Abstract
The hypoxia-inducible transcription factor-1α (HIF-1α) is a major regulator of angiogenesis, carcinogenesis, and various processes by which cells adapt to hypoxic conditions. Therefore, the identification of critical players regulating HIF-1α is not only important for the understanding of angiogenesis and different cancer phenotypes, but also for unraveling new therapeutic options. We report a novel mechanism by which HIF-1α is degraded after glycogen synthase kinase-3 (GSK-3)–induced phosphorylation and recruitment of the ubiquitin ligase and tumor suppressor F-box and WD protein Fbw7. Further, experiments with GSK-3β and Fbw7-deficient cells revealed that GSK-3β and Fbw7-dependent HIF-1α degradation can be antagonized by ubiquitin-specific protease 28 (USP28). In agreement with this, Fbw7 and USP28 reciprocally regulated cell migration and angiogenesis in an HIF-1α–dependent manner. Therefore, we have identified a new pathway that could be targeted at the level of GSK-3, Fbw7, or USP28 to influence HIF-1α–dependent processes like angiogenesis and metastasis.
Introduction
Adequate functioning of mammalian cells requires the presence of oxygen. When oxygen availability is limited, cells initiate processes allowing adaptation to these hypoxic conditions. Hypoxia-inducible transcription factors (HIFs) are crucial regulators of these adaptational responses. Of the 3 HIF family members known to date, HIF-1 is the best characterized, and is known to regulate the expression of more than 100 genes,1 products of which control angiogenesis, oxygen transport, glucose metabolism, vascular tone, and cell proliferation and survival.2,3 HIF-1 exists as a dimer composed of the proper hypoxia-inducible α subunit (HIF-1α) and the constitutively expressed β-subunit, which is also known as arylhydrocarbon receptor nuclear translocator (ARNT). Hypoxia is also a major feature of several diseases, and almost all solid tumors contain hypoxic regions in which the oxygen concentrations are greatly reduced compared with the surrounding tissue. Further, several studies from various tumor entities showed that increased levels of HIFs, especially HIF-1α, are associated with a poor prognosis.4,5
The protein stability of HIF-1α is crucially regulated by the hydroxylation of 2 proline residues (P402 and P564)6-8 located in the oxygen-dependent degradation domain, whereas the transactivity can be additionally influenced by the hydroxylation of an asparagine residue (N803) in the C-terminal transactivation domain.9 These reactions are carried out by at least 4 proline hydroxylase domain–containing enzyme family members.6,8-12 The hydroxylation of HIF-1α is the prerequisite for the recruitment of the tumor suppressor protein von Hippel-Lindau (VHL),13-17 which acts as an E3 ubiquitin ligase and targets HIF-1α for ubiquitylation and proteasomal degradation.8,15,18,19 In this way, the VHL βdomain binds directly to HIF-1α, whereas the α-domain binds to elongin C, which in turn nucleates a complex that contains elongin B, Cul2, and Rbx1/Roc1.16,17 Moreover, recent work from several groups, including our own, showed that HIF-1α can also be degraded in a VHL-independent manner, whereas the mechanisms by which the Hsp90 inhibitor 17-AAG and the receptor of activated protein kinase C (RACK1)20 or the hypoxia-associated factor (HAF)21 are recruited to HIF-1α remained unknown. However, our own investigations suggested that phosphorylation of HIF-1α by glycogen synthase kinase-3 (GSK-3) could be a signal for the recruitment of another as-yet-unknown E3 ubiquitin ligase.22 Interestingly, it has been shown that other short-living proteins such as c-Myc, Jun, Notch, and cyclin E could be ubiquitylated in a GSK-3–dependent manner by a SCF (Skp1, Cul1, and F-box protein) complex containing the F-box protein Fbw7 (also called hCdc4, Sel-10, or Fbxw7).23,24 Similar to VHL, F-box proteins constitute the substrate-recognition component that, together with S-phase kinase-associated protein 1 (SKP1), cullin 1 (CUL1), and RING box 1 (RBX1, also called ROC1 or HRT1) form the so called SCF complex multi-subunit E3 ubiquitin ligases.
There are 3 known Fbw7 isoforms (Fbw7α, Fbw7β, and Fbw7γ) produced by alternative splicing that localize to the nucleoplasm, cytoplasm, and nucleolus, respectively.23,25,26 The importance of the fbw7 gene for proliferative processes was indicated by several studies. The heterozygous inactivation of the fbw7 gene was found to be associated with malignant transformation, especially in ovarian cells and T cells,27 in breast cancer cells,28 and later also in human colorectal cancers,29 suggesting that Fbw7 may be a tumor suppressor associated with chromosomal instability and some types of malignancy.
Because hypoxia and HIF-1α were found to be critical determinants of cell proliferation, growth, and apoptosis, we hypothesized that Fbw7 may target HIF-1α for ubiquitination and degradation after it became phosphorylated by GSK-3. In addition, the action of Fbw7 on c-Myc can be counteracted partially by the ubiquitin-specific protease USP28.30 Therefore, the aim of the present study was to investigate the potential of Fbw7 and USP-28 to regulate HIF-1α protein stability and whether they influence cell migration, invasion, and proliferation in an HIF-1α–dependent manner. Using cells deficient for GSK3β (Fbw7−/−) and by performing knockdown studies with Fbw7 shRNA and USP28 shRNA, we found that Fbw7 can mediate ubiquitylation of HIF-1α when it was phosphorylated by GSK-3 and that the Fbw7-mediated reduction in the HIF-1α protein half-life was counteracted by USP28. The negative regulation of HIF-1α by Fbw7 was independent of the VHL-recruiting hydroxylation sites, but mutation of the GSK-3 phosphorylation sites abolished the Fbw7 effects. In agreement with this, the GSK-3β and Fbw7-dependent down-regulation of HIF-1α is important for angiogenesis, because overexpression of Fbw7 in endothelial cells affected hypoxia-dependent tube formation. Conversely, overexpression of USP28 increased tube formation, and knocking down HIF-1α reversed these effects. Our present results provide a novel mechanism by which GSK-3β–dependent phosphorylation of HIF-1α can result in the Fbw7-dependent but VHL-independent degradation of HIF-1α, which is reversibly regulated by USP28.
Methods
Materials
All biochemicals and enzymes were of analytical grade and were obtained from commercial suppliers.
Cell culture
HepG2, HeLa, and HEK 293 cells were cultured under normoxia (16% O2, 79% N2, and 5% CO2 by volume) in MEM supplemented with 10% FCS. Human microvascular endothelial cells (HMEC-1) were cultured under the same atmosphere in MCDB131 medium (Gibco) supplemented with 10% FCS. Mouse embryonic fibroblasts GSK-3β+/+ and GSK-3β−/− were cultured in DMEM with 10% FCS and were a gift from J. Woodgett (Samuel Lunenfeld Research Institute, Toronto, ON). We obtained HCT116 Fbw7+/+ and HCT116 Fbw7−/− cells29 from B. Vogelstein (Johns Hopkins University, Baltimore, MD) and grew them under normoxia in McCoy 5a medium with 10% FCS. For protein extraction, cells were seeded into 10-cm dishes and, when necessary, transfections were performed the next day for 12 hours. The medium was then changed and cells were further cultured either under normoxia or hypoxia (5% O2, 90% N2, and 5% CO2 by volume).
Plasmid constructs
The construct for shHIF-1α,31 has been described previously. The various constructs for full-length HIF-1α with mutations in proline 402, serine 551, threonine 555, serine 589, proline 564, and asparagine 803 were generated using the QuickChange mutagenesis kit (Promega).
The constructs for the Fbw7 expression vectors (p3XFlag-Fbw7α, p3XFlag-Fbw7β, p3XFlag-Fbw7γ, and p3XFlag-Fbw7WD) were obtained from M. Welcker and B. Clurman (both Fred Hutchinson Cancer Research Center, Seattle, WA) and have been described previously.24
The constructs for pRetrosuper-Fbw7 shRNA-1, pRetrosuper-Fbw7 shRNA-2, pDZ-Flag-USP28, pRetrosuper-USP28 shRNA-1, and pRetrosuper-USP28 shRNA-3 were described previously.30
RNA preparation and northern blot analysis
The isolation of total RNA and northern blot analysis was performed essentially as described previously.32 Digoxigenin-labeled antisense RNAs, generated by in vitro transcription from pCRII-HIF-800 and pBS-actin using T7 polymerase, served as hybridization probes. Blots were quantified with a video densitometer (Biotech Fischer).
Western blot analysis, HIF-1α protein half-life studies, and HIF-1α immunoprecipitation and ubiquitylation assays
Western blot analysis was carried out as described previously.32 In brief, lysates or culture medium from HepG2, HEK 293, GSK-3β+/+, GSK-3β−/−, HCT116 Fbw7+/+, and HCT116 Fbw7−/− cells were collected and 100 μg of protein was loaded onto a 7.5% or 10% SDS-PAGE gel. After electrophoresis and electroblotting onto a nitrocellulose membrane, proteins were detected with mAbs against human HIF-1α (1:2000; BD Biosciences), c-Myc (1:500; Santa Cruz Biotechnology), Flag M2 (1:1000; Sigma-Aldrich), V5 tag (1:5000; Invitrogen), GSK-3α/β (1:1000; Santa Cruz Biotechnology), human PAI-1 (1:200; American Diagnostica), or hemagglutinin (1:500; Santa Cruz Biotechnology). Polyclonal Abs were against HIF-1α (1:1000; Novus Biologicals), ubiquitin (1:500; Santa Cruz Biotechnology), albumin (1:500; Nordic Immunology), or Golgi membrane (1:1000; Bioscience). The secondary Ab was either an anti–mouse or an anti–rabbit IgG conjugated to HRP (1:5000; Bio-Rad). The ECL system (Amersham) was used for detection.
For half-life studies, HEK 293 cells were transfected with expression vectors encoding Flag-tagged Fbw7γ, hemagglutinin-tagged GSK-3β, or Flag-tagged USP28. After 24 hours, cycloheximide (10 μg/mL; Sigma-Aldrich) was added to the medium, cells were harvested at the indicated time points and protein levels were measured by immunoblot analysis. For immunoprecipitation and ubiquitylation assays, the cells were treated with the proteasome inhibitor MG 132 (50μM; Calbiochem). Four hours later, cells were scraped in lysis buffer (50mM Tris/HCl, pH 7.5, 150mM NaCl, 1% Triton X-100, 2mM EDTA, 2mM EGTA, 1mM PMSF, and complete protease inhibitor cocktail tablet; Roche). After scraping, lysates were incubated with continuous shaking at 4°C for 20 minutes and then centrifuged at 12 000g at 4°C for 15 minutes. To recover anti-V5 immunoprecipitates, 150 μg of protein was incubated with 2 μg of Ab for 1 hour at 4°C before Sepharose beads (30 μL per reaction mixture) were added for 12 hours. Thereafter, the beads were washed 5 times with lysis buffer and recovered, pellets were dissolved in 2× Laemmli buffer, loaded onto a 7.5% SDS gel, blotted, and detected with Abs against ubiquitin and the Flag epitope.
Cell proliferation assay
When indicated, GSK-3β+/+ and GSK-3β−/− cells were transfected with expression vectors for USP28, shHIF-1α, shUSP28, and shFbw7 or with the respective control vectors using Metafectene Pro for 6 hours. Cells were seeded onto 96-well plates at a density of 10 000 cells per well and allowed to settle overnight. After a medium change, the cells were labeled with bromodeoxyuridine (BrdU) for 24 hours and the BrdU cell proliferation kit (Calbiochem) was used for detection.
Protein phosphorylation
GSK-3β+/+ and GSK-3β−/− cells were treated with MG 132 (50μM; Calbiochem) for 4 hours, scraped, and 500 μg of total protein was incubated for 1 hour at 4°C with 2 μg of Ab to recover either endogenous HIF-1α or Gal4-HIF fusion proteins. Next, Sepharose beads (30 μL per reaction mixture) were added overnight at 4°C. Thereafter, the beads were washed 5 times and recovered pellets were dissolved in 2× Laemmli buffer and loaded onto a 7.5% or 10% SDS gel. Thereafter, gels were fixed 2 times in 100-mL fixation solution (50% methanol and 10% acetic acid) for 30 minutes and washed 3 times with 100 mL of distilled water for 10 minutes. Phosphoproteins were visualized with the Pro-Q Diamond Phosphoprotein gel stain (Invitrogen); gels were incubated in 60 mL of Pro-Q Diamond stain in the dark for 90 minutes and then destained 3 times in a solution containing 20% acetonitrile, 50mM sodium acetate, pH 4, for 30 minutes, followed by washing 2 times with distilled water for 5 minutes. Images of the stained gels were acquired using a Typhoon imager (Molecular Dynamics). Phosphoprotein stained gels were poststained with Coomassie blue.
Anchorage-independent growth in soft agar
A mixture of 25 μL of prewarmed (37°C) 2× DMEM (DMEM/F12; Invitrogen) containing 20% FCS and 25 μL of prewarmed (56°C) 1.2% agar (DNA grade) were plated onto each well of a 96-well plate to serve as a prelayer for the assay. Ten microliters of a GSK-3β+/+ or GSK-3β−/− cell suspension containing 2 × 103 cells was mixed with 20 μL of 2× DMEM/F12 and 30 μL of 0.8% agar (DNA grade) and transferred into the 96-well plate. Semisolid feeder layers were then prepared by mixing 25 μL of 2× DMEM/F12 and 25 μL of 1.2% agar (DNA grade) and layered on top of the solidified cell layers. The cells were allowed to grow in a humidified 37°C incubator with 5% CO2 for 1-2 weeks. Cell growth was measured using the Alamar blue assay (Invitrogen) and fluorescence was recorded in a Fluroskan Ascent FL type 374 (Thermo Scientific) with an excitation wavelength of 530 nm and emission at 590 nm.
Cell migration assay
Cell migration was measured using medium-treated 24-well Transwell chambers (BD Biosciences) with 8.0-μm polycarbonate membranes. The bottom chamber was filled with 600 μL of medium containing 10% FCS; cells were seeded into the top chamber at a density of 1 × 104 cells per well in 100 μL of serum-free medium. After incubation in a humidified incubator with 5% CO2 at 37°C for 16 hours, cells were fixed with 4% paraformaldehyde and the nonmigratory cells were scraped off from the top of the Transwell with a cotton swab. The migrated cells attached to the bottom chamber were stained with 0.0075% crystal violet.
In vitro Matrigel angiogenesis assay
HMEC-1 cells were transfected with expression vectors for Fbw7, USP28, shFbw, shUSP, and shHIF-1α or with the respective control vectors. Cells were seeded at a density of 50 000 cells per well on a 96-well plate mounted with growth factor–reduced Matrigel (BD Biosciences). The cells were allowed to settle for 2 hours in normoxia and were then cultured for 6 hours at 1% oxygen. After incubation, the cells were stained with the fluorescent dye Calcein AM (Mobitec). The formation of capillary-like structures was assessed by light microscopy and quantified using ImageJ Version 1.457 software at the Wright Cell Imaging Facility in Toronto, ON.
Statistical analysis
Densitometry data were plotted as the fold induction of relative density units, with the zero value absorbance in each figure set arbitrarily to 1 or 100%. Statistical comparisons of absorbance differences were performed by the Mann-Whitney test (Statview Version 4.5 software; Abacus Concepts), and P ≤ .05 was considered significant. Luc values presented are means ± SEM. Results were compared by ANOVA for repeated Luc measurements followed by the Newman-Keuls test.
Results
GSK-3β–dependent phosphorylation targets HIF-1α for ubiquitylation
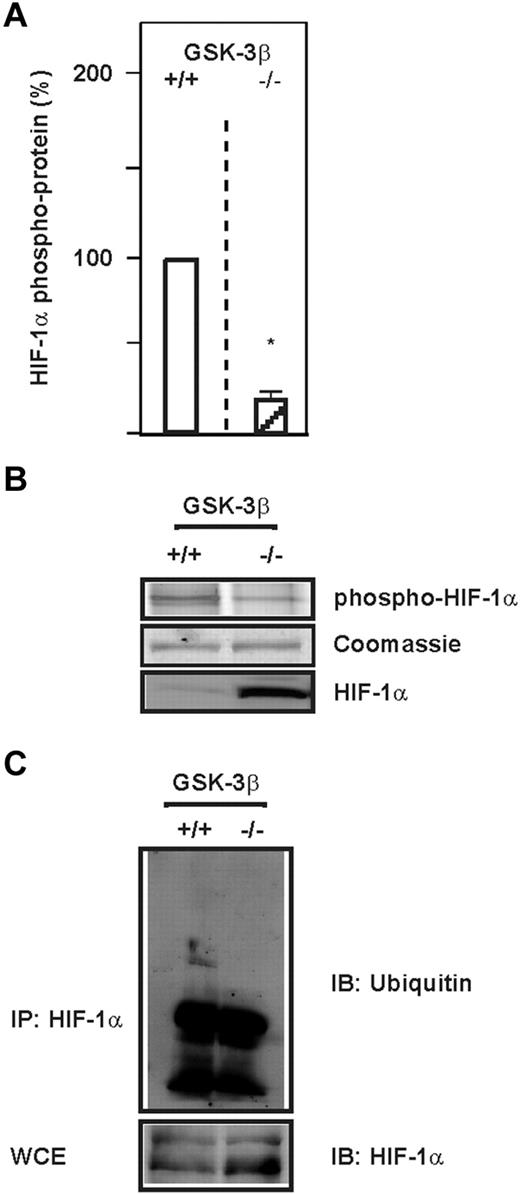
GSK-3 has been found to phosphorylate many proteins playing important roles in a variety of cellular processes, including cell proliferation, differentiation, and apoptosis.33 To investigate whether GSK-3β can also phosphorylate HIF-1α in intact cells and whether this contributes to HIF-1α ubiquitylation, we used GSK-3β–deficient mouse embryonic fibroblasts (GSK-3β−/− MEFs). We found that total HIF-1α protein levels and phospho HIF-1α levels were reciprocally regulated; in GSK-3β−/− cells, total HIF-1α levels were high and phospho HIF-1α levels were decreased by approximately 80% compared with the wild-type cells (Figure 1A-B). This is in agreement with our previous findings showing that HIF-1α phosphorylation by GSK-3β was lost when S551, T555, and S589 were substituted with alanine.22 We next investigated whether the phosphorylation by GSK-3β can promote HIF-1α ubiquitylation on the endogenous level. When lysates from GSK-3β−/− cells were immunoprecipitated with an HIF-1α Ab and probed with an ubiquitin Ab, we found that GSK-3β−/− MEFs contained less ubiquitylated HIF-1α then wild-type MEFs (Figure 1C). These data indicate that GSK-3β phosphorylates HIF-1α and promotes ubiquitylation of the HIF-1α protein.
GSK-3β phosphorylates HIF-1α and induces its ubiquitylation. (A) HIF-1α was immunoprecipitated from GSK-3β+/+ and GSK-3β−/− cells and phospho HIF-1α protein levels were measured with the Pro-Q Diamond Phosphoprotein Gel Stain. The phospho HIF-1α protein levels from GSK-3β+/+ cells were set to 100%. Values are means ± SEM of 3 independent experiments. *Significant difference between GSK-3β+/+ versus GSK-3β−/−. (B) Representative Pro-Q Diamond Phosphoprotein Gel Stain analysis. (C) HIF-1α was immunoprecipitated (IP) from GSK-3β+/+ and GSK-3β−/− cells and ubiquitylation was analyzed by immunoblotting (IB) with ubiquitin Abs. WCE indicates whole-cell lysates.
GSK-3β phosphorylates HIF-1α and induces its ubiquitylation. (A) HIF-1α was immunoprecipitated from GSK-3β+/+ and GSK-3β−/− cells and phospho HIF-1α protein levels were measured with the Pro-Q Diamond Phosphoprotein Gel Stain. The phospho HIF-1α protein levels from GSK-3β+/+ cells were set to 100%. Values are means ± SEM of 3 independent experiments. *Significant difference between GSK-3β+/+ versus GSK-3β−/−. (B) Representative Pro-Q Diamond Phosphoprotein Gel Stain analysis. (C) HIF-1α was immunoprecipitated (IP) from GSK-3β+/+ and GSK-3β−/− cells and ubiquitylation was analyzed by immunoblotting (IB) with ubiquitin Abs. WCE indicates whole-cell lysates.
GSK-3 recruits Fbw7 to down-regulate HIF-1α
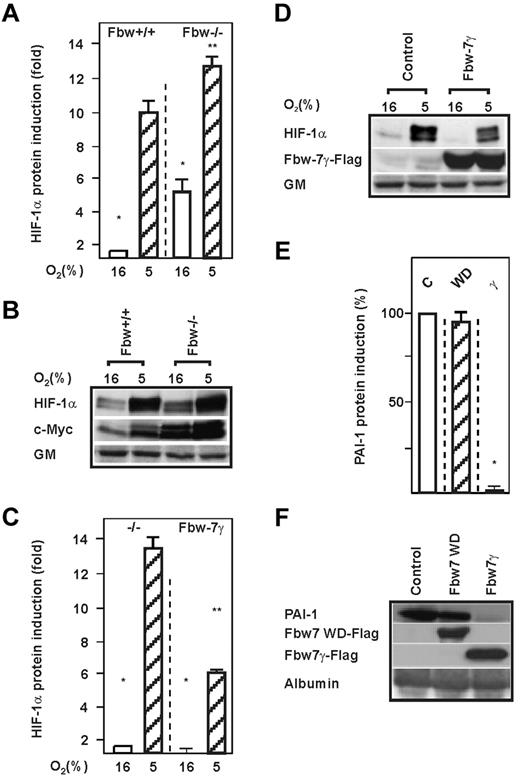
Fbw7 was found to be recruited and to mediate ubiquitylation and degradation of short-living proteins such as c-Myc after they were phosphorylated by GSK-3.24 To examine whether the absence or presence of Fbw7 affects endogenous HIF-1α protein levels, we used HCT116 Fbw7+/+ and HCT116 Fbw7−/− cells, exposed them to normoxia or hypoxia, and measured HIF-1α protein levels by Western blot. We found that loss of Fbw7 enhanced HIF-1α protein levels by approximately 5-fold under normoxia and by approximately 12-fold under hypoxia compared with Fbw7+/+ cells (Figure 2A-B). To further ascertain that this increase was mediated by the Fbw7 pathway, we reintroduced Flag-tagged Fbw7γ into Fbw7−/− cells and determined the expression of endogenous HIF-1α. Reexpression of Fbw7γ in the Fbw7−/− cells abrogated the increase in HIF-1α levels, demonstrating that Fbw7 is involved in HIF-1α degradation (Figure 2C-D). In agreement with this, knockdown of Fbw7 using 2 different Fbw7 shRNA constructs increased HIF-1α protein levels by approximately 10-fold (supplemental Figure 1E-F, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Lack of Fbw7 induces HIF-1α protein levels. (A,C) HCT116 Fbw7+/+ and HCT116 Fbw7−/− cells were cultured under normoxia (16% O2) for 24 hours and then further cultured for 4 hours under normoxic or hypoxic (5% O2) conditions. The HIF-1α protein levels measured by Western blot under normoxia (16% O2) were set to 1. Values are means ± SEM of 3 independent experiments. *Significant difference between 16% O2 versus 5% O2; **significant difference between 5% O2 Fbw7+/+ or 5% O2 Fbw7−/− versus 5% O2 + Fbw7. (B,D) Representative Western blot analysis. One hundred micrograms of total protein from HCT116 Fbw7+/+ and HCT116 Fbw7−/− cells was analyzed with Abs against HIF-1α, c-Myc, Flag M2, and Golgi membrane (GM). (E) HepG2 cells were transfected with an expression vector for either FlagM2-tagged Fbw7γ (γ), Fbw7WD (WD), or control vector, and PAI-1 levels were measured by Western blot. The PAI-1 protein level in the control was set to 100%. Values are means ± SEM of 3 independent experiments. *Significant difference in the control vector versus Fbw7γ and Fbw7WD. (F) Representative Western blot. One hundred micrograms of protein from the medium and 100 μg of total protein extract from HepG2 cells transfected as in panel E were analyzed with the hPAI-1, FlagM2, and albumin Abs, respectively.
Lack of Fbw7 induces HIF-1α protein levels. (A,C) HCT116 Fbw7+/+ and HCT116 Fbw7−/− cells were cultured under normoxia (16% O2) for 24 hours and then further cultured for 4 hours under normoxic or hypoxic (5% O2) conditions. The HIF-1α protein levels measured by Western blot under normoxia (16% O2) were set to 1. Values are means ± SEM of 3 independent experiments. *Significant difference between 16% O2 versus 5% O2; **significant difference between 5% O2 Fbw7+/+ or 5% O2 Fbw7−/− versus 5% O2 + Fbw7. (B,D) Representative Western blot analysis. One hundred micrograms of total protein from HCT116 Fbw7+/+ and HCT116 Fbw7−/− cells was analyzed with Abs against HIF-1α, c-Myc, Flag M2, and Golgi membrane (GM). (E) HepG2 cells were transfected with an expression vector for either FlagM2-tagged Fbw7γ (γ), Fbw7WD (WD), or control vector, and PAI-1 levels were measured by Western blot. The PAI-1 protein level in the control was set to 100%. Values are means ± SEM of 3 independent experiments. *Significant difference in the control vector versus Fbw7γ and Fbw7WD. (F) Representative Western blot. One hundred micrograms of protein from the medium and 100 μg of total protein extract from HepG2 cells transfected as in panel E were analyzed with the hPAI-1, FlagM2, and albumin Abs, respectively.
We also investigated whether Fbw7-mediated regulation of HIF-1α affects the expression of the HIF-1 target gene plasminogen activator inhibitor-1 (PAI-1).32 Indeed, overexpression of Fbw7γ decreased PAI-1 protein levels, whereas the dominant negative mutant lacking the WD domain (Fbw7WD) had no effect on PAI-1 (Figure 2E-F).
To verify that the Fbw7 effects were not because of transcriptional regulation, we measured HIF-1α mRNA levels in Fbw7-overexpressing cells. Northern blot analyses with RNA probes against HIF-1α and β-actin showed that neither Fbw7 isoform affected HIF-1α mRNA levels (supplemental Figure 1A-B). In agreement with this, when HIF-1α translation was measured, we found that Fbw7 had no effect on HIF-1α protein synthesis (supplemental Figure 1D). Therefore, these data suggest that Fbw7 contributes to HIF-1α protein degradation.
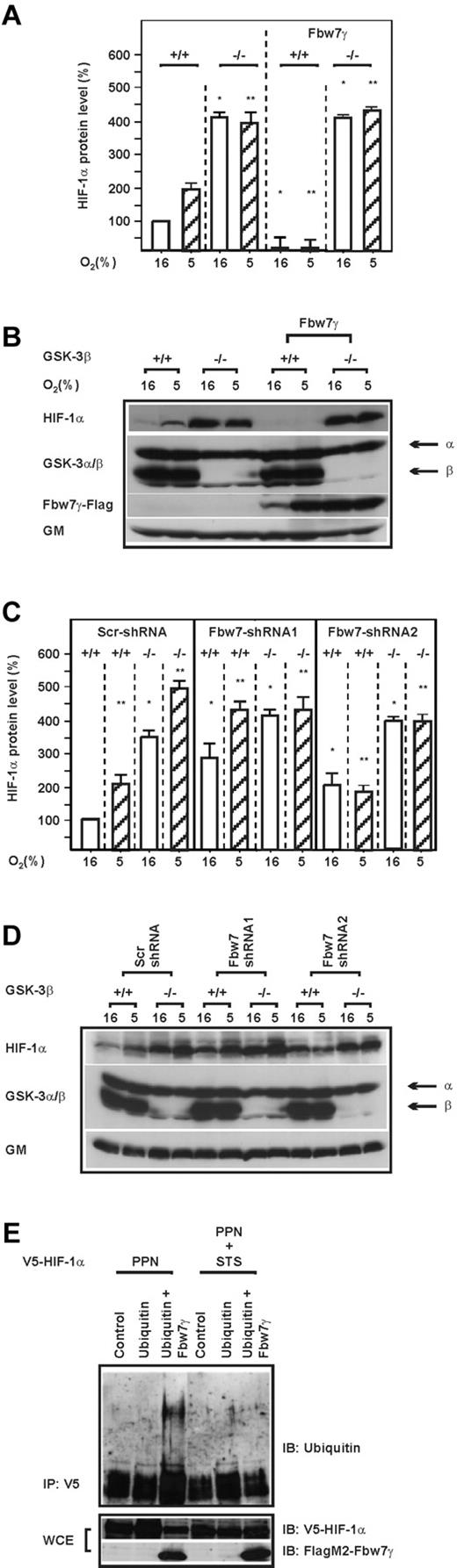
Next we investigated whether down-regulation of HIF-1α protein levels by Fbw7 is dependent on GSK-3β. To determine this, we used GSK-3β+/+ and GSK-3β−/− cells, cultured them under normoxia and hypoxia, and measured the endogenous HIF-1α protein levels by Western blot. We found that loss of GSK-3β increased endogenous HIF-1α protein levels by approximately 400% under both normoxia and hypoxia (Figure 3A-B). Overexpression of Fbw7γ in GSK-3β−/− cells had no effect on HIF-1α under either normoxia or hypoxia, whereas expression of Fbw7γ in GSK-3β+/+ cells dramatically reduced HIF-1α protein levels (Figure 3A-B). Therefore, these data show that down-regulation of HIF-1α by Fbw7 is dependent on GSK-3β.
The destabilization and ubiquitylation of HIF-1α by Fbw7 is dependent on GSK-3β. (A,C) HIF-1α protein levels were measured by Western blot in GSK-3β+/+ and GSK-3β−/− cells or GSK-3β+/+ and GSK-3β−/− cells transfected with either an expression vector for FlagM2-tagged Fbw7γ or with vectors allowing expression of scrambled (Scr) shRNA or Fbw7 shRNA1 or Fbw7 shRNA2. The HIF-1α protein levels in GSK-3β+/+ cells were set to 100%. Values are means ± SEM of 3 independent experiments. *Significant difference between shRNA versus scrambled control. (B,D) Representative Western blot analysis. One hundred micrograms of total protein from GSK-3β+/+ and GSK-3β−/− cells was analyzed with Abs against HIF-1α, GSK-3α/β, Flag M2, and Golgi membrane (GM). (E) Immunoblot (IB) analysis of anti-V5 immunoprecipitates (IP) and whole-cell lysates (WCE) from HEK 293 cells cotransfected with expression vectors for the V5-tagged full-length HIF-1α mutant HIF-1α P402A/P564A/N803A (PPN), HIF-1α P402A/S551A/T555V/P564A/S589A/ N803A (PPNSTS), and Flag-tagged Fbw7γ (Fγ) or hemagglutinin-tagged ubiquitin.
The destabilization and ubiquitylation of HIF-1α by Fbw7 is dependent on GSK-3β. (A,C) HIF-1α protein levels were measured by Western blot in GSK-3β+/+ and GSK-3β−/− cells or GSK-3β+/+ and GSK-3β−/− cells transfected with either an expression vector for FlagM2-tagged Fbw7γ or with vectors allowing expression of scrambled (Scr) shRNA or Fbw7 shRNA1 or Fbw7 shRNA2. The HIF-1α protein levels in GSK-3β+/+ cells were set to 100%. Values are means ± SEM of 3 independent experiments. *Significant difference between shRNA versus scrambled control. (B,D) Representative Western blot analysis. One hundred micrograms of total protein from GSK-3β+/+ and GSK-3β−/− cells was analyzed with Abs against HIF-1α, GSK-3α/β, Flag M2, and Golgi membrane (GM). (E) Immunoblot (IB) analysis of anti-V5 immunoprecipitates (IP) and whole-cell lysates (WCE) from HEK 293 cells cotransfected with expression vectors for the V5-tagged full-length HIF-1α mutant HIF-1α P402A/P564A/N803A (PPN), HIF-1α P402A/S551A/T555V/P564A/S589A/ N803A (PPNSTS), and Flag-tagged Fbw7γ (Fγ) or hemagglutinin-tagged ubiquitin.
We further substantiated our data by shRNA-mediated knockdown of Fbw7 in GSK-3β+/+ and GSK-3β−/− cells. When quantifying endogenous HIF-1α abundance, we found that knockdown of Fbw7 with 2 different shRNAs increased endogenous HIF-1α protein levels in GSK-3β+/+ cells under both normoxia and hypoxia compared with the controls. In contrast, knocking-down Fbw7 in GSK-3β−/− cells had no significant influence on endogenous HIF-1α protein levels (Figure 3C-D). In agreement with this, we could demonstrate that mutation of all 3 GSK-3 phosphorylation sites within HIF-1α and use of a dominant-negative Fbw7 mutant (Fbw7WD) abolished the GSK-3β– and Fbw7γ–mediated reduction of HIF-1α protein levels (supplemental Figure 2A-B). In addition, half-life measurements showed that the GSK-3β– and Fbw7-initiated degradation of HIF-1α occurred independently of the hydroxylation and VHL-mediated degradation (supplemental Figure 2C-D)
Because we demonstrated that Fbw7-driven HIF-1α down-regulation requires phosphorylation of HIF-1α at the GSK-3 sites, we investigated whether Fbw7 can ubiquitinate full-length HIF-1α where the hydroxylation sites and the GSK-3 sites were mutated either alone (PPN) or in combination (PPNSTS). We found that Fbw7 could ubiquitylate HIF-1α when the hydroxylation sites (PPN) were mutated. However, the Fbw7-mediated ubiquitylation could not be detected when the GSK-3 sites were additionally mutated (PPNSTS) indicating that Fbw7 can mediate a VHL-independent ubiquitination of HIF-1α and that the GSK-3 sites are required for these effects (Figure 3E).
These data indicate that phosphorylation of HIF-1α by GSK-3β recruits Fbw7, which then mediates HIF-1α ubiquitination and proteasomal degradation independently of VHL.
USP28 stabilizes HIF-1α by antagonizing the action of Fbw7
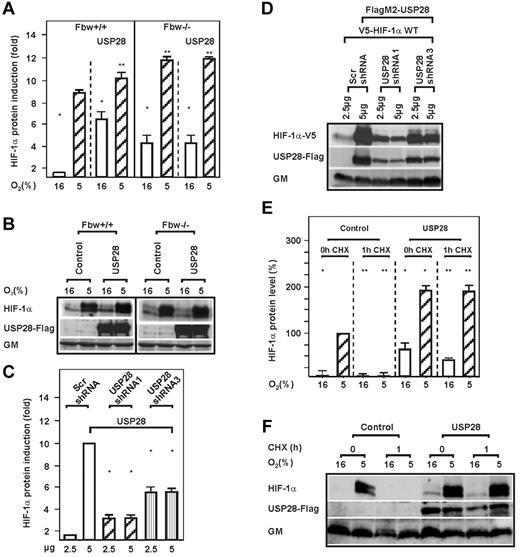
Previous findings showed that the action of Fbw7 could be antagonized by USP28.34 This prompted us to investigate whether USP28 would induce HIF-1α and counteract its Fbw7-dependent degradation. Indeed, overexpression of USP28 increased HIF-1α under both normoxia and hypoxia, but only in Fbw7+/+ cells (Figure 4A-B and E-F). In agreement with this, knockdown of USP28 using 2 different USP28 shRNA constructs reduced HIF-1α protein levels by approximately 80% and 50%, respectively (Figure 4C-D). Further, cycloheximide experiments revealed that USP28 prolonged the half-life of endogenous HIF-1α under both normoxia and hypoxia (Figure 4E-F and supplemental Figure 3). Interestingly, USP28 induced HIF-1α only in Fbw7+/+ cells, whereas overexpression of USP28 in Fbw7−/− cells had no effect on HIF-1α levels (Figure 4A-B).
USP28 prolongs the half-life of HIF-1α but requires Fbw7. (A) HCT116 Fbw7+/+ and HCT116 Fbw7−/− cells were transfected with expression vectors for FlagM2-tagged USP28 and cultured for 4 hours under normoxic (16% O2) or hypoxic (5% O2) conditions. HIF-1α protein levels were measured by Western blot analysis. The HIF-1α protein levels under normoxia (16% O2) were set to 1. Values are means ± SEM of 3 independent experiments. *Significant difference between 16% O2 versus 5% O2; **significant difference between 5% O2 Fbw7+/+ or 5% O2 Fbw7−/− versus 5% O2 + Fbw7 or USP28. (C) HEK 293 cells were transfected with vectors allowing expression of scrambled (Scr) shRNA or USP28 shRNA1 or shRNA3. HIF-1α protein levels were measured by Western blot analysis. The HIF-1α protein levels in the controls were set to 1. *Significant difference between shRNA versus scrambled control. (E) HepG2 cells transfected with the USP28 expression vector were cultured under normoxia (16% O2) or hypoxia (5% O2) and after inhibition of protein synthesis with cycloheximide (CHX; 10 μg/mL), the HIF-1α protein half-life was measured by Western blot analysis. The HIF-1α protein levels at 0 hours were set to 100%. Values are means ± SEM of 3 independent experiments. *Significant difference between HIF-1α versus CHX; **significant difference between USP28 versus CHX. (B,D,F) Representative Western blot analyses. One hundred micrograms of total protein from HCT116 Fbw7+/+, HCT116 Fbw7−/−, HEK 293, and HepG2 cells were analyzed with Abs against HIF-1α, c-Myc, Flag M2, V5, and Golgi membrane (GM).
USP28 prolongs the half-life of HIF-1α but requires Fbw7. (A) HCT116 Fbw7+/+ and HCT116 Fbw7−/− cells were transfected with expression vectors for FlagM2-tagged USP28 and cultured for 4 hours under normoxic (16% O2) or hypoxic (5% O2) conditions. HIF-1α protein levels were measured by Western blot analysis. The HIF-1α protein levels under normoxia (16% O2) were set to 1. Values are means ± SEM of 3 independent experiments. *Significant difference between 16% O2 versus 5% O2; **significant difference between 5% O2 Fbw7+/+ or 5% O2 Fbw7−/− versus 5% O2 + Fbw7 or USP28. (C) HEK 293 cells were transfected with vectors allowing expression of scrambled (Scr) shRNA or USP28 shRNA1 or shRNA3. HIF-1α protein levels were measured by Western blot analysis. The HIF-1α protein levels in the controls were set to 1. *Significant difference between shRNA versus scrambled control. (E) HepG2 cells transfected with the USP28 expression vector were cultured under normoxia (16% O2) or hypoxia (5% O2) and after inhibition of protein synthesis with cycloheximide (CHX; 10 μg/mL), the HIF-1α protein half-life was measured by Western blot analysis. The HIF-1α protein levels at 0 hours were set to 100%. Values are means ± SEM of 3 independent experiments. *Significant difference between HIF-1α versus CHX; **significant difference between USP28 versus CHX. (B,D,F) Representative Western blot analyses. One hundred micrograms of total protein from HCT116 Fbw7+/+, HCT116 Fbw7−/−, HEK 293, and HepG2 cells were analyzed with Abs against HIF-1α, c-Myc, Flag M2, V5, and Golgi membrane (GM).
We also attempted to examine whether USP28 can antagonize the effect of all 3 Fbw7 isoforms (α, β, and γ) and found that all 3 strongly decreased HIF-1α protein levels. In contrast, when USP28 was overexpressed, this counteracted the Fbw7 effects (supplemental Figure 4). These data show that USP28 stabilizes HIF-1α and antagonizes the action of Fbw7.
HIF-1α interacts with Fbw7 but not with USP28
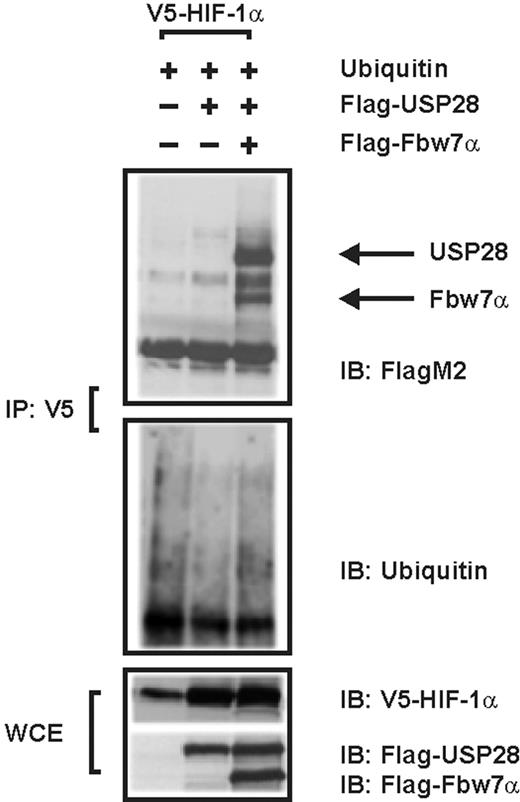
Because our studies with the Fbw7-deficient cells showed that Fbw7 is required for USP28 to exert its effects on HIF-1α, we next investigated whether USP28 forms complexes with HIF-1α to promote deubiquitination. To do this, we performed coimmunoprecipitation and ubiquitination assays in cells and found that USP28 does not interact with HIF-1α directly. In contrast, when lysates were used from cells also expressing Fbw7, we could recover USP28. In addition, the presence of Fbw7 enhanced ubiquitin levels, which could be detected when the same precipitated lysates were probed with the ubiquitin Ab (Figure 5). These data indicate that Fbw7 interacts with HIF-1α and mediates the antagonizing action of USP28.
USP28 interacts with Fbw7 but not HIF-1α and counteracts Fbw7-dependent HIF-1α ubiquitylation. Immunoblot (IB) analysis of anti-V5 immunoprecipitates (IP) and whole-cell lysates (WCE) from HEK 293 cells cotransfected with expression vectors for V5-tagged full-length HIF-1α, FlagM2-tagged USP28, and Flag-tagged Fbw7α. Blots from IPs were probed with either FlagM2 or ubiquitin Abs; WCEs were probed with V5 or FlagM2 Abs.
USP28 interacts with Fbw7 but not HIF-1α and counteracts Fbw7-dependent HIF-1α ubiquitylation. Immunoblot (IB) analysis of anti-V5 immunoprecipitates (IP) and whole-cell lysates (WCE) from HEK 293 cells cotransfected with expression vectors for V5-tagged full-length HIF-1α, FlagM2-tagged USP28, and Flag-tagged Fbw7α. Blots from IPs were probed with either FlagM2 or ubiquitin Abs; WCEs were probed with V5 or FlagM2 Abs.
GSK-3β affects cell migration in an HIF-1α–dependent manner
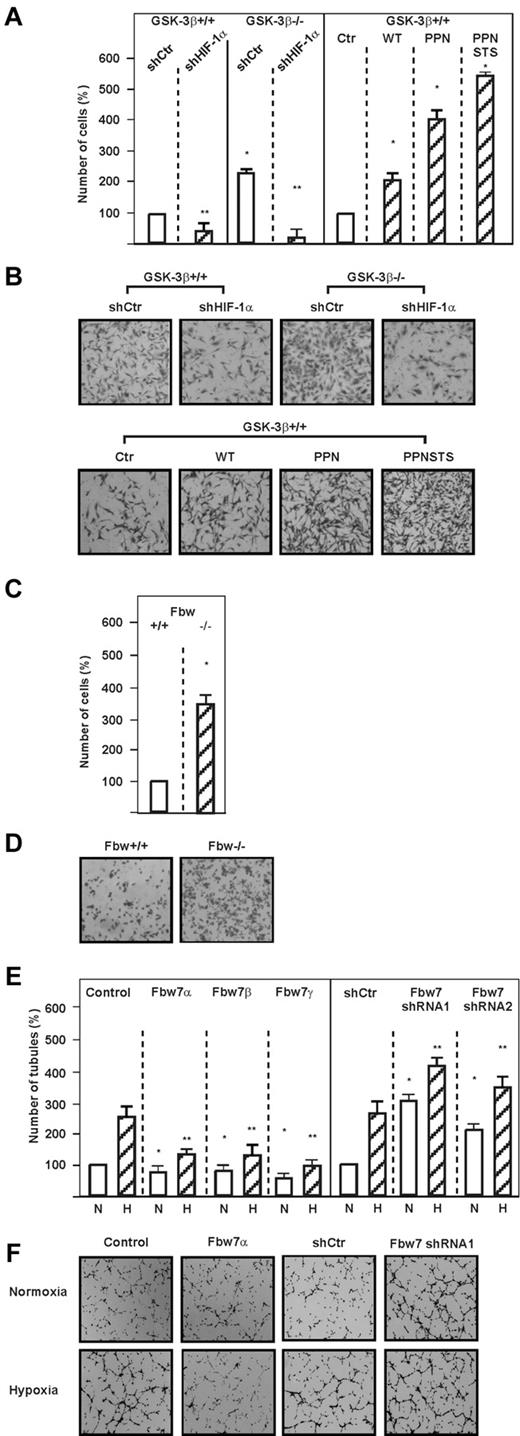
To determine whether the GSK-3–mediated effects on HIF-1α affect the invasive cellular potential, we performed Transwell migration assays with GSK-3β+/+ and GSK-3β−/− cells. A significant increase in migration could be detected with GSK-3β−/− cells compared with GSK-3β+/+ cells (Figure 6A-B). These effects were indeed mediated to a large extent by HIF-1α, because transfection of shRNA against HIF-1α abolished the increased migration of GSK-3β−/− cells (Figure 6A-B). In agreement with this, overexpression of the nondegradable HIF-1α mutants in GSK-3β+/+ cells also increased the migration significantly (Figure 6A-B). Likewise, loss of Fbw7 had the same impact on cell migration as depletion of GSK-3β (Figure 6C-D). These data show that lack of GSK-3β promotes invasive and anchorage-independent cellular growth in an HIF-1α–dependent manner
Loss of GSK-3β induces cell migration via HIF-1α. (A) GSK-3β+/+ and GSK-3β−/− cells were transfected with vectors allowing expression of scrambled control shRNA (shCtr) or shRNA against HIF-1α (shHIF-1α). In addition, GSK-3β+/+ cells were transfected with expression vectors for the V5-tagged full-length HIF-1α (WT) or either the hydroxylation resistant V5-tagged HIF-1α P402A/P564A/N803A (PPN) or HIF-1α containing additional mutations in the GSK-3 sites P402A/S551A/T555V/ P564A/S589A/N803A (PPNSTS) or the empty vector (Ctr). Cells were seeded into Transwell chambers and the number of migrated cells was counted and quantified using ImageJ software. Data represent the number of cells relative to the control, which was set to 100%. *Significant difference between GSK-3β+/+ or GSK-3β+/+ + shRNA control versus GSK-3β−/− or GSK-3β−/− + shRNA control, as well as between control vector versus HIF-1α WT, HIF-1α PPN, or HIF-1α PPNSTS. **Significant difference between shRNA control versus HIF-1α shRNA. (B) Photographs from a representative Transwell chamber experiment. (C) HCT116 Fbw7+/+ and HCT116 Fbw7−/− cells were cultured in Transwell chambers for 16 hours and the number of migrated cells was counted and quantified using ImageJ Version 1.457 software. Data represent the number of cells relative to the control, which was set to 100%. *Significant difference between HCT116 Fbw7+/+ versus HCT116 Fbw7−/−. (D) Photographs from a representative Transwell chamber experiment. (E) HMEC-1 cells were either transfected with empty control vector (Control), expression vectors for Fbw7α, Fbw7β, Fbw7γ, or with vectors allowing expression of scrambled control shRNA (shCtr) or shRNA1 or shRNA2 against Fbw7. Transfected cells were seeded onto Matrigel-coated wells for 2 hours, and then exposed to normoxia (N) or hypoxia (H) for 6 hours. Formation of capillary-like structures was assessed by counting the number of tubules using ImageJ software. Data represent the number of tubules relative to the control, which was set to 100%. *Significant difference between the control vector versus Fbw7 isoforms under normoxic conditions; **significant difference between the control vector versus Fbw7 isoforms under hypoxic conditions. (F) Photographs from representative examples of an in vitro angiogenesis experiment.
Loss of GSK-3β induces cell migration via HIF-1α. (A) GSK-3β+/+ and GSK-3β−/− cells were transfected with vectors allowing expression of scrambled control shRNA (shCtr) or shRNA against HIF-1α (shHIF-1α). In addition, GSK-3β+/+ cells were transfected with expression vectors for the V5-tagged full-length HIF-1α (WT) or either the hydroxylation resistant V5-tagged HIF-1α P402A/P564A/N803A (PPN) or HIF-1α containing additional mutations in the GSK-3 sites P402A/S551A/T555V/ P564A/S589A/N803A (PPNSTS) or the empty vector (Ctr). Cells were seeded into Transwell chambers and the number of migrated cells was counted and quantified using ImageJ software. Data represent the number of cells relative to the control, which was set to 100%. *Significant difference between GSK-3β+/+ or GSK-3β+/+ + shRNA control versus GSK-3β−/− or GSK-3β−/− + shRNA control, as well as between control vector versus HIF-1α WT, HIF-1α PPN, or HIF-1α PPNSTS. **Significant difference between shRNA control versus HIF-1α shRNA. (B) Photographs from a representative Transwell chamber experiment. (C) HCT116 Fbw7+/+ and HCT116 Fbw7−/− cells were cultured in Transwell chambers for 16 hours and the number of migrated cells was counted and quantified using ImageJ Version 1.457 software. Data represent the number of cells relative to the control, which was set to 100%. *Significant difference between HCT116 Fbw7+/+ versus HCT116 Fbw7−/−. (D) Photographs from a representative Transwell chamber experiment. (E) HMEC-1 cells were either transfected with empty control vector (Control), expression vectors for Fbw7α, Fbw7β, Fbw7γ, or with vectors allowing expression of scrambled control shRNA (shCtr) or shRNA1 or shRNA2 against Fbw7. Transfected cells were seeded onto Matrigel-coated wells for 2 hours, and then exposed to normoxia (N) or hypoxia (H) for 6 hours. Formation of capillary-like structures was assessed by counting the number of tubules using ImageJ software. Data represent the number of tubules relative to the control, which was set to 100%. *Significant difference between the control vector versus Fbw7 isoforms under normoxic conditions; **significant difference between the control vector versus Fbw7 isoforms under hypoxic conditions. (F) Photographs from representative examples of an in vitro angiogenesis experiment.
Fbw7 and USP28 affect hypoxia- and HIF-1α–dependent angiogenesis
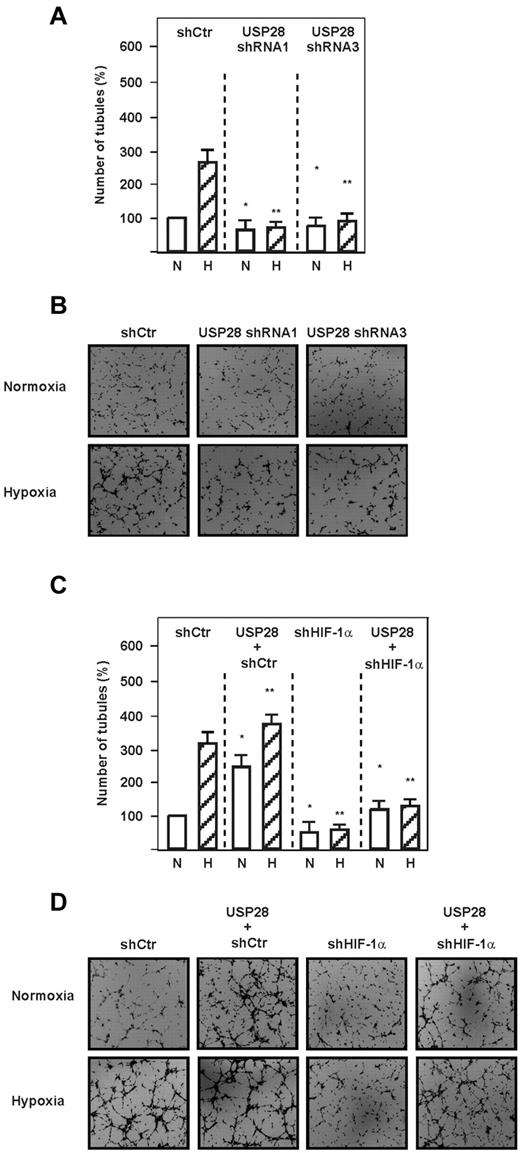
Because HIF-1α is associated with the regulation of angiogenesis, we next investigated whether the Fbw7-dependent down-regulation of HIF-1α affects capillary formation in an in vitro Matrigel assay. Hypoxia and transfection of endothelial cells with 2 different Fbw7 shRNAs induced the formation of capillary-like structures; hypoxia exerted an increase of approximately 250% compared with normoxia, whereas Fbw7 shRNA-1 or Fbw7 shRNA-2 increased the formation of capillary-like structures by approximately 300% and 200% under normoxia and by approximately 400% and 350% under hypoxia, respectively (Figure 6E-F). In contrast, overexpression of all Fbw7 isoforms decreased the formation of capillary-like structures under both normoxia and hypoxia (Figure 6E-F). We next investigated whether USP28 may also act as Fbw7 antagonist in the angiogenic response and found that USP28 shRNA-1 and shRNA-3 decreased formation of capillary like structures by approximately 200% and 190%, respectively, under hypoxia (Figure 7A-B). In agreement with this, we found that up-regulation of angiogenesis by USP28 is mediated by HIF-1α, because knocking down HIF-1α abolished the USP28-mediated formation of capillary-like structures under both normoxia and hypoxia (Figure 7C-D). These data indicate that reciprocal regulation of HIF-1α stability by Fbw7 and USP28 contributes to endothelial angiogenic responses under normoxia and hypoxia.
USP28 is involved in hypoxia and HIF-1–dependent capillary formation. (A) HMEC-1 cells were transfected with vectors expressing USP28 shRNA1 or USP28 shRNA3 or control shRNA (shCtr). Cells were seeded onto Matrigel-coated wells for 2 hours and then exposed to hypoxia (H) or normoxia (N) for 6 hours. The number of tubules from each well was counted using ImageJ Version 1.457 software. Data represent the number of tubules relative to the control, which was set to 100%. *Significant difference between the shRNA control vector versus USP28 shRNA vectors under normoxia. **Significant difference between the shRNA control vector versus USP28 shRNA vectors under hypoxia. (B) Photographs from a representative in vitro angiogenesis experiment. (C) HMEC-1 cells were transfected with vectors for control shRNA (shCtr) or shRNA against HIF-1α (shHIF-1α) alone or in combination with a vector expressing USP28. Cells were plated onto Matrigel-coated wells as described in panel A. *Significant difference between the control vector versus USP28 and shRNA under normoxia; **significant difference between the control vector versus USP28 and shRNA under hypoxia. (D) Photographs from a representative in vitro angiogenesis experiment.
USP28 is involved in hypoxia and HIF-1–dependent capillary formation. (A) HMEC-1 cells were transfected with vectors expressing USP28 shRNA1 or USP28 shRNA3 or control shRNA (shCtr). Cells were seeded onto Matrigel-coated wells for 2 hours and then exposed to hypoxia (H) or normoxia (N) for 6 hours. The number of tubules from each well was counted using ImageJ Version 1.457 software. Data represent the number of tubules relative to the control, which was set to 100%. *Significant difference between the shRNA control vector versus USP28 shRNA vectors under normoxia. **Significant difference between the shRNA control vector versus USP28 shRNA vectors under hypoxia. (B) Photographs from a representative in vitro angiogenesis experiment. (C) HMEC-1 cells were transfected with vectors for control shRNA (shCtr) or shRNA against HIF-1α (shHIF-1α) alone or in combination with a vector expressing USP28. Cells were plated onto Matrigel-coated wells as described in panel A. *Significant difference between the control vector versus USP28 and shRNA under normoxia; **significant difference between the control vector versus USP28 and shRNA under hypoxia. (D) Photographs from a representative in vitro angiogenesis experiment.
The GSK-3β–, Fbw7-, and USP28-dependent HIF-1α regulation contributes to proliferation and colony formation
We next investigated whether GSK-3β–, Fbw7-, and USP28-mediated HIF-1α regulation is involved in cell proliferation. We first used GSK-3β+/+ cells and GSK-3β−/− cells in which Fbw7, USP28, or HIF-1α were knocked-down alone or in combination and measured BrdU incorporation into newly synthesized DNA. We found that knockdown of Fbw7 increased cell proliferation by approximately 2-fold in GSK-3β+/+ cells, whereas USP28 knockdown had no effect. In contrast, neither the Fbw7 shRNA nor USP28 shRNA affected BrdU incorporation in GSK-3β−/− cells (supplemental Figure 5A).
Next we investigated whether the GSK-3β–dependent, Fbw7/USP28–mediated effects on BrdU incorporation involved HIF-1α. We found that USP28 increased BrdU incorporation in GSK-3β+/+ cells, whereas knocking down HIF-1α abolished these effects. In contrast, neither USP28 nor HIF-1α shRNA caused changes in BrdU incorporation in GSK-3β−/− cells (supplemental Figure 5B).
Because our data demonstrated that USP28 is critical for HIF-1α–dependent proliferation, we investigated whether the USP28–HIF-1α axis affects also anchorage-independent cell growth. Indeed, we found that knockdown of HIF-1α abolished the USP28-mediated increase in HEK293 cell colony formation (supplemental Figure 5C-D). These data suggest that Fbw7 and USP28 can reciprocally regulate cell proliferation via HIF-1α in a GSK-3β–dependent manner.
Discussion
In the present study, we identified a novel mechanism by which Fbw7 can induce ubiquitinylation and degradation of HIF-1α when it is phosphorylated by GSK-3. The ubiquitinylation of HIF-1α by Fbw7 was independent of VHL and could be antagonized by USP28.
The ubiquitylation of proteins is carried out by the successive action of ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-protein ligase (E3) enzymes.35 The rate-limiting step within this process is carried out by E3s, the function of which is to link target substrates with cognate E2s, thus enabling transfer of ubiquitin to primarily lysine residues within the target protein. Because E3s bind directly to substrates, this indicates that they provide specificity in ubiquitination reactions. Depending on its substrate specificity, an E3 ligase can act as either a tumor promoter or suppressor. Within that context, it was found that the VHL protein interacts with elongin B, elongin C (in yeast Skp1), cullin2 (in yeast cdc53), and Rbx1 (also called Hrt1 or ROC1),16,17 thus forming a complex that belongs to the SCF family of E3s. One of the best known substrates of this E3 complex is HIF-1α, which is bound to the substrate recognition component VHL.13-17 A prerequisite for VHL binding to HIF-1α is proline hydroxylation of the latter, which is carried out by members of a family of proline hydroxylases.8,10-12
We reported previously that GSK-3 can initiate a VHL-independent degradation of HIF-1α, which suggested the involvement of an E3 ligase that can be recruited to HIF-1α after phosphorylation. In the present study, we continued and extended those studies and obtained several new findings with respect to HIF-1α regulation. First, we identified Fbw7 as an E3-ubiquitin ligase that can be recruited to HIF-1α after its phosphorylation by GSK-3, indicating that Fbw7 is a critical component for the degradation of HIF-1α. Second, loss of GSK-3β and Fbw7 affected cell migration, colony, and capillary formation in an HIF-1α–dependent manner, which suggests that loss-of-function mutations in GSK-3β and Fbw7 can lead to tumorigenesis and malignancy.
Fbw7 is a member of a growing family of F-box proteins; 68 human and 74 mouse genes encoding F-box proteins have been described thus far. All of them are assumed to be a part of the SCF complex constituting an E3 ubiquitin ligase.35,36 Within that complex, the F-box protein constitutes the variable component that serves as a receptor for target proteins37,38 and thereby determines target specificity, whereas the components Skp1, Cul1, and Rbx1 are the invariable subunits.39 Among the many F-box proteins that have been identified, Fbw1/β-TrCP and Fbw7 (also called hCdc4 in yeast, hSel10 in C elegans, or Ago in Drosophila) have been shown to control the abundance of proteins with key roles in cell division, growth, and differentiation. Fbw1 ubiquitylates β-catenin and IκB and various other proteins, such as cyclin E, Notch1, c-Myc, Myb, SREBP, c-Jun, and PGC-1α.24,28,38,40-43 In agreement with this, our study extends this list and shows that Fbw7 but not Fbw1 (data not shown) can interact with HIF-1α. The Fbw7 protein binds to substrates via its WD40 repeats, but often this requires substrate phosphorylation within motifs termed Cdc4 phosphodegrons.28,40,44 Because Fbw7 is frequently deleted or mutated in human cancers and is involved in the degradation of oncogenic proteins such as cyclin E, c-Myc, and c-Jun, Fbw7 was thought to be considered a tumor-suppressor gene. In agreement with this, a study investigating more than 1500 human tumors found that approximately 6% of tumors displayed mutations in the Fbw7 coding region. The highest mutation rate was found in cholangiocarcinomas (35%), T-cell acute lymphocytic leukemia (T-ALL: 31%), and endometrial (9%), colon (9%), and stomach (6%) cancers.45 In fact, 43% of all these mutations resulted in amino acid substitutions within the WD40 domain that are shared by all 3 isoforms, suggesting that all 3 might collectively contribute to its tumor-suppressive action.45 These data are consistent with our findings showing that all 3 Fbw7 isoforms can target HIF-1α for proteasomal degradation and that loss of the WD domain antagonizes GSK-3 initiated degradation, leading to higher HIF-1α levels, which has been found to be associated with several tumors.3-5 In addition, our data showing that loss of GSK-3β and/or Fbw7 enhances cell proliferation, colony formation, and angiogenesis in an HIF-1α–dependent manner provide a mechanistic explanation for the observations seen in the patient material. Further, the link between HIF-1α and Fbw7 is not only important for tumorigenesis but also for appropriate development. This is underlined by the findings that Fbw7-null mice,46 like HIF-1α-deficient mice,47 die approximately 8-10 days after birth because of severe defects in hematopoietic, vascular, and neuronal development. Concomitantly, the accumulation of cyclin E and Notch1 proteins was found in Fbw7 (−/−) embryos. It has been reported that Fbw7 targets Notch1, which controls c-Myc transcriptionally.48 Both Notch48 and c-Myc49 have been shown to cross-couple with HIF-1α and to be important in mediating hypoxia-induced tumor cell migration and invasion and cell-cycle progression, respectively. These previous data, together with the findings of the present study, indicate that Fbw7 becomes an integral part of the complex molecular pathway in the hypoxic response controlling DNA damage, genetic alterations, and malignant progression.
Our study also provides an additional new control mechanism that affects degradation of HIF-1α by Fbw7. This mechanism is provided by USP28, which associates with Fbw7 and antagonizes the effect of Fbw7 on HIF-1α protein stability.
Previous studies have identified examples in which a ubiquitin-specific protease interacts directly with a ubiquitin ligase: USP47 interacts with Fbw1/β-TRCP, the E3 ligase that is involved in cell-cycle progression, DNA damage, and apoptosis; USP7 interacts with Mdm2, the E3 ligase that degrades p53; and USP33 interacts with VHL, which targets HIF-1α.50 Therefore, the removal of ubiquitin chains by deubiquitylating enzymes plays a critical role in regulating the half-life and activity of many cellular proteins. The co-immunoprecipitation experiments in the present study show that USP28 can form a ternary complex with HIF-1α, but this requires association of Fbw7 to HIF-1α. This is underlined by the experiments in the Fbw7-lacking HCT116 cells, in which USP28 alone did not affect HIF-1α protein levels. Interestingly, USP28 could counteract the down-regulation of HIF-1α by all Fbw7 isoforms. This mechanism appears to be different from that observed with c-Myc, in which only Fbw7α recruited c-Myc. In addition, USP28 did not form a complex with c-Myc and the nucleolar isoform of Fbw7, Fbw7γ, providing an explanation of why c-Myc is selectively degraded by Fbw7γ.30
Expression of USP28 is strongly elevated in human colon and breast carcinomas, suggesting that it may play an important role for tumor angiogenesis, tumor-cell survival, and proliferation. This is in agreement with our findings showing that Fbw7 isoforms down-regulate hypoxia-dependent angiogenesis in endothelial cells and that overexpression of USP28 increases angiogenesis. Because this effect was completely abolished using the shRNA against HIF-1α and USP28, this supports the regulatory role of both Fbw7 and USP28 for HIF-1α protein stability and HIF-1–dependent angiogenesis.
The results of this study extend our understanding of the mechanisms mediating the degradation of HIF-1α and introduce a new mechanism for HIF-1α regulation that is not limited by the presence of oxygen and is therefore independent of HIF-1α hydroxylation and the VHL-E3 ligase complex. The degradation of HIF-1α by the GSK-3/Fbw7/USP28 system may constitute a novel adaptive response of a cell to allow HIF-1α function in response to various physiologic and nonphysiologic signals that affect cell division, growth, differentiation, and apoptosis. Overall, these findings offer the potential to affect HIF-1α–dependent processes by targeting GSK-3, Fbw7, or USP28.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG GO709/4-4), the 7th Framework of the European Union (Metoxia; to A.G.), the Fondation Leducq (to A.G. and T.K.), Fonds der Chemischen Industrie, the Finnish Academy of Science, the Sigrid Juselius Foundation, and the Biocenter Oulu (to T.K.).
Authorship
Contribution: D.F. performed most of the experimental work, analyzed the data, and wrote the manuscript; A.G. designed the experimental work, analyzed the data, and wrote the manuscript; and T.K. planned the study, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Kietzmann, MD, Department of Biochemistry and Biocenter Oulu, University of Oulu, P O Box 3000, FI-90014 Oulu, Finland; e-mail: tkietzm@gwdg.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal