Abstract
Tissue factor pathway inhibitor (TFPI) produces factor Xa-dependent feedback inhibition of factor VIIa/tissue factor-induced coagulation. Messages for 2 isoforms of TFPI have been identified. TFPIα mRNA encodes a protein with an acidic N-terminus, 3 Kunitz-type protease inhibitor domains and a basic C-terminus that has been purified from plasma and culture media. TFPIβ mRNA encodes a form in which the Kunitz-3 and C-terminal domains of TFPIα are replaced with an alternative C-terminus that directs the attachment of a glycosylphosphatidylinositol (GPI) anchor, but whether TFPIβ protein is actually expressed is not clear. Moreover, previous studies have suggested that the predominant form of TFPI released from cells by phosphatidylinositol-specific phospholipase C (PIPLC) treatment is TFPIα, implying it is bound at cell surfaces to a separate GPI-anchored coreceptor. Our studies show that the form of TFPI released by PIPLC treatment of cultured endothelial cells and placental microsomes is actually TFPIβ based on (1) migration on SDS-PAGE before and after deglycosylation, (2) the lack of a Kunitz-3 domain, and (3) it contains a GPI anchor. Immunoassays demonstrate that, although endothelial cells secrete TFPIα, greater than 95% of the TFPI released by PIPLC treatment from the surface of endothelial cells and from placental microsomes is TFPIβ.
Introduction
Tissue factor pathway inhibitor (TFPI) is the key endogenous regulator of TF-initiated coagulation. TFPI inhibits factor Xa (FXa) directly, and, in a FXa-dependent manner, inhibits factor VIIa/tissue factor (FVIIa/TF) activity.1 The human TFPI gene encodes for 2 dominant messages that are generated by alternative intron/exon splicing (GenBank NM_006287.4, GenBank NM_001032281.2). TFPIα, the originally described message, encodes a signal peptide that is removed by processing, followed by a 276-amino acid protein that consists of an acidic N-terminal region, 3 tandem Kunitz-type protease inhibitor domains, and a basic C-terminal region.2 Functional studies reveal that the Kunitz-2 domain mediates binding to and inhibition of FXa, whereas the Kunitz-1 domain is responsible for recognition and inhibition of the TF/FVIIa complex.3 The Kunitz-3 domain lacks protease inhibitor activity; however, it and the C-terminal basic region of TFPIα are required for protein S enhancement of inhibition of FXa.4,5 The TFPIβ message lacks the Kunitz-3 and C-terminal exons of TFPIα, and in their place is an exon that encodes a 42-amino acid C-terminal sequence following residue 181 of TFPIα. This new C-terminal sequence is predicted to direct the proteolytic cleavage of the peptide following N193 with the attachment of a GPI anchor. The protein mass of TFPIβ is less than that of TFPIα, but it migrates on SDS-PAGE at a molecular weight similar to TFPIα (46 kDa) apparently because of greater sialylation of its O-linked carbohydrate.6
In humans, TFPI circulates in plasma at approximately 70 ng/mL (1.6nM), with 80% being a C-terminal truncated form that is lipoprotein associated and the remaining 20% being full-length and near full-length TFPIα forms.7,8 Platelets contain TFPIα and, at the site of injury TFPIα levels, increase dramatically.9,10 The parenteral administration of heparin to humans increases the circulating levels of total TFPI in plasma to approximately 2.5-fold baseline levels.11,12 The form of TFPI that is released is full-length TFPIα.13 Heparin also increases the release of TFPI from endothelial cells in culture,6,14 even though treatment with heparin does not perceptibly reduce surface TFPI.6,15,16
The endothelium appears to be the major source of TFPI in vivo. In human endothelial cell cultures, the ratio of TFPIα/TFPIβ mRNA varies between 5 and 10.17 Human TFPIα protein has been identified in endothelial cell culture media and purified from HepG2 cells and plasma,13,18,19 but human TFPIβ protein has previously only been identified indirectly in ECV304 cells.6,15,17
A substantial fraction of the TFPI produced by endothelial cells remains at the cell surface and associated with caveolae.16,20 Phosphatidylinositol-specific phospholipase C (PIPLC) treatment, which cleaves GPI-anchored proteins, releases approximately 80% of cell surface TFPI, and the remaining TFPI can be removed by heparin treatment.15,17,21 The form of TFPI released from the cell surface by PIPLC has been presumed to be TFPIα, based largely on interpretation of a placental microsome study.22 It has been reported that TFPIβ is not produced by human endothelial EAhy926 cells.23 The cell surface association of TFPIα was thought to involve its interaction with a separate, as yet unidentified, GPI-anchored coreceptor(s)15-17,20-23 that may control its cellular trafficking and surface expression.21 As we initiated studies intended to identify the putative GPI-anchored coreceptor(s) for TFPIα, our results led to the realization that TFPIβ, rather than TFPIα, is the dominant PIPLC-releasable isoform on endothelial cells and placental microsomes.
Methods
Materials
Unless otherwise stated, reagents were from Sigma-Aldrich, including protease inhibitor cocktail, deglycosylation kit (EDEGLY-1KT), and HRP-conjugated goat anti–rabbit IgG. DMEM, penicillin/streptomycin, SDS-PAGE gels, pRSETA vector, and BL21(DE3)pLysS cells were from Invitrogen. Monoclonal anti-TFPI Mab2H8 and Mab2B12, and rabbit polyclonal anti-TFPI have been described.24 Affi-gel matrix and molecular weight protein standards were from Bio-Rad. HRP was conjugated to antibody using Lightning-Link reagent (Novus Biologics). Western blotting was to nitrocellulose (Millipore), and Lumi-light substrate was from Roche Diagnostics. Purified human recombinant TFPIα, a gift from Monsanto, was used to generate the standard curves for all ELISAs. Fluorescently labeled Aerolysin (FLAER) was from Pinewood Scientific/Protox Biotech (Cedarlane Lab) and anti–AlexaFluor-488 from Invitrogen.
Expression, purification, and characterization of rPIPLC
Using conventional methods, the Bacillus thuringienis (bacteria kindly provided by Wayne Barnes, Washington University, St Louis, MO) PIPLC gene was cloned into a modified pRSETA vector, expressed in BL21(DE3)pLysS cells, and rPIPLC released from cells by osmotic shock. The rPIPLC was isolated using diethylaminoethyl-Sepharose chromatography, followed by size exclusion chromatography to yield a single approximately 21 kDa band on SDS-PAGE and PIPLC specific activity greater than 1000 U/mg as determined by rate of cleavage of radioactive phosphatidylinositol versus commercially available PIPLC (Invitrogen).
Cell line
The human liver adenocarcinoma derived SK–Hep-1 line (ATCC HTB-52), of endothelial origin,25 and the human cell line ECV304(ATCC CRL-1998), which possesses some endothelial cell characteristics, were obtained from ATCC. HUVEC-derived cell line EA.hy926 was a kind gift from Cora-Jean Edgell (University of North Carolina, Chapel Hill, NC), and HUVECs (CC-2517) were purchased from Lonza Walkersville. Chinese hamster ovary (CHO) cell lines expressing human recombinant TFPIα (rTFPIα) and recombinant TFPIβ (rTFPIβ) and C127 cells expressing rTFPIα have been previously described.6,15
Cell culture, harvest, and PIPLC treatment
All cells were cultured at 37°C with 5% CO2, in T150 cm2 flasks. HUVECs were grown as prescribed in Clonetics EGM Endothelial Cell Growth Medium Bullet Kit (CC-3124). All other cell lines were grown in DMEM supplemented with 10% heat-inactivated FCS and 1% penicillin/streptomycin. On reaching more than 80% confluence, media was decanted; cells were washed with PBS, and 20 mL of serum-free media (EGM for HUVECs, DMEM for all other cells) was added. Cells were cultured for 24 hours, at which time media was collected and filtered through 0.22-μm filters and stored at −20°C. Cells were detached by dissociation buffer (Invitrogen), counted, collected by low-speed centrifugation (2000g, 5 minutes), and washed thrice by suspension and 10 mL PBSE (PBS + 1mM EDTA) per wash and centrifugation. Final cell pellets were suspended in 1 mL PBSI (PBS, 1mM EDTA with 1% protease inhibitor cocktail) per T150 flask of starting cells. To obtain PIPLC releasates, cells were incubated with 2 units PIPLC per milliliter for 1 hour at room temperature. Cells were removed by centrifugation for 2 minutes at 6000g, and releasates were analyzed for the TFPI isoform or used in affinity purification of TFPI. Where indicated, EAhy926 cells were treated with 10 units heparin per milliliter for 1 hour at room temperature, and the releasate was analyzed for TFPI isoforms. Posttreatment cell pellets were washed twice with PBSI, resuspended in 1 mL PBSI, and subjected to 2 freeze-thaw cycles. Cell lysates were clarified by centrifugation, and resulting supernatants were analyzed.
Preparation of placental microsomes
Human placenta from a full-term pregnancy was obtained with permission. All procedures were performed on ice or at 4°C. The placenta was washed with PBSE; large thrombi and connective tissue were removed by dissection; remaining tissue was cut into approximately 10-g sections, blotted with paper towels, and frozen at −80°C. Frozen tissue was ground into a fine powder using an electronic meat grinder. Two volumes of PBSE were added to the tissue powder followed by homogenization using a polytron (Tekmar). The homogenate was centrifuged at 6000g for 10 minutes, and the resulting supernatant was decanted and subjected to centrifugation at 25 000g for 20 minutes. The pellet containing crude microsomal material was suspended in 5 volumes of PBSI; the suspension was homogenized using the polytron, followed by centrifugation at 6000g for 10 minutes to remove any remaining particulates, with the resulting supernatant subjected to centrifugation at 25 000g for 20 minutes. The resulting microsomal pellet was washed twice more by repeated suspension into 5 volumes of PBSI, homogenization, and centrifugation at 25 000g for 20 minutes. The final microsomal pellet was suspended in 1 mL of PBSI per 10-g placental starting material. For TFPI isoform analysis, microsomes were treated with no additions, 4 U/mL PIPLC alone, or PIPLC plus 10 U/mL heparin for 1 hour at room temperature. After clarification by centrifugation at 25 000g for 20 minutes, PIPLC-released TFPI was analyzed by ELISA and Western blot analysis. For purification of placental TFPI, microsomes were treated with 4 U/mL PIPLC, and TFPI was purified from the releasate by 2H8-affinity chromatography.
Immunoaffinity isolation of TFPI
TFPI was purified from (1) serum-free conditioned media of C127 cells and CHO cells expressing human rTFPIα, (2) PIPLC releasate from CHO cells expressing TFPIβ, (3) SK–Hep-1 PIPLC releasate, and (4) placental microsomal PIPLC releasate. Briefly, Mab2H8 was linked at 5 mg/mL to Affi-gel matrix (Bio-Rad). Media or releasate was passed at 0.5 mL/minute over a 5-mL 2H8-Affigel column equilibrated in 100mM NaCl2, 20mM HEPES, 0.02% sodium azide, pH 7.4. In each case, the column was washed with 20 volumes of start buffer and eluted with 0.1M glycine, pH 2.5; 2-mL fractions were collected into tubes containing one-tenth volume of 1M Tris-HCl, pH 8.0. Fractions containing protein were pooled, concentrated, and buffer-exchanged into PBSE using 10 000 molecular weight cutoff Millipore concentrators.
Deglycosylation and Western analysis of TFPI
TFPI samples were subjected to deglycosylation using the EDEGLY-1KT system as described by the manufacturer using the combination of PNGase F, O-glycosidase, and α-2(3,6,8,9)–neuraminidase unless otherwise noted. After the treatment, samples were evaluated by Western blot analysis with chemiluminescent detection. HRP-conjugated rabbit polyclonal anti-TFPI antibody was used to detect TFPI.6 To detect GPI anchors, Western blots were incubated for 1 hour with 50nM FLAER in 0.5% casein buffer (0.5% casein, 150mM NaCl, 50mM Tris, pH 9), followed by 2 washes with 0.5% casein buffer and incubation with 2 μg/mL rabbit anti–Alexa-488 antibody for 1 hour. After 2 washes with 0.5% casein buffer, the blot was incubated for 1 hour with HRP-conjugated goat anti–rabbit antibody, and after 4 washes with PBS plus 0.1% Tween-20 (PBST) detection was achieved by chemiluminescence.
ELISAs to quantify TFPIα and TFPIβ
Two ELISA assays were used. The R&D Systems Human TFPI Quantikine ELISA Kit uses an anti-K3 monoclonal antibody for capture and polyclonal anti-TFPI antibody for detection. It recognizes TFPIα that contains a Kunitz-3 domain, but neither truncated TFPIα that lacks a Kunitz 3 domain nor TFPIβ. This assay was performed as recommended, except that human recombinant TFPIα expressed in Escherichia coli was used as standard. The TFPItotal immunoassay uses mAbs 2H8 (anti-K1) and 2B12 (anti-K2) and recognizes both TFPIα and TFPIβ and has been described.6 TFPItotal and TFPIα immunoassays were run side by side with shared sample dilutions and standards. As no appreciable amounts of truncated forms of TFPI were detected by Western blot, TFPIβ was calculated as TFPItotal minus TFPIα. Data in figures are mean ± SE, for a minimum of 3 experiments.
Results
Distribution of rTFPI isoforms
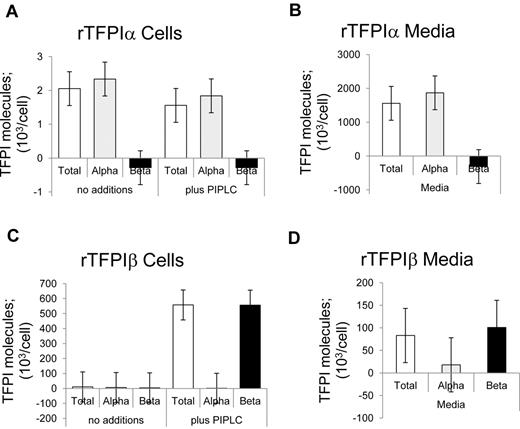
The distributions of TFPI expressed in media and released from isolated cells on treatment without and with PIPLC were determined using immunoassays. The TFPItotal ELISA recognized both rTFPIα and rTFPIβ isoforms (Figure 1). By contrast, the TFPIα-specific ELISA, which uses a Kunitz 3-specific capture antibody, recognized rTFPIα but did not detect rTFPIβ. With respect to rTFPIα, incubation of isolated cells for 1 hour, under PIPLC reaction conditions, in the presence or absence of PIPLC enzyme, resulted in the detection of soluble TFPIα levels that were approximately 0.1% of that expressed in the media over a 24-hour period, and PIPLC had no effect on release of rTFPIα. With respect to rTFPIβ, PIPLC treatment resulted in significant release of TFPI from CHO cells as detected by the TFPItotal assay, but the TFPIα-specific ELISA did not recognize this TFPI. Although TFPIβ is directed to be GPI-anchored, an appreciable level of TFPIβ was identified in rTFPIβ overexpressing CHO cell media.
Distribution of recombinant TFPI isoforms. (A-B) CHO cells expressing rTFPIα. (C-D) CHO cells expressing rTFPIβ. (A,C) Washed cells treated with PIPLC. (B,D) Serum-free conditioned media. Samples were analyzed for TFPI isoforms using immunoassays.
Distribution of recombinant TFPI isoforms. (A-B) CHO cells expressing rTFPIα. (C-D) CHO cells expressing rTFPIβ. (A,C) Washed cells treated with PIPLC. (B,D) Serum-free conditioned media. Samples were analyzed for TFPI isoforms using immunoassays.
Distribution of TFPI isoforms in endothelial cell lines
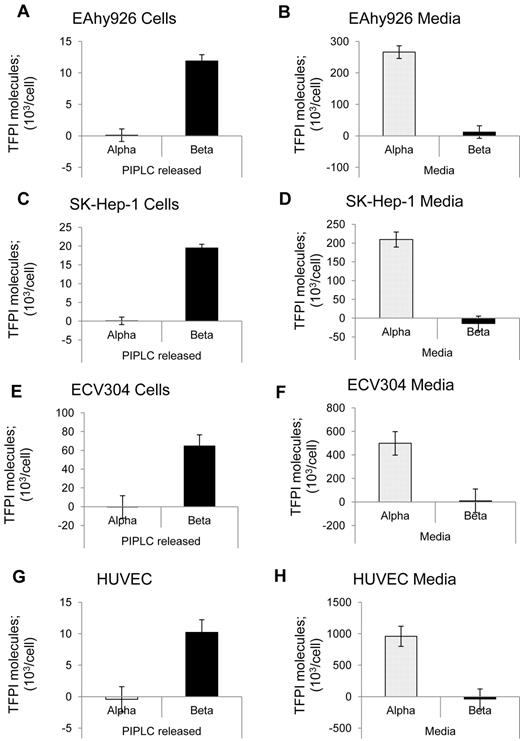
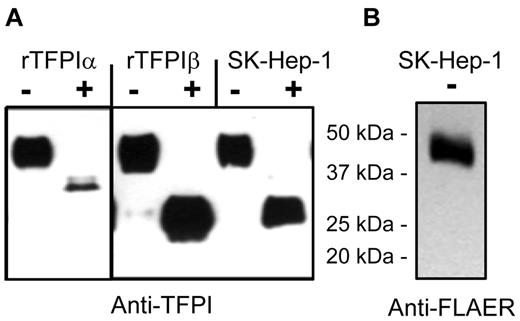
The distribution of TFPI isoforms expressed into media and released by PIPLC from cells was evaluated using the endothelial cell lines EAhy926, SK–Hep-1, and ECV304 and primary HUVECs. Figure 2 shows that these 4 cell types express TFPIα as the dominant isoform in conditioned media, with no significant TFPIβ detected in the media. By contrast, PIPLC treatment of cells released TFPI that was recognized by the TFPItotal ELISA but not recognized by the TFPIα-specific ELISA. The PIPLC-released TFPI migrated as a single 46-kDa band for each cell type, as shown for SK–Hep-1 cells (Figure 3). FLAER recognizes GPI-anchor moieties after PIPLC treatment, and FLAER recognized the 46-kDa band in the affinity-purified TFPI from SK–Hep-1 PIPLC releasate.
Distribution of TFPI isoforms for various endothelial cell lines. (A-B) EAhy926 cells. (C-D) SK–Hep-1 cells. (E-F) ECV304 cells. (G-H) HUVECs. (A,C,E,G) Washed cells were treated without or with PIPLC and analyzed for TFPI isoforms using immunoassays. PIPLC release was calculated by subtracting the without-PIPLC levels from the with-PIPLC levels. (B,D,F,H) Serum-free conditioned media were analyzed for TFPI isoforms using immunoassays.
Distribution of TFPI isoforms for various endothelial cell lines. (A-B) EAhy926 cells. (C-D) SK–Hep-1 cells. (E-F) ECV304 cells. (G-H) HUVECs. (A,C,E,G) Washed cells were treated without or with PIPLC and analyzed for TFPI isoforms using immunoassays. PIPLC release was calculated by subtracting the without-PIPLC levels from the with-PIPLC levels. (B,D,F,H) Serum-free conditioned media were analyzed for TFPI isoforms using immunoassays.
Western blots and identification of TFPI isoforms by deglycosylation for SK–Hep-1 endothelial cells. (A) Recombinant TFPIα, PIPLC-released rTFPIβ, and PIPLC-released SK–Hep-1 TFPI were affinity purified and then treated without (−) or with (+) degylcosylation enzymes and probed using anti-TFPI polyclonal antibodies. (B) PIPLC-released SK–Hep-1 TFPI probed with FLAER and anti-FLAER.
Western blots and identification of TFPI isoforms by deglycosylation for SK–Hep-1 endothelial cells. (A) Recombinant TFPIα, PIPLC-released rTFPIβ, and PIPLC-released SK–Hep-1 TFPI were affinity purified and then treated without (−) or with (+) degylcosylation enzymes and probed using anti-TFPI polyclonal antibodies. (B) PIPLC-released SK–Hep-1 TFPI probed with FLAER and anti-FLAER.
Although TFPIα and TFPIβ each migrate on SDS-PAGE as 46-kDa gylcoproteins, TFPIα and TFPIβ isoforms can be distinguished by deglycosylation followed by Western blot analysis, as shown in Figure 3. Deglycosylated rTFPIα and rTFPIβ migrate as 36-kDa and 28-kDa proteins, respectively. Deglycosylated TFPI purified from SK–Hep-1 PIPLC-releasate consisted of a single band at 28 kDa. Taken together, these data demonstrate that TFPIβ is the dominant PIPLC-releasable form on endothelial cells.
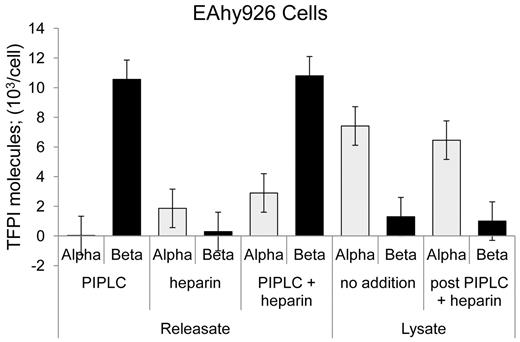
Heparin has been shown to release a small fraction of cellular TFPI, and FACS analysis indicated that this occurs with no diminution in cell surface TFPI expression.15,17,21 TFPItotal and TFPIα-specific ELISAs were performed for EAhy926 cells treated with PIPLC, heparin, or both. Heparin treatment released only TFPIα at a modest level of approximately 15% that of PIPLC-released TFPI (Figure 4). EAhy926 cells treated with no additions or heparin and PIPLC and then subjected to lyses by 2 freeze-thaw cycles released predominantly TFPIα. Similar amounts were released irrespective of “pre”-treatment and at a level of approximately 65% of the TFPIβ amount released from cell surface by PIPLC treatment (Figure 4).
Release of TFPI isoforms from EAhy926 cells by various treatments. EAhy926 cells were treated as indicated with no additions, PIPLC, and PIPLC plus heparin, and releasates were analyzed using immunoassays. Cells treated with no addition and PIPLC plus heparin were collected by centrifugation, washed once, and lysed by 2 freeze-thaw cycles. Lysates were clarified by centrifugation and supernatants analyzed for TFPI isoforms by immunoassays.
Release of TFPI isoforms from EAhy926 cells by various treatments. EAhy926 cells were treated as indicated with no additions, PIPLC, and PIPLC plus heparin, and releasates were analyzed using immunoassays. Cells treated with no addition and PIPLC plus heparin were collected by centrifugation, washed once, and lysed by 2 freeze-thaw cycles. Lysates were clarified by centrifugation and supernatants analyzed for TFPI isoforms by immunoassays.
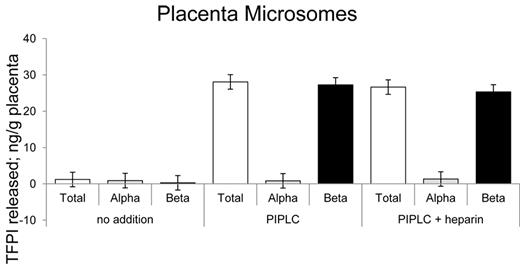
TFPIβ is the PIPLC-released isoform on human placental microsomes
A preliminary PIPLC dose-response curve revealed that treatment of placental microsomes with 2 units of PIPLC per milliliter released maximal levels of TFPI within 1 hour of incubation with no additional effects with up to 16 units of PIPLC (data not shown). On that basis, placental microsome preparations were routinely treated with 4 units per milliliter PIPLC to assure maximal TFPI release. Shown in Figure 5 are the ELISA results for placental microsomes treated with no additions, with PIPLC and with PIPLC plus heparin. Greater than 95% of the PIPLC-dependent released TFPI was recognized by the TFPItotal ELISA and not the TFPIα-specific ELISA. Heparin treatment showed no additional change in TFPI levels than that released by PIPLC treatment.
Release of TFPI isoforms from placental microsomes. Placental microsomes were treated as indicated with no additions, PIPLC, and PIPLC plus heparin, and releasates were analyzed using immunoassays.
Release of TFPI isoforms from placental microsomes. Placental microsomes were treated as indicated with no additions, PIPLC, and PIPLC plus heparin, and releasates were analyzed using immunoassays.
Western blots with anti-TFPI antibodies showed that the predominant form (> 95% by densitometry) of TFPI released by PIPLC treatment of placental microsomes is a 46-kDa isoform consistent in size with being either full-length TFPIα or TFPIβ (Figure 6A). Deglycosylation of affinity-purified TFPI from placental PIPLC releasate yielded an approximately 28-kDa band that runs similarly to deglycosylated rTFPIβ but not rTFPIα. Western analysis using FLAER and anti-FLAER antibody for the detection of GPI anchor-reactive moieties also identified the 46-kDa band of affinity-purified TFPI from placental microsomes and the 28-kDa deglycosylated form when rPNGase F (kindly provided by Evan Sadler, Washington University, St Louis, MO) was used in the deglycosylation reaction (Figure 6B). Notable, commercially available PGNase F (that is not recombinant) destroyed the FLAER recognition site (data not shown), presumably because of a contaminating activity. Taken together, these results demonstrate that the PIPLC-releasable TFPI isoform of placental microsomes is TFPIβ.
Western blots of placental microsomal TFPI. (A) Recombinant TFPIα, PIPLC-released rTFPIβ, and PIPLC-released TFPI from placental microsomes were affinity purified and then treated without (−) or with (+) degylcosylation enzymes and probed using anti-TFPI polyclonal antibodies. (B) Placental microsomal TFPI treated without (−) or with (+) degylcosylation enzymes and probed with FLAER and anti-FLAER.
Western blots of placental microsomal TFPI. (A) Recombinant TFPIα, PIPLC-released rTFPIβ, and PIPLC-released TFPI from placental microsomes were affinity purified and then treated without (−) or with (+) degylcosylation enzymes and probed using anti-TFPI polyclonal antibodies. (B) Placental microsomal TFPI treated without (−) or with (+) degylcosylation enzymes and probed with FLAER and anti-FLAER.
Discussion
The human TFPI gene is putatively expressed as 2 isoforms from mRNAs generated by alternative intron/exon splicing. Based on canonical signal sequences, one would expect TFPIα to be secreted and TFPIβ to be directly GPI-anchored to cell surfaces. Indeed, recombinant TFPIα expressed by CHO cells is predominantly secreted into media, whereas recombinant TFPIβ is largely associated with cell surfaces and released by PIPLC treatment. Although TFPIβ mRNA has been detected in endothelial cells, before this report evidence for the expression of the human TFPIβ protein was limited to indirect identification in ECV304 cells.6,15,17 Consistent with the expected distribution of native TFPI isoforms based on encoded signal sequences, we demonstrate here the presence of TFPIβ on endothelial cells and in placental microsomes and show that GPI-anchored TFPIβ is the predominant, and perhaps sole, TFPI isoform released by PIPLC treatment.
It has been reported that TFPIβ was not produced by EAhy926 endothelial cells.21 By contrast, we show that EAhy926 cells (and other endothelial cells) express TFPIβ that is released by PIPLC treatment. In HUVECs and endothelial-like cell lines (EAhy926, ECV304), the ratio of TFPIα/TFPIβ mRNA varies between 5 and 10.17 In proportion to this ratio, our data suggest that, over a 24 hour period, endothelial cell cultures secrete from 10 to 50 times more TFPIα into media compared with the amount of PIPLC-releasable GPI-anchored TFPI found on the cell surfaces and that the soluble portion of cellular lysates contain predominantly TFPIα.
The initial characterization of the association of TFPI with human placenta concluded that GPI-anchored TFPIα was the dominant form on placental endothelium, based on the observation that both anti-TFPI polyclonal antibody and antibody directed to the C-terminus of TFPIα recognized a PIPLC-releasable TFPI approximately 41-kDa band on Western blots.22 Perhaps due in part to the lack of a means to quantify TFPIα and TFPIβ isoforms at the time, several groups, including ours, presumed that TFPIα was the dominant PIPLC-releasable TFPI isoform in placenta and on endothelial surfaces. With the limited TFPIβ-specific peptide sequence (predicted to be just 12 residues with an attached GPI-anchor) and possible glycosylation, we and others have been unable to generate potent TFPIβ-specific peptide antibodies that directly recognize TFPIβ protein. However, using presently available ELISAs that can discriminate between TFPI that contains Kunitz-3 domain from TFPI that lacks a Kunitz-3 domain, and deglycosylation coupled to Western blot analysis that distinguishes TFPIα from TFPIβ isoforms, we are able to report here that TFPβ, not TFPIα, is the dominant GPI-anchored TFPI in human placenta and on endothelial cell surfaces. Our data show that PIPLC treatment of placental microsomes (extensively washed with PBS containing 1mM EDTA and protease inhibitor cocktail) releases TFPI that (1) migrates on SDS-PAGE at a size of 46 kDa, and on deglycosylation migrates at 28 kDa; (2) lacks a Kunitz-3 domain based on a TFPIα-specific ELISA; and (3) contains a GPI-anchor based on recognition by FLAER. These characteristics are consistent with only TFPIβ. The notion of a GPI-anchored “coreceptor” for the localization of TFPIα at the cell surface appears unnecessary.
Maroney et al have reported that adult mouse plasma and detergent-lysed mouse tissues contain predominantly TFPIβ, whereas detergent-lysed mouse placenta contains predominantly TFPIα.26 Whether the TFPIα in murine placenta is cell-surface associated, PIPLC releasable, or intracellular was not determined.
Although heparin treatment elevates TFPIα plasma levels and releases TFPIα from endothelial cell cultures, for placental microsomes we found no significant release of TFPI by heparin. This is similar to that reported by Mast et al, where a very limited release of TFPI by heparin was observed, and they estimated that there is 10- to 100-fold more GPI-anchored TFPI than heparin-releasable TFPI on PBS/EDTA washed placental fragments.22
We estimate the quantity of endothelial cell surface TFPIβ in humans to be 170 nmol based on the fact that PIPLC releases 10 000 molecules of TFPIβ per endothelial cell in HUVEC culture, assuming that all endothelial cells express this number of molecules on average and assuming that there are 1012 endothelial cells in the body.27 Based on the average circulating plasma level of 1.6nM TFPI and using a plasma volume of 3 L,28 the normal amount of circulating plasma TFPI can be calculated to be 4.8 nmol, of which approximately 80% lacks a Kunitz-3 domain. Heparin infusion releases an additional approximately 7.2 nmol of full-length TFPIα into plasma with its source presumed to be the endothelium.11-13 Using these assumptions, approximately 95% of readily available mature TFPI in the body would appear to be GPI-anchored TFPIβ.
There is probably an additional, intraendothelial cell reservoir of TFPI that is not released by heparin or PIPLC (which together deplete surface TFPI)15,19 but is released by cell lysis. Extrapolating from our results with EAhy926 cells, this repository could represent an additional approximately 100 nmol of intracellular TFPI that is predominantly TFPIα. This intracellular TFPI may be immature, partially processed TFPI, TFPI within a degradation pathway, or a storage form of TFPI that can be released by stimuli other than heparin.29
The TF/FVIIa complex triggers coagulation and, along with the transient TF/FVIIa/FXa complex, also signals proinflammatory responses through cell surface protease-activated receptors.30 Only TFPIα containing a Kunitz-3 domain and C-terminus is a highly potent inhibitor of FXa. By comparison, full-length TFPIα, truncated 2-Kunitz domain TFPIα, and TFPIβ each effectively inhibit the TF/FVIIa/FXa complex. The location of TFPIβ GPI-anchored on endothelial cell surfaces suggests a plausible role for TFPIβ regulation of TF-mediated inflammatory responses via protease-activated receptor signaling. Additional investigation of the relative roles that TFPI isoforms play in modulating coagulation and inflammatory responses is warranted.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jeremy P. Wood and Paula B. Tracy of the University of Vermont for sharing their methodology for identification of GPI anchors using the FLAER and the rabbit anti–Alexa-488.
This work was supported by the National Institutes of Health (grant 5R01HL077193).
National Institutes of Health
Authorship
Contribution: G.J.B., T.J.G., and E.T. designed the research, performed experiments, and analyzed results; and G.J.B. and T.J.G. wrote the paper and made the figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: George J. Broze Jr, Washington University School of Medicine, Division of Hematology, Box 8125, 660 S Euclid Ave, St Louis, MO 63110; e-mail: gbroze@im.wustl.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal