CD4+ or CD8+ T-cell clones were obtained by limiting dilution assays. Ag-specific proliferation of T-cell clones was assessed after coculture of responder CD4+ and CD8+ T cells with BDCA-3+ DCs pulsed with A fumigatus conidia or fungal RNA and measured by 3H-thymidine incorporation. Shown are the percentages of CD4+ or CD8+ T-cell clones/106 cells. (D) Purified T-cell subsets were stimulated with 0.1% phytohemagglutinin in the presence of autologous PBMCs. DNA synthesis was measured by 3H-thymidine uptake (cpm, counts per minute). “None” indicates untreated DCs; and Ct, T cells alone. Data are mean values ± SD of at least 3 independent samples for each +95C/A genotype assessed in triplicate. *P < .05; **P < .01; and ***P < .001 for treated versus untreated (None) BDCA-3+ DCs.
The +95C/A SNP in TLR3 associates with defective CD8+ T cells by peripheral DCs. Human CD141+ (BDCA-3+) DCs were isolated from peripheral blood of healthy volunteers bearing distinct TLR3 +95C/A genotypes (CC, CA, and AA) and were either left untreated or pulsed with poly(I:C), A fumigatus conidia, or fungal RNA. Relative expression of TLR3 (A) and type I IFNs (IFNA1 and IFNB1; B) was assessed by RT-PCR. (C) Frequencies of Ag-specific CD4+ or CD8+ T-cell clones were obtained by limiting dilution assays. Ag-specific proliferation of T-cell clones was assessed after coculture of responder CD4+ and CD8+ T cells with BDCA-3+ DCs pulsed with A fumigatus conidia or fungal RNA and measured by 3H-thymidine incorporation. Shown are the percentages of CD4+ or CD8+ T-cell clones/106 cells. (D) Purified T-cell subsets were stimulated with 0.1% phytohemagglutinin in the presence of autologous PBMCs. DNA synthesis was measured by 3H-thymidine uptake (cpm, counts per minute). “None” indicates untreated DCs; and Ct, T cells alone. Data are mean values ± SD of at least 3 independent samples for each +95C/A genotype assessed in triplicate. *P < .05; **P < .01; and ***P < .001 for treated versus untreated (None) BDCA-3+ DCs.
Discussion
The ability of exogenous fungal Ags to activate memory-protective CD8+ cells without CD4+ T-cell help (“helpless”) is of relevance for both basic and clinical mycology. This demands a better definition of cells and molecules that control CD8+ T-cell priming and differentiation into effector and memory cells in vivo. The major findings to emerge from the present study are that TLR3 signaling in DCs pulsed with A fumigatus conidia or RNA efficiently prime resting CD8+ T cells in vivo and in vitro for memory responses to the fungus by a mechanism that appears to involve acidic endosomal vesicles and cytosolic processing. Susceptibility to aspergillosis was increased in conditions of TLR3 deficiency in both mice and humans, a finding pointing to a previously unrecognized role for the TLR3-dependent Ag-recognition pathway in aspergillosis. Similarly to what was observed with other fungal pathogens,20,21,40 by producing effector cytokines such as IFN-γ and exhibiting cytotoxic activity against fungus-laden cells or the fungus itself, CD8+ T cells provided vaccine-induced resistance likely by restricting the fungal growth and terminating the cellular response in infection.
Both CD4+ T cells13-15 and CD8+ T cells13,25,41 are present in the murine and human T-cell repertoire to the fungus. However, the role of CD8+ T cells in infection, the specific fungal Ags they recognize, and mechanisms underlying their activation are unknown. Our study provides evidence that both CD4+ and CD8+ T cells mediate memory responses to the fungus, contingent on the nature of the fungal vaccine. Indeed, the cell wall glucanase Crf1p, known to be presented by 3 common human MHC class II alleles and to induce memory CD4+ Th1 cells to the fungus,42 activated antifungal memory CD4+ Th1 cells independently of TLR3. We have evidence that recognition of Crf1p and CD4+ Th1-cell activation occurs via MyD88 (A.D.L. and L.R., unpublished observations, 2011), a finding suggesting that TLRs other than TLR3 are involved in fungal Ag sampling and signaling for activation of the CD4 arm of the memory immune response to the fungus. However, the finding that Tlr3−/− mice failed to mount an efficient memory response to the fungus while retaining the ability to activate memory fungal Ag–specific CD4+ T-cell responses suggests either a superior activity of the CD8-dependent mechanisms in immune memory to the fungus or a dysfunctional fungal processing in conditions of TLR3 deficiency.
TLR3 is a crucial “danger” signaling receptor that senses endogenous mRNA released by necrotic cells.3 Therefore, it is possible that TLR3 is activated in infection by sensing host damage. However, in this case, CD8α+ DCs are solely responsible for cross-presenting Ags from apoptotic host cells.43 In addition, we have shown recently that TLR3 restrained rather than promoted danger-dependent inflammation in aspergillosis.17 Therefore, sensing fungal RNA is the likely function of TLR3 in infection. By migrating from early endosomes to LAMP1+ endosomes after stimulation with dsRNA, TLR3 mediates the activation of DCs for the cross-presentation of exogenous Ags to CD8+ T cells.6 Therefore, TLR3-specific adjuvants are used to tailor CD8+ T-cell immune responses independently of CD4 help.44 We have tested the adjuvant activity of TLR3 and found that administration of Crf1p with poly(I:C) skewed T-cell memory from class II–dependent CD4+ T cells—obtained with CpG known to block cross-presentation43 —to class I–dependent CD8+ T cells (data not shown). This finding highlights the importance of adjuvants in skewing antifungal memory responses to candidate subunit fungal vaccines. However, the function of fungal RNA may go beyond its adjuvant activity, because RNA-transfected DCs also originate fungal Ags.26 As in viral infection,45 the requirement of host translational machinery could enhance the efficiency of class I presentation. In the present study, we show that these Ags are processed via both the TLR3-dependent endosomal/lysosomal pathway and the class I–dependent proteasomal degradation pathway. It is also likely that direct presentation of fungal Ags, newly synthesized from endogenous RNA produced after fungal internalization and degradation, occurs in infection through cytosolic receptors other than the endosomal TLR3. Consistent with the findings obtained with conidia, many routes of Ag sampling and presentation are apparently used by DCs confronted with the fungus.
TLR3 may further optimize CD8+ T-cell activation by facilitating CCR7 expression on DCs through unimpaired phagocytosis. Although CCR7 deficiency does not impair fungal clearance in primary aspergillosis,46 CCR7+ DCs are required for the priming of CD8+ T cells in the draining lymph nodes, and this finding highlights the interesting difference with CD4+ T-cell anti–A fumigatus priming that was reported to be mediated by migratory CCR2+ inflammatory monocytes.47 However, the role of TLR3 may go beyond regulation of adaptive memory to include regulation of inflammation and tolerance. The defective IDO activation likely contributed to the inability of these mice to promote tolerogenic responses to the fungus that are required to limit inflammation in infection.18 Indeed, IL-17A–producing Th17 cells were expanded in Tlr3−/− mice and likely contributed to inflammatory pathology, as suggested previously.33 In addition to the ability of type I IFNs to inhibit Th17 development,48 it is interesting that by disabling lymph node priming, as occurs in CCR7 deficiency,49 promoted IL-17A–mediated lung pathology via abnormal T-cell sensitization in ectopic lymph node formation.50 Both defective Ccr7 expression and peribronchiolar lymphoid tissues were observed in Tlr3−/− mice.
In conclusion, the present study discloses the contribution of TLR3 to CD8+ T-cell memory responses to A fumigatus and identifies a TLR3 SNP leading to a loss-of-function phenotype of DCs that is associated with susceptibility to infection and concomitant failure to activate antifungal CD8+ T cells in HSCT patients. Our findings may offer a plausible explanation for the increased susceptibility to aspergillosis after CMV reactivation in HSCT patients, and predict that, in addition to TLR9,51 TLR3 SNPs are also a likely risk factor for infection by CMV. The ability to subvert endogenous MHC class I pathway via TRIF degradation is a trait shared with other members of the herpesvirus family.52 Regardless of possible differences between viral and fungal cross-presentation pathways, our findings suggest that a defective Ag cross-presentation pathway may predispose to both viral and fungal infections in HSCT patients and suggest novel therapeutic interventions.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Cristina Massi Benedetti for digital art and editing and Dr Alain S. Bell for collection and processing of DNA samples from human patients.
These studies were supported by the Specific Targeted Research Project ALLFUN (FP7-HEALTH-2009 contract number 260338 to L.R.), by SYBARIS (FP7-HEALTH-2009 contract number 242220 to L.R.), and by the Italian Project AIDS 2010 by the Istituto Superiore di Sanità (contract number 40H40 to L.R.). A.C. and C.C. were supported by fellowships from Fundação para a Ciência e Tecnologia, Portugal (contracts SFRH/BPD/46292/2008 and SFRH/BD/65962/2009, respectively).
Authorship
Contribution: A.C., J.-P. L., A.V., F.A., and L.R. designed the research; A.D.L., S.B., C.C., C.D., S.M., K.P, R.G.I., F.F., and A.P. performed the experiments; and A.C. and L.R. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Luigina Romani, Microbiology Section, Department of Experimental Medicine and Biochemical Sciences, University of Perugia, Via del Giochetto, 06126 Perugia, Italy; e-mail: lromani@unipg.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal