Abstract
Mixed lineage leukemia (MLL) is a key epigenetic regulator of normal hematopoietic development and chromosomal translocations involving MLL are one of the most common genetic alterations in human leukemia. Here we show that ASB2, a component of the ECSASB E3 ubiquitin ligase complex, mediates MLL degradation through interaction with the PHD/Bromodomain region of MLL. Forced expression of ASB2 degrades MLL and reduces MLL transactivation activity. In contrast, the MLL-AF9 fusion protein does not interact with ASB2 and is resistant to ASB2 mediated degradation. Increased expression of ASB2 during hematopoietic differentiation is associated with decreased levels of MLL protein and down-regulation of MLL target genes. Knockdown of ASB2 leads to increased expression of HOXA9 and delayed cell differentiation. Our data support a model whereby ASB2 contributes to hematopoietic differentiation, in part, through MLL degradation and HOX gene down-regulation. Moreover, deletion of the PHD/Bromo region renders MLL fusion proteins resistant to ASB2-mediated degradation and may contribute to leukemogenesis.
Introduction
The histone H3 lysine 4 (H3K4) methyltransferase mixed lineage leukemia (MLL) is necessary for the maintenance of HOX patterning and essential for normal hematopoiesis. Full-length MLL is a 3968 amino acid multi-domain protein, which is proteolytically cleaved into a 320 kDa N-terminal fragment (MLLN) and a 180 kDa C-terminal fragment (MLLC) that noncovalently associate to form a stable complex.1 MLLN contains several DNA-binding domains including 3 AT-hooks and a CxxC domain, as well as a poorly understood PHD/Bromodomain (PHD/Bromo) region that contains 4 plant homeodomain fingers (PHD1-4), and a bromodomain between PHD3 and PHD4. MLLC contains a transactivation domain and a SET domain with intrinsic H3K4 methyltransferase activity (Figure 1A). MLL positively regulates target gene expression through methylation of H3K4, an epigenetic mark closely associated with transcriptional activation. Genome-wide analysis has identified a large number of genes that are regulated by MLL, including homeobox (HOX) genes and their cofactors, such as MEIS1.2 HOX genes are a group of transcription factors that specify segment identity and cell fate during development, and play essential roles during hematopoiesis.3 MLL is responsible for maintaining expression of HOX and MEIS1 through H3K4 methylation in hematopoietic stem cells and progenitors, which is required for stem cell self-renewal and progenitor expansion.2,4,5 Mll-null mice are defective in maintaining Hox gene expression resulting in embryonic lethality by E10.5, whereas reexpression of Hox genes in Mll-deficient progenitors rescues hematopoietic colony formation.6,7 HOX expression decreases concurrent with hematopoietic differentiation. This is crucial for normal hematopoiesis, as constitutive activation of HOX genes is associated with leukemia and other malignancies.3 In fact, HOXA9 was identified as the most highly correlated gene for poor prognosis in acute myeloid leukemia (AML).8
Chromosomal translocations involving MLL are one of the most common genetic alterations in human leukemia, accounting for up to 80% of infant leukemia and approximately 5%-10% of adult leukemia overall.9,10 Most of the leukemogenic MLL fusion proteins contain the N-terminus of MLL fused in frame to the C-terminus of a translocation partner, generally a transcription activator or a dimerizing protein, thus forming a chimeric protein with abnormal transactivation ability.10 Both in vitro and in vivo studies have demonstrated that these MLL fusion proteins induce leukemogenesis mainly through constitutive activation of HOXA9 and MEIS1.11-13 Notably, translocations of MLL invariably occur within the breakpoint cluster region (BCR), which leads to the deletion or disruption of the PHD/Bromo region.14 Further, insertion of PHD/Bromo into MLL-AF9 and MLL-ENL fusion proteins abolishes their transformation ability, suggesting that this region may be important for the regulation of MLL.15,16 Recent studies discovered that reciprocal MLL fusion proteins containing the N-terminus of the fusion partner and the C-terminus of MLL can also have oncogenic properties. For example, the AF4-MLL fusion protein induces ALL in mice independent of MLL-AF4, and a NUP98-MLL fusion was discovered in 2 AML cases.17,18 However, current observations indicate that these fusion proteins transform through mechanisms independent of HOXA9 activation.17,18
The ankyrin repeat and suppressor of cytokine signaling (SOCS) box-containing (ASB) protein family contains 18 members (ASB1-18), which function as the substrate recognition module in the ECSASB (Elongin B/C-Cullin–SOCS box protein) E3 ubiquitin ligase complex.19 Through interaction with Elongin C (EloC), the ASB proteins associate with Elongin B (EloB), Cullin5 (Cul5) and Rbx2, and target the substrate for ubiquitination.19 Several studies have shown that ASB proteins function in a wide range of biologic processes.20-23 ASB2 was discovered as a gene induced by all-trans retinoid acid (ATRA) via a retinoid receptor (RAR) binding element present in the ASB2 promoter. ASB2 expression is rapidly induced during ATRA induced differentiation of leukemia cell lines, including acute promyelocytic leukemia (APL) cells containing the PML-RARα fusion protein, as well as HL60 and NB4 cell lines, whereas ectopic expression of ASB2 promotes growth inhibition and cell differentiation.24,25 Filamins A and B are 2 known targets of ASB2.26 ASB2 expression is also activated by Notch signaling, which leads to degradation of Jak2 directly and degradation of Jak3 and E2A indirectly by bridging the formation of a Cul1-Cul5 dimeric E3 ligase complex.27,28 Besides a role in hematopoiesis, ASB2 can also regulate muscle differentiation.29
In this study, we explored the function of the MLL PHD/Bromo region and identified ASB2 as a novel regulator of MLL stability during hematopoietic differentiation.
Methods
Cell culture
Human embryonic kidney 293 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS. Hoxa9-ER cells were cultured in Iscove modified Dulbecco medium (IMDM) supplemented with 15% FBS (StemCell Technologies) and 100nM 4-OHT. NB4 and K562 cells were cultured in RPMI-1640 medium supplemented with 10% FBS. MLL-AF9, MLL-ENL and E2A-HLF cells were cultured in IMDM supplemented with 15% FBS (StemCell Technologies) and 10 ng/mL IL3.
Vector construction
pCXN2-Flag-MLL, and pCXN2-Flag-MLL-AF9 have been previously described.30 Flag-MLL-AF9 was cloned into MSCV vector through digestion with restriction enzyme and ligation. CxxC, CxxC-PHD/Bromo, and the deletion constructs were cloned from full-length MLL, Flag or Myc tag and the nuclear localization signals were added by PCR reaction. The fragments were then ligated to MigR1 or pCMV vector (Clontech). pCMV-ASB2-3XFlag, pCMV-ASB6-3XFlag, and pCMV-ASB7-3XFlag expression vectors were kindly provided by Dr Junya Kohroki (Tokyo University of Science, Tokyo, Japan). The Flag tag was replaced with HA tag by PCR-based mutagenesis. ASB2 deletion constructs were cloned from pCMV-ASB2-HA and ligated into the pCMV vector. pcDNA5/FRT-Flag-EloB, pcDNA5/FRT-Flag-Cullin5, and pCI-neo-EloC expression vectors were kindly provided by Dr Joan Conaway (Stowers Institute for Medical Research, Kansas City, MO).
Immunoprecipitation and Western blotting
Immunoprecipitation and Western blotting were performed as previously described.16 293 cells were transfected with FuGene 6 (Roche) according to the manufacturer's instructions and cultured for 48 hours. Cells were then lysed with BC-300 lysis buffer (20mM Tris-HCl [pH 7.4], 10% glycerol, 300mM KCl, 0.1% NP-40) and incubated with agarose affinity beads overnight at 4°C. Beads were washed 3 times with BC-300 buffer. Proteins were eluted by boiling in SDS-loading buffer, resolved by SDS-PAGE, and detected by Western blotting. Primary antibodies against MLL, ASB2, EloB, SIII p15 (EloC) and β-actin were obtained from Bethyl and Upstate, Abcam and Imgenex, Santa Cruz biotechnology, BD Transduction Laboratories, and Sigma-Aldrich, respectively. Antibodies against Myc and HA tag were obtained from Abcam. Antibody against Flag tag and M2 anti-Flag agarose affinity beads were purchased from Sigma-Aldrich. Anti-Myc agarose affinity beads were purchased from Clontech.
Protein identification by LC-tandem mass spectroscopy
293 cells were transfected with empty vector or Flag tagged CxxC or CxxC-PHD/Bromo. Cells were lysed and immunoprecipitations were performed as described in “Immunoprecipitation and Western blotting.” Bound material was eluted with Flag peptide and concentrated with a Micron YM-30 centrifugal filter column (Millipore). Proteins were then separated by SDS-PAGE. Mass spectroscopy analysis was performed using a LTQ-Orbitrap XL mass spectrometer (ThermoFisher). Proteins were identified by searching the data against Human IPI database (Version 3.41; 72 254 entries) appended with decoy (reverse) sequences using X!Tandem/Trans-Proteomic Pipeline (TPP) software suite (Version 4.5).31 All proteins with a ProteinProphet probability score of > 0.9 (error rate < 2%) were considered positive identifications.
Dual luciferase assay
293 cells were transfected with MLL or MLL-AF9, ASB2, Renilla luciferase reporter (internal control), and Hoxa9-LUC reporter with FuGene 6 according to the manufacturer's instructions. Cells were serum starved in 0.5% FBS in OPTI-MEM media for 48 hours. Luciferase assays were performed using the Dual Luciferase Assay Kit (Promega) according to the manufacturer's instructions. Emission was detected using a Monolight 3010 Luminometer (BD Biosciences).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as previously described.32 Antibodies against Flag tag and H3K4me3 were obtained from Sigma-Aldrich and Abcam, respectively. Primary antibody against MLL was kindly provided by Dr Yali Dou (University of Michigan, Ann Arbor, MI). qPCR was performed on the precipitated DNAs with TaqMan primers and probes from Applied Biosystems. Binding was quantitated as follows: ΔCT = CT(input)−CT(Chromatin IP), % total = 2ΔCT. Primer and probe sequences are listed in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Real-time quantitative reverse transcription PCR
RNA was extracted from cells using TRIzol reagent (Invitrogen) and cDNA was synthesized using Superscript III Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. Real-time quantitative reverse transcription PCR (RT-qPCR) was performed with Taqman gene expression assays (Applied Biosystems) and ABI 7500 PCR Detection System. Data were analyzed using comparative ΔΔCt method (described in ABI Prism 7700 Sequence Detection System User Bulletin No. 2). Taqman primer probe sets for mouse Asb2, Mll, Hoxa9, Meis1, and human ASB2, MLL, HOXA9, and MEIS1 were purchased from Applied Biosystems.
shRNA knockdown and characterization of cell differentiation
pSM2 shRNA against human ASB2 (clone ID V2HS_97797) and pTRIPZ shRNA against human ASB2 (clone ID V2THS_97800) were purchased from Open Biosystems. Lentiviral vector pTRIPZ shRNAs were packaged by the University of Michigan Vector Core. Retroviral vector pSM2 shRNAs were packaged in Plat-E cells as previously described.33 Cells were transduced by spinoculation and selected with puromycin at 0.5 μg/mL (for Hoxa9-ER cells) and 0.4 μg/mL (for NB4 cells).
To evaluate cell differentiation, pTRIPZ shRNA-transduced NB4 cells were treated with 0.5 μg/mL doxycycline for 24 hours to induce shRNA expression. Then ATRA or DMSO was added to the medium at 1μM to induce differentiation. For flow cytometry analysis, cells were collected and washed with PBS, and stained with APC-mouse anti–human CD11b antibody or isotype control (BD Pharmingen) for 30 minutes. Cells were then washed with standard buffer (1× PBS, 0.1% sodium azide, 1% heat inactivated FBS) twice and resuspended in standard buffer. FACS data were collected on an LSRII (BD Pharmingen) and analyzed with FlowJo Version 9.3.3 flow cytometry analysis software. For nitroblue tetrazoleum (NBT) assay, 2 × 105 cells were suspended in 200 μL medium with 1 mg/mL NBT and 30 ng/mL 12-O-tetradecanoylphorbol-13-acetate (Sigma-Aldrich), and incubated at 37°C for 30 minutes. Cytospins were then prepared and the percentage of NBT-positive cells was determined by microscopically counting at least 500 cells per experimental condition.
Results
ASB2 interacts with the PHD/Bromo region of MLL
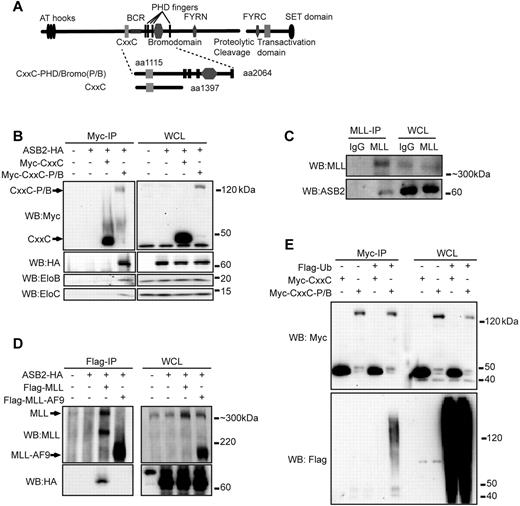
The PHD/Bromo region of MLL is located downstream of the BCR and invariably deleted from MLL fusion proteins (Figure 1A). To elucidate its function, we performed mass spectroscopy analysis to identify proteins that associate with the PHD/Bromo region of MLL. A Flag tagged MLL construct that encompasses the CxxC-PHD/Bromo region containing a nuclear localization signal (NLS) was transiently expressed in human embryonic kidney 293 cells and immunoprecipitated with M2 anti-Flag agarose beads (Figure 1A). Coeluted proteins were resolved by SDS-PAGE and subjected to mass spectroscopy analysis. An empty vector containing the Flag tag and NLS, and Flag tagged CxxC with NLS were immunoprecipitated in parallel as controls to exclude nonspecific binding and proteins that associate with the CxxC domain. Multiple peptides corresponding to components of the ECSASB E3 ubiquitin ligase complex were identified that specifically interact with CxxC-PHD/Bromo, including Cullin 5 (Cul5), EloC, and several ASB proteins, such as ASB7, ASB10, ASB14, and ASB18 (supplemental Figure 1).
PHD/Bromo region interacts with ASB2 and mediates MLL ubiquitination. (A) Schematic diagram of the structure of wild-type MLL. The CxxC and CxxC-PHD/Bromo fragment used in immunoprecipitation are shown with the first and last MLL amino acid retained in the constructs indicated. (B) Myc tagged CxxC or CxxC-PHD/Bromo (CxxC-P/B) was coexpressed in 293 cells with HA tagged ASB2. Cells were treated with MG132 for 6 hours. Immunoprecipitation of CxxC or CxxC-PHD/Bromo followed by Western blotting shows that ASB2 and endogenous EloB and EloC specifically interact with CxxC-PHD/Bromo. (C) NB4 cells were treated with ATRA for 48 hours and with MG132 for 16 hours. MLL was immunoprecipitated with an anti-MLL antibody, and Western blotting using an anti-ASB2 antibody shows that endogenous ASB2 coprecipitates with MLL. (D) Flag tagged MLL or MLL-AF9 was coexpressed in 293 cells with HA tagged ASB2. After MG132 treatment, anti-Flag immunoprecipitation was performed followed by Western blotting, showing that ASB2 interacts with MLL but not with MLL-AF9. (E) Myc tagged CxxC or CxxC-PHD/Bromo and Flag tagged ubiquitin were coexpressed in 293 cells. Immunoprecipitation of CxxC or CxxC-PHD/Bromo was performed after MG132 treatment. Western blotting shows that CxxC-PHD/Bromo is conjugated with ubiquitin.
PHD/Bromo region interacts with ASB2 and mediates MLL ubiquitination. (A) Schematic diagram of the structure of wild-type MLL. The CxxC and CxxC-PHD/Bromo fragment used in immunoprecipitation are shown with the first and last MLL amino acid retained in the constructs indicated. (B) Myc tagged CxxC or CxxC-PHD/Bromo (CxxC-P/B) was coexpressed in 293 cells with HA tagged ASB2. Cells were treated with MG132 for 6 hours. Immunoprecipitation of CxxC or CxxC-PHD/Bromo followed by Western blotting shows that ASB2 and endogenous EloB and EloC specifically interact with CxxC-PHD/Bromo. (C) NB4 cells were treated with ATRA for 48 hours and with MG132 for 16 hours. MLL was immunoprecipitated with an anti-MLL antibody, and Western blotting using an anti-ASB2 antibody shows that endogenous ASB2 coprecipitates with MLL. (D) Flag tagged MLL or MLL-AF9 was coexpressed in 293 cells with HA tagged ASB2. After MG132 treatment, anti-Flag immunoprecipitation was performed followed by Western blotting, showing that ASB2 interacts with MLL but not with MLL-AF9. (E) Myc tagged CxxC or CxxC-PHD/Bromo and Flag tagged ubiquitin were coexpressed in 293 cells. Immunoprecipitation of CxxC or CxxC-PHD/Bromo was performed after MG132 treatment. Western blotting shows that CxxC-PHD/Bromo is conjugated with ubiquitin.
The ASB protein family contains 18 members. As multiple ASB proteins were identified by mass spectroscopy, we focused our attention on ASB2 because of its specific activity against MLL protein (see Figure 2A) and known roles in hematopoiesis. ASB2 is expressed in hematopoietic cells and has been shown to promote both differentiation and cell cycle arrest in certain myeloid leukemia cell lines, consistent with a function in hematopoiesis.24,25 To test the interaction between the PHD/Bromo region and the ECSASB2 complex, Myc-tagged CxxC or CxxC-PHD/Bromo and HA tagged ASB2 were coexpressed in 293 cells in the presence of the proteasome inhibitor MG132. ASB2 coprecipitated specifically with CxxC-PHD/Bromo but not CxxC confirming an interaction dependent on the PHD/Bromo region. Moreover, endogenous EloB and EloC also coprecipitated with CxxC-PHD/Bromo (Figure 1B). To determine whether MLL and ASB2 interact under physiologic conditions, NB4 cells were treated with ATRA to induce ASB2 expression and with MG132 to inhibit MLL degradation. Immunoprecipitation of endogenous MLL showed that ASB2 coprecipitated with MLL, further supporting a physiologic interaction in hematopoietic cells (Figure 1C). As MLL fusion proteins generally lack the PHD/Bromo region, we tested whether the interaction is disrupted in the context of MLL-AF9 by coexpressing MLL or MLL-AF9 with ASB2 in 293 cells. ASB2 coprecipitated with full-length MLL but not the MLL-AF9 fusion protein, again suggesting that the PHD/Bromo region is necessary for ASB2 interaction with MLL (Figure 1D). Based on these observations, we predicted that ubiquitination of MLL would be at least partially dependent on the presence of the PHD/Bromo region. To this end, CxxC or CxxC-PHD/Bromo was coexpressed with ubiquitin in 293 cells. Immunoprecipitation revealed an ubiquitin smear associated with the CxxC-PHD/Bromo but not the CxxC domain alone (Figure 1E). These data indicate that the PHD/Bromo region interacts with the ECSASB2 E3 ubiquitin ligase and results in the ubiquitination of MLL.
ASB2 degrades MLL and reduces MLL transactivation ability
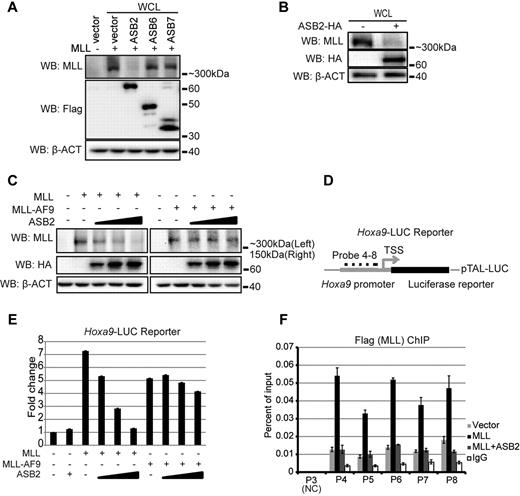
To test the effect of ASB2 on MLL protein turnover, full-length MLL was coexpressed in 293 cells with ASB2 or 2 other ASB proteins, ASB6 and ASB7. Western blotting with whole cell lysate revealed that overexpression of ASB2 led to significant degradation of MLL, whereas overexpression of ASB6 and ASB7 did not affect MLL protein levels (Figure 2A). The effect of ASB2 was also confirmed on endogenous MLL as ASB2 expression in 293 cells led to reduced endogenous MLL protein level detected by Western blotting (Figure 2B). Because ASB2 interacts with wild-type MLL but not MLL-AF9 fusion protein (Figure 1D), we predicted that overexpression of ASB2 would degrade MLL, although largely not affecting MLL-AF9 protein levels. To test this, equimolar amounts of plasmids expressing MLL or MLL-AF9 were cotransfected into 293 cells with an increasing dosage of ASB2. Western blotting revealed that ASB2 led to degradation of MLL in a dose-dependent manner, whereas the MLL-AF9 levels were largely unaffected (Figure 2C). As ASB2 functions in the ECS E3 ligase complex that contains multiple subunits, we tested the effect of the other components on the stability of MLL and MLL-AF9. EloB, EloC, or Cul5 was coexpressed with MLL or MLL-AF9 in 293 cells. Western blotting with whole cell lysate revealed that Cul5 did not affect MLL stability and EloB and EloC had a limited effect on the degradation of MLL (supplemental Figure 2A), demonstrating the importance of substrate recognition by ASB2. In contrast, the stability of MLL-AF9 was not significantly changed (supplemental Figure 2A).
ASB2 leads to reduced MLL protein level and transactivation ability. (A) MLL was coexpressed with Flag tagged ASB2, 6 and 7 in 293 cells. Western blotting with whole cell lysate shows that ASB2 specifically degrades MLL. β-actin blot shows equal loading of the samples. (B) Expression of ASB2 in 293 cells followed by Western blotting with whole cell lysate shows that ASB2 leads to degradation of endogenous MLL. (C) Equimolar amounts of MLL or MLL-AF9 expression plasmids were cotransfected with ASB2 at the ratio of 6:1, 2.5, and 4 in 293 cells. Western blotting with whole cell lysate shows that MLL degradation is ASB2 dose-dependent, wherease the levels of MLL-AF9 are not affected. β-actin blot indicates equal loadings. (D) Schematic diagram of the Hoxa9-LUC reporter. (E) Dual luciferase assay was performed in 293 cells with Hoxa9-LUC reporter. Lanes 3 through 6 show expression of MLL with increasing dosage of ASB2, and lane 7 through 10 show expression of MLL-AF9 with increasing dosage of ASB2. The ratios between MLL or MLL-AF9 and ASB2 were the same as in panel C. All changes are normalized to lane 1, which includes Hoxa9-LUC and an empty expression vector. Error bars indicate SD. Results of 1 of more than 3 representative experiments performed are shown. (F) ChIP assay was performed in 293 cells transfected with Hoxa9-LUC, MLL, and ASB2. The ratio between MLL and ASB2 was 6:4. Probes 4 through 8 cover the promoter region of Hoxa9-LUC. Probe3 recognizes a region that exists in the endogenous Hoxa9 promoter but is not included in Hoxa9-LUC, and serves as a negative control. The nomenclature is consistent with Figure 6E, which shows the position of the probes on endogenous Hoxa9 locus. Error bars indicate SD. Results of 1 of more than 3 representative experiments performed are shown.
ASB2 leads to reduced MLL protein level and transactivation ability. (A) MLL was coexpressed with Flag tagged ASB2, 6 and 7 in 293 cells. Western blotting with whole cell lysate shows that ASB2 specifically degrades MLL. β-actin blot shows equal loading of the samples. (B) Expression of ASB2 in 293 cells followed by Western blotting with whole cell lysate shows that ASB2 leads to degradation of endogenous MLL. (C) Equimolar amounts of MLL or MLL-AF9 expression plasmids were cotransfected with ASB2 at the ratio of 6:1, 2.5, and 4 in 293 cells. Western blotting with whole cell lysate shows that MLL degradation is ASB2 dose-dependent, wherease the levels of MLL-AF9 are not affected. β-actin blot indicates equal loadings. (D) Schematic diagram of the Hoxa9-LUC reporter. (E) Dual luciferase assay was performed in 293 cells with Hoxa9-LUC reporter. Lanes 3 through 6 show expression of MLL with increasing dosage of ASB2, and lane 7 through 10 show expression of MLL-AF9 with increasing dosage of ASB2. The ratios between MLL or MLL-AF9 and ASB2 were the same as in panel C. All changes are normalized to lane 1, which includes Hoxa9-LUC and an empty expression vector. Error bars indicate SD. Results of 1 of more than 3 representative experiments performed are shown. (F) ChIP assay was performed in 293 cells transfected with Hoxa9-LUC, MLL, and ASB2. The ratio between MLL and ASB2 was 6:4. Probes 4 through 8 cover the promoter region of Hoxa9-LUC. Probe3 recognizes a region that exists in the endogenous Hoxa9 promoter but is not included in Hoxa9-LUC, and serves as a negative control. The nomenclature is consistent with Figure 6E, which shows the position of the probes on endogenous Hoxa9 locus. Error bars indicate SD. Results of 1 of more than 3 representative experiments performed are shown.
Next, we examined the effect of ASB2 on MLL-mediated transcriptional transactivation. Dual luciferase assays were performed with a firefly luciferase reporter driven by the murine Hoxa9 promoter (Hoxa9-LUC reporter; Figure 2D). Expression of MLL led to a greater than 7-fold activation of reporter expression (Figure 2E lane 3). Although ASB2 alone had no effect on reporter expression, coexpressed ASB2 reduced MLL mediated transactivation on the Hoxa9-LUC reporter in a dose-dependent manner (Figure 2E lanes 2,4-6). Only a slight decrease in the reporter expression was observed when MLL-AF9 was coexpressed with ASB2 (Figure 2E lanes 7-10). We also tested the recruitment of MLL to the promoter region of Hoxa9-LUC on expression of ASB2. 293 cells were transfected with MLL, ASB2, and the Hoxa9-LUC reporter, and ChIP assay was performed with probes that specifically recognize the murine Hoxa9 promoter (Figure 2D). Consistent with the luciferase assay, MLL showed robust binding to the Hoxa9 promoter, which decreased to background levels on expression of ASB2 (Figure 2F).
The bromodomain and 4th PHD finger of MLL mediate interaction with ASB2
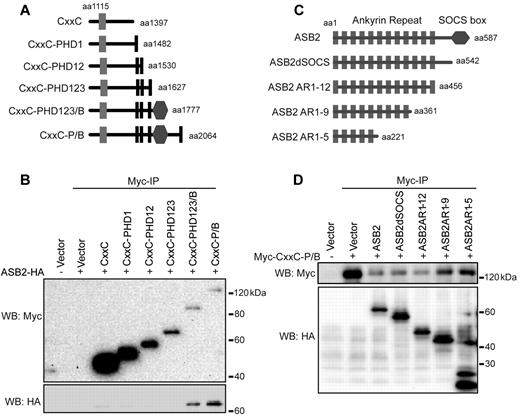
The PHD/Bromo region of MLL contains 4 PHD fingers (PHD1-4) and an atypical bromodomain between PHD3 and PHD4 (Figure 1A). To further characterize the MLL-ASB2 interaction and identify which part of the PHD/Bromo region mediates the interaction with ASB2, we made a series of deletion constructs spanning the CxxC-PHD/Bromo region (Figure 3A), and performed coimmunoprecipitation experiments with full-length ASB2. Deletion beyond the bromodomain led to disruption of the MLL-ASB2 interaction (Figure 3B). A more robust interaction was observed when PHD4 was included (Figure 3B compare lanes 7-8) suggesting that both the Bromodomain and PHD4 are needed for efficient ASB2 binding. Consistent with this, MG132 treatment led to stabilization of the 2 deletions containing Bromo or Bromo-PHD4, supporting that this region is important for ubiquitination (supplemental Figure 3A). To investigate whether MLL and ASB2 directly associate, in vitro binding assays were performed with bacterially expressed and purified His-GST tagged ASB2-HA and His-MBP tagged PHD3-Bromo-PHD4 (PHD3-4). ASB2 specifically bound to PHD3-4, but not to PHD3, suggesting a direct interaction (supplemental Figure 3B-C). In addition, to test whether Bromo-PHD4 can serve as a substrate of ASB2, in vitro ubiquitination assays were performed with immunopurified ECSASB2 complex from 293 cells and bacterially purified His-MBP-PHD3-4. Purified ECSASB2 complex showed ATP-dependent auto-ubiquitination activity in vitro (supplemental Figure 3D lane 2). An enhanced ubiquitin smear was observed on addition of PHD3-4 but not PHD3 alone, indicating that ECSASB2 requires the Bromo/PHD4 region of MLL for ubiquitination in vitro (supplemental Figure 3D).
The Bromodomain/PHD4 of MLL and Ankyrin Repeat 1-5 of ASB2 mediate the interaction between MLL and ASB2. (A) Schematic diagram of the CxxC-PHD/Bromo serial deletions. The first and last MLL amino acid retained in the constructs are indicated. (B) CxxC-PHD/Bromo deletion constructs described in panel A were cotransfected with ASB2 into 293 cells followed by anti-Myc immunoprecipitation. Binding of ASB2 was detected by Western blotting with anti-HA antibody. (C) Schematic of HA tagged ASB2 serial deletions. The first and last ASB2 amino acid retained in the constructs are indicated. (D) ASB2 deletion constructs described in panel C were cotransfected with Myc-CxxC-PHD/Bromo in 293 cells followed by anti-Myc immunoprecipitation. Binding of ASB2 was detected by Western blotting with anti-HA antibody.
The Bromodomain/PHD4 of MLL and Ankyrin Repeat 1-5 of ASB2 mediate the interaction between MLL and ASB2. (A) Schematic diagram of the CxxC-PHD/Bromo serial deletions. The first and last MLL amino acid retained in the constructs are indicated. (B) CxxC-PHD/Bromo deletion constructs described in panel A were cotransfected with ASB2 into 293 cells followed by anti-Myc immunoprecipitation. Binding of ASB2 was detected by Western blotting with anti-HA antibody. (C) Schematic of HA tagged ASB2 serial deletions. The first and last ASB2 amino acid retained in the constructs are indicated. (D) ASB2 deletion constructs described in panel C were cotransfected with Myc-CxxC-PHD/Bromo in 293 cells followed by anti-Myc immunoprecipitation. Binding of ASB2 was detected by Western blotting with anti-HA antibody.
ASB2 contains 12 N-terminal ankyrin repeats which mediate protein-protein interactions, and a C-terminal SOCS box, which is necessary for the interaction with EloC.20 To map the interaction region on ASB2, we made serial deletions of ASB2 that contain different numbers of ankyrin repeats (Figure 3C). Coimmunoprecipitation experiments with Myc-CxxC-PHD/Bromo showed that the first 5 ankyrin repeats are sufficient for the interaction with MLL (Figure 3D).
Increased ASB2 expression during hematopoietic differentiation is associated with decreased MLL protein levels and MLL target gene expression
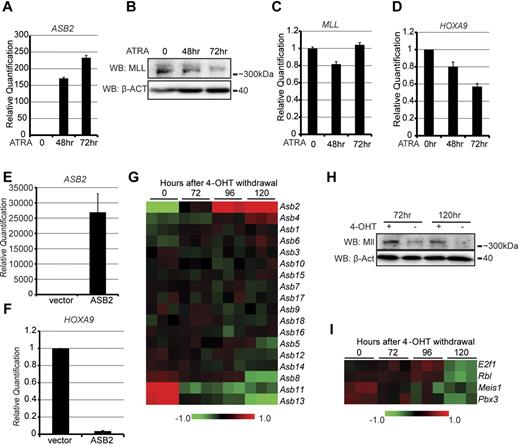
Previous studies have shown that ASB2 expression is up-regulated during differentiation of myeloid leukemia cell lines and that ectopically expressed ASB2 induces myeloid growth arrest and differentiation.24,25 To further explore the biologic significance of MLL regulation by ASB2, we examined the levels of ASB2, MLL, and MLL target genes during differentiation in both human and murine leukemia cell lines. First, the NB4 human leukemia cell line was treated with ATRA to induce cell differentiation. Consistent with previous publications,24 we observed a significant increase of ASB2 expression on ATRA treatment (Figure 4A). MLL protein levels decreased gradually and coordinately with ASB2 up-regulation (Figure 4B). Notably, MLL transcription did not significantly change with differentiation as examined by RT-qPCR (Figure 4C), indicating that the decrease in MLL protein is post-transcriptional. RT-qPCR was also performed to test the expression levels of HOXA9, a major target of MLL, which decreased approximately 40% after 72 hours of treatment (Figure 4D). The expression of ASB2, MLL, and another MLL target gene, MEIS1, was examined in ATRA treated K562 leukemia cells. Similar to NB4 cells, ASB2 expression increased during ATRA-induced cell differentiation, along with decreased MLL protein levels and MEIS1 expression, whereas the transcription of MLL did not change significantly (supplemental Figure 4A-E).
ASB2 expression leads to MLL degradation and MLL target gene down-regulation during hematopoietic differentiation. (A-D) NB4 cells were treated with ATRA for 0, 48, and 72 hours to induce differentiation. The expression of ASB2 (A), the protein levels (B) and transcription levels (C) of MLL, and the expression of HOXA9 (D) were measured by RT-qPCR or Western blotting. Transcription is shown relative to the level at 0 hours. Error bars indicate SD. β-ACTIN blot indicates equal loading. (E-F) ASB2 expression vector was transfected into NB4 cells by electroporation, and the expression levels of ASB2 (E) and HOXA9 (F) were measured by RT-qPCR. Expression is shown relative to empty vector transfected control cells. Error bars indicate SD. (G-I) Expression of Asb2, Mll, and Mll target genes during 4-OHT withdrawal induced differentiation of the Hoxa9-ER cell line. (G) Heat map generated from gene expression array data collected at different time points after 4-OHT withdrawal. The Asb genes were clustered according to their expression change. Data are shown in triplicate for each time point. (H) Mll protein level in Hoxa9-ER cells was determined by Western blotting at 72 and 120 hours with or without 4-OHT withdrawal. (I) Heat map of the Mll target gene expression generated from gene expression array data as in panel G.
ASB2 expression leads to MLL degradation and MLL target gene down-regulation during hematopoietic differentiation. (A-D) NB4 cells were treated with ATRA for 0, 48, and 72 hours to induce differentiation. The expression of ASB2 (A), the protein levels (B) and transcription levels (C) of MLL, and the expression of HOXA9 (D) were measured by RT-qPCR or Western blotting. Transcription is shown relative to the level at 0 hours. Error bars indicate SD. β-ACTIN blot indicates equal loading. (E-F) ASB2 expression vector was transfected into NB4 cells by electroporation, and the expression levels of ASB2 (E) and HOXA9 (F) were measured by RT-qPCR. Expression is shown relative to empty vector transfected control cells. Error bars indicate SD. (G-I) Expression of Asb2, Mll, and Mll target genes during 4-OHT withdrawal induced differentiation of the Hoxa9-ER cell line. (G) Heat map generated from gene expression array data collected at different time points after 4-OHT withdrawal. The Asb genes were clustered according to their expression change. Data are shown in triplicate for each time point. (H) Mll protein level in Hoxa9-ER cells was determined by Western blotting at 72 and 120 hours with or without 4-OHT withdrawal. (I) Heat map of the Mll target gene expression generated from gene expression array data as in panel G.
Because ATRA has a wide range of biologic effects, we examined whether the down-regulation of HOXA9 and MEIS1 is a direct consequence of ASB2 expression. To test this, ASB2 was transfected into NB4 cells and K562 cells by electroporation. ASB2 expression led to a significant decrease of HOXA9 expression in NB4 cells and MEIS1 expression in K562 cells suggesting that the changes in gene expression are a direct result of ASB2 expression (Figure 4E-F, supplemental Figure 4F).
Asb2 expression was also examined in a differentiation system using murine bone marrow cells transformed with an inducible Hoxa9-estrogen receptor (Hoxa9-ER) construct and maintained in 4-hydroxytamoxifen (4-OHT). Withdrawal of 4-OHT causes Hoxa9-ER translocation to the cytoplasm leading to cell differentiation and cell cycle arrest, which is largely complete within 5 days.33 Microarray gene expression profiling was previously performed on Hoxa9-ER cells in the presence of 4-OHT and at 72, 96, and 120 hours after 4-OHT withdrawal,33 and the 18 Asb family members were clustered according to their expression level changes. Of these, Asb2 was the most highly up-regulated gene upon differentiation (Figure 4G, supplemental 4G). Along with Asb2 up-regulation, we observed a decrease in Mll protein levels at both 72 and 120 hours after 4-OHT withdrawal compared with cells grown in the presence of 4-OHT independent of changes in Mll transcription (Figure 4H, supplemental Figure 4H). Moreover, multiple Mll target genes, including E2f1, Rbl, Meis1, and Pbx3 were down-regulated consistent with up-regulated Asb2 degrading Mll protein (Figure 4I).
To further evaluate the role of ASB2 on MLL target gene expression and cell differentiation, we generated a stable NB4 cell line expressing an inducible pTRIPZ shRNA construct against ASB2 for knockdown studies. After doxycycline treatment, shRNA expression was activated, which was reflected by expression of red fluorescent protein (RFP) on the pTRIPZ vector, leading to knockdown of ASB2 (Figure 5A, supplemental Figure 5A). With ASB2 knockdown, we observed an increase in MLL protein level, and a 60% up-regulation of HOXA9 expression (Figure 5B-C). Further, MLL turnover was monitored with cycloheximide treatment. Prolonged MLL degradation was observed in ASB2 knockdown cells compared with control scrambled shRNA-transduced cells (supplemental Figure 5B). The effect of ASB2 on cell differentiation was determined by monitoring the CD11b surface marker expression and nitroblue tetrazoleum (NBT) assay after ATRA treatment. ASB2 knockdown led to a delay in ATRA-induced cell differentiation, as lower CD11b expression and a lower percentage of NBT-positive cells were detected at both 24 and 48 hours (Figure 5D-E). We also observed that ASB2 knockdown resulted in faster growth compared with cells expressing control shRNA. Moreover, ATRA treatment led to a rapid growth arrest in control cells, whereas ASB2 knockdown cells retained the ability to grow, albeit at a slower rate (Figure 5F). As further confirmation, a constitutive pSM2 shRNA targeting another region of ASB2 was stably transduced into NB4 cells. Similar up-regulation of HOXA9 was observed with ASB2 knockdown (supplemental Figure 5C-D).
Knockdown of ASB2 up-regulates HOXA9 and delays cell differentiation. (A) RT-qPCR confirmation of ASB2 knockdown in NB4 cells after doxycycline treatment for 24 hours. Expression is shown relative to scrambled shRNA-transduced control cells. Error bars indicate SD. (B) Protein level of MLL was detected by Western blotting with whole cell lysate, which shows an increase after doxycycline induced ASB2 knockdown. (C) Expression of HOXA9 was measured by RT-qPCR after doxycycline induced ASB2 knockdown. Expression is shown relative to scrambled shRNA-transduced control cells. Error bars indicate SD. (D-E) NB4 cells were treated with doxycycline for 24 hours to induce ASB2 knockdown and then with ATRA to induce differentiation. (D) Expression of CD11b was determined by flow cytometry at 24 and 48 hours. (E) NBT assays were performed at 24 and 48 hours and the percentage of positive cells is shown. Error bars indicate SD calculated from 3 independent experiments. (F) Control and ASB2 knockdown NB4 cells were grown in liquid culture with or without ATRA treatment. A proliferation advantage was observed from ASB2 knockdown cells in both conditions. Error bars indicate SD.
Knockdown of ASB2 up-regulates HOXA9 and delays cell differentiation. (A) RT-qPCR confirmation of ASB2 knockdown in NB4 cells after doxycycline treatment for 24 hours. Expression is shown relative to scrambled shRNA-transduced control cells. Error bars indicate SD. (B) Protein level of MLL was detected by Western blotting with whole cell lysate, which shows an increase after doxycycline induced ASB2 knockdown. (C) Expression of HOXA9 was measured by RT-qPCR after doxycycline induced ASB2 knockdown. Expression is shown relative to scrambled shRNA-transduced control cells. Error bars indicate SD. (D-E) NB4 cells were treated with doxycycline for 24 hours to induce ASB2 knockdown and then with ATRA to induce differentiation. (D) Expression of CD11b was determined by flow cytometry at 24 and 48 hours. (E) NBT assays were performed at 24 and 48 hours and the percentage of positive cells is shown. Error bars indicate SD calculated from 3 independent experiments. (F) Control and ASB2 knockdown NB4 cells were grown in liquid culture with or without ATRA treatment. A proliferation advantage was observed from ASB2 knockdown cells in both conditions. Error bars indicate SD.
ASB2 reduced colony formation ability of MLL fusion transformed cells
Previous studies showed that expression of wild-type MLL is required for MLL fusion protein-mediated transformation.34 As our data suggest that ASB2 leads to efficient degradation of MLL, we predicted that ASB2 expression would diminish MLL fusion-mediated transformation through degradation of wild-type MLL. To test this, we established a murine cell line transformed with Flag tagged MLL-AF9. ASB2 was cloned into the MigR1 retroviral expression vector, and transduced into MLL-AF9 cells. Positively-transduced cells were sorted by flow cytometry according to green fluorescent protein (GFP) expression on the MigR1 vector, and colony formation ability was examined by methylcellulose replating assays. ASB2 expression led to a more than 60% decrease in colony number compared with empty vector-transduced control cells (Figures 6A-B, supplemental Figure 6A), as well as reduced cell proliferation in liquid culture (Figure 6C). Consistent with this, a 40% decrease of Hoxa9 expression was observed in ASB2-transduced cells compared with vector control (Figure 6D), despite no detectable change in Mll transcription (supplemental Figure 6B). Notably, an ASB2 construct containing a deletion of the SOCS box still led to degradation of MLL, a phenomenon similar to that reported for ASB4,35 and led to decreased colony formation when expressed in MLL-AF9 cells (supplemental Figure 6C-F). Decreased colony formation and cell proliferation was also observed using MLL-ENL cells transduced with ASB2 (supplemental Figure 6G-I). Importantly, expression of ASB2 in E2A-HLF cells, which transform in an MLL and Hoxa9 independent manner, had no effect on either colony formation or cell proliferation, indicating that the effect of ASB2 is specific for MLL-fusion transformed cells (supplemental Figure 6J-L).
ASB2 reduces colony formation of MLL-AF9 transformed cells. (A) Colony numbers of MLL-AF9 cells transduced with empty vector or ASB2 from methylcellulose replating assay. Error bars indicate SD from 3 independent experiments. (B) P-iodonitro-tetrazolium violet staining of the colonies. (C) MLL-AF9 cells transduced with empty vector or ASB2 were grown in liquid culture. Significantly reduced cell proliferation was observed for ASB2-transduced cells. Error bars indicate SD. (D) RT-qPCR was performed to measure the expression of Hoxa9 in MLL-AF9 cells transduced with empty vector or ASB2. Expression is shown relative to empty vector-transduced control cells. Error bars indicate SD. (E) Schematic diagram showing the endogenous Hoxa9 locus and probes used in the ChIP assay. TSS: transcription start site. (F-H) ChIP assays were performed to determine the recruitment of Mll (F), the H3K4me3 level (G), and the recruitment of MLL-AF9 (H) on the Hoxa9 locus with probes shown in (E). Antibodies against MLL, H3K4me3 and Flag were used, respectively. Blue lines are data from MLL-AF9 cells transduced with empty vector; red lines are data from MLL-AF9 cells transduced with ASB2; and gray lines are IgG controls for each cell line. Error bars indicate SD.
ASB2 reduces colony formation of MLL-AF9 transformed cells. (A) Colony numbers of MLL-AF9 cells transduced with empty vector or ASB2 from methylcellulose replating assay. Error bars indicate SD from 3 independent experiments. (B) P-iodonitro-tetrazolium violet staining of the colonies. (C) MLL-AF9 cells transduced with empty vector or ASB2 were grown in liquid culture. Significantly reduced cell proliferation was observed for ASB2-transduced cells. Error bars indicate SD. (D) RT-qPCR was performed to measure the expression of Hoxa9 in MLL-AF9 cells transduced with empty vector or ASB2. Expression is shown relative to empty vector-transduced control cells. Error bars indicate SD. (E) Schematic diagram showing the endogenous Hoxa9 locus and probes used in the ChIP assay. TSS: transcription start site. (F-H) ChIP assays were performed to determine the recruitment of Mll (F), the H3K4me3 level (G), and the recruitment of MLL-AF9 (H) on the Hoxa9 locus with probes shown in (E). Antibodies against MLL, H3K4me3 and Flag were used, respectively. Blue lines are data from MLL-AF9 cells transduced with empty vector; red lines are data from MLL-AF9 cells transduced with ASB2; and gray lines are IgG controls for each cell line. Error bars indicate SD.
To further confirm that the effect of ASB2 on colony formation is through wild-type Mll degradation, we examined the recruitment of Mll and MLL-AF9 to the Hoxa9 locus. ChIP assays were performed with a series of probes spanning the Hoxa9 promoter region and exons 1 and 2 (Figure 6E). The recruitment of endogenous Mll was determined using an Mll antibody that recognizes a region that is lost in MLL-AF9 fusion protein. Mll showed a robust binding to the Hoxa9 locus in control cells, whereas the binding was significantly reduced on ASB2 expression (Figure 6F). Consistent with this, histone H3 lysine 4 tri-methylation (H3K4me3) was reduced in ASB2-transduced cells compared with control cells (Figure 6G). In contrast, recruitment of MLL-AF9, which was detected using Flag antibody, did not change significantly with ASB2 expression (Figure 6H). These data indicate that ASB2 reduces the amount of wild-type Mll at target genes critical for leukemia, but does not alter MLL-AF9 fusion protein binding.
Discussion
Post-translational modifications, such as phosphorylation, ubiquitination, and acetylation, provide a rapid and flexible way to regulate protein activity in response to intrinsic and extrinsic cellular signals. As one of the major mechanisms of controlling protein abundance, ubiquitination has been shown to regulate a wide range of biologic processes, including cell cycle progression, cell proliferation, and differentiation. Recent studies have shown that several histone modifying enzymes are regulated through ubiquitination. For example, the E3 ligase CUL4-DDB1 regulates the histone H4K20 monomethylase PR-Set7/Set8 during S phase, which is important for replication licensing, cell cycle progression, and DNA damage response.36-39 Moreover, 2 components of the MLL complex, WDR5 and RBBP5, have been shown to be regulated by CUL4-DDB1, suggesting a strict regulation of H3K4 trimethylation by E3 ubiquitin ligases.40 Indeed, previous studies have shown that MLL stability is regulated during cell cycle progression by the E3 ligase SCFSkp2 and APCCdc20. Further work has shown that ATR mediated phosphorylation of MLL because of genotoxic stress disrupts the interaction with SCFSkp2, linking MLL to the mammalian S phase checkpoint.41,42
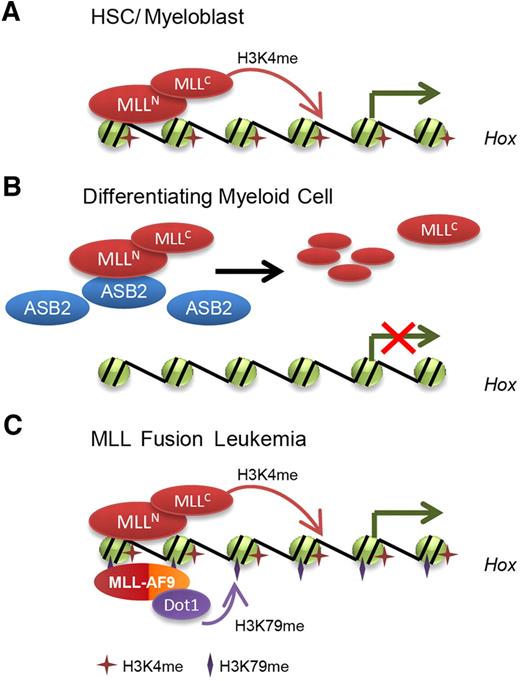
In this study, we identified ASB2 as a novel regulator of MLL, and linked HOX gene down-regulation during hematopoiesis to MLL ubiquitination and degradation through the ECSASB2 ubiquitin ligase complex. ASB2 expression leads to degradation of MLL and reduces its transactivation ability (Figure 2). A significant increase in ASB2 expression was observed during differentiation of both human leukemia (NB4 and K562) cell lines and the murine leukemia (Hoxa9-ER) cell line, displaying a reciprocal pattern to MLL protein levels and MLL target gene expression (Figure 4). Furthermore, expression of ASB2 leads to HOXA9 and MEIS1 down-regulation, whereas knockdown of ASB2 up-regulates HOXA9 and renders the cells resistant to differentiation stimuli (Figures 4–5). These findings support a model where MLL is regulated post-translationally through ASB2 up-regulation during hematopoietic differentiation leading to an active ECSASB2 E3 ubiquitin ligase complex that rapidly degrades MLL protein and down-regulates HOX genes (Figure 7A-B). In contrast, loss of the PHD/Bromo region and escape from ASB2-mediated degradation may contribute to MLL fusion-mediated leukemogenesis (Figure 7C). Our study adds to the list of regulators of MLL and suggests that SCFSkp2 and APCCdc20 act coordinately to control MLL protein turnover during cell cycle progression, whereas ECSASB2 regulates MLL during hematopoietic differentiation. Together, these data suggest that MLL protein turnover is tightly controlled by several E3 ubiquitin ligase complexes during various biologic processes.
Model for the regulation of MLL degradation and HOX gene expression by ASB2 during normal and malignant hematopoiesis. (A) In hematopoietic stem cell and progenitor compartments, MLL maintains HOX gene expression. (B) During differentiation, ASB2 expression is induced, which leads to MLL ubiquitination and degradation. Less MLL is localized to HOX gene loci, resulting in inhibition of HOX gene transcription. (C) In MLL fusion protein-mediated leukemia, MLL fusion proteins lack the PHD/Bromo region that interacts with ASB2 and escape from ASB2-mediated degradation. Moreover, low expression of ASB2 leads to stabilization of wild-type MLL, which may also contribute to the constitutive activation of HOX genes and leukemogenesis.
Model for the regulation of MLL degradation and HOX gene expression by ASB2 during normal and malignant hematopoiesis. (A) In hematopoietic stem cell and progenitor compartments, MLL maintains HOX gene expression. (B) During differentiation, ASB2 expression is induced, which leads to MLL ubiquitination and degradation. Less MLL is localized to HOX gene loci, resulting in inhibition of HOX gene transcription. (C) In MLL fusion protein-mediated leukemia, MLL fusion proteins lack the PHD/Bromo region that interacts with ASB2 and escape from ASB2-mediated degradation. Moreover, low expression of ASB2 leads to stabilization of wild-type MLL, which may also contribute to the constitutive activation of HOX genes and leukemogenesis.
Our experiments show that the interaction between ASB2 and MLL is mediated through the PHD/Bromo region (Figures 1,Figure 2–3). This region contains 4 PHD fingers and a bromodomain between PHD3 and PHD4. The third PHD finger of MLL has been reported to bind di/tri-methylated histone H3K4 as well as the cyclophilin Cyp33 simultaneously, switching MLL from an activator to a repressor,43-46 and loss of MLL PHD3 is required for MLL-ENL–mediated transformation.16 The first and fourth PHD fingers function together with the phenylalanine/tyrosine-rich (FYRN) domain to mediate the intramolecular interaction between MLLN and MLLC.47 In our experiments, inclusion of the bromodomain is sufficient to mediate the interaction with ASB2, whereas the addition of PHD4 enhances binding (Figure 3). Several bromodomains have been demonstrated to mediate protein-protein interaction through recognizing acetylated lysines.48 Interestingly, although exhibiting a typical bromodomain fold, the bromodomain of MLL does not recognize acetylated histones, perhaps because of loss of a conserved asparagine found in acetyl-lysine binding bromodomains.44 Moreover, a sequence analysis of various human bromodomains defines the MLL bromodomain as an outlier, because of significant sequence variation.48,49 Further work is needed to determine whether the interaction between ASB2 and the MLL bromodomain is mediated through acetylation.
A recent study reports that the wild-type allele of MLL is required for MLL-AF9–mediated leukemogenesis. Wild-type Mll is recruited to the Hoxa9 locus in MLL-AF9 transformed cells, and plays a crucial role for the maximal methylation of both H3K79 and H3K4, as well as the expression of Hoxa9. Knockdown or excision of wild-type Mll in MLL-AF9 transduced cells inhibits cell proliferation, reduces colony formation, and inhibits the development of leukemia in vivo.34 Based on these observations, the authors propose that MLL fusion-transformed leukemia cells may be more sensitive to the inhibition of wild-type MLL, as they have a reduced cellular level of MLL, and wild-type MLL is a potential target for leukemia therapy.34 This is consistent with our findings, as ASB2 expression in MLL-AF9–transformed cells phenocopies knockdown or excision of wild-type MLL, including decreased H3K4 methylation, decreased Hoxa9 expression, and reduced cell proliferation and colony formation (Figure 6). Notably, ASB2 expression is rapidly induced by ATRA.24,25 This suggests that ATRA treatment, which is already in use for treatment of promyelocytic leukemia, would have activity against MLL associated leukemia. Indeed, 2 leukemia cell lines harboring the MLL-AF9 fusion protein, THP-1 and MOLM-14 cells, have been shown to be sensitive to ATRA-induced differentiation.50
In summary, our work has established a novel ubiquitination pathway for MLL degradation during hematopoietic differentiation by ECSASB2. This represents a mechanism for HOX gene down-regulation during blood cell maturation by degradation of MLL, and provides a rationale for targeting the wild-type MLL protein for therapy of MLL associated leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Yali Dou (University of Michigan, Ann Arobr, MI) for providing the ChIP grade antibody against MLL; Dr Junya Kohroki (Tokyo University of Science, Tokyo, Japan) for providing ASB2, ASB6, and ASB7 expression vectors; and Dr Joan Conaway (Stowers Institute for Medical Research, Kansas City, MO) for providing EloB, EloC,and Cul5 expression vectors. They also thank Dr Sean J. Morrison and Shenghui He (University of Michigan, Ann Arbor, MI) for kindly sorting the murine hematopoietic cell populations.
This work was supported in part by an ASH scholar award (A.G.M.), the National Institutes of Health grants 1K99 CA158136-01 (A.G.M.) and R01 CA151425-01 (J.L.H), and a SCOR grant from Lymphoma & Leukemia Society (J.L.H).
National Institutes of Health
Authorship
Contribution: J.W. and A.G.M. performed experiments. J.W. made the figures and wrote the paper. J.L.H. and A.G.M. designed the experiments, reviewed the data and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew G. Muntean, Dept of Pathology, University of Michigan Medical School, Ann Arbor, MI 48109; e-mail: andrewmu@umich.edu; or Jay L. Hess, Dept of Pathology, University of Michigan Medical School, Ann Arbor, MI; e-mail: jayhess@umich.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal