Abstract
The impact of HIV-1 Nef-mediated HLA-I down-regulation on CD8+ cytotoxic T lymphocytes (CTLs) varies by epitope, but the determining factors have not been elucidated. In the present study, we investigated the impact of Nef on the antiviral efficiency of HIV-1–specific CTLs targeting 17 different epitopes to define properties that determine susceptibility to Nef. The impact of Nef was not correlated with the presenting HLA-I type or functional avidity of CTLs, but instead was related directly to the kinetics of infected cell clearance. Whereas Gag-specific CTLs generally were less susceptible to Nef than those targeting other proteins, this was determined by the ability to eliminate infected cells before de novo synthesis of viral proteins, which was also observed for CTLs targeting a Nef epitope. This very early clearance of infected cells depended on virus inoculum, and the required inoculum varied by epitope. These results suggest that whereas Gag-specific CTLs are more likely to recognize infected cells before Nef-mediated HLA-I down-regulation, this varies depending on the specific epitope and virus inoculum. Reduced susceptibility to Nef therefore may contribute to the overall association of Gag-specific CTL responses to better immune control if a sufficient multiplicity of infection is attained in vivo, but this property is not unique to Gag.
Introduction
Multiple studies have demonstrated a major contribution of CD8+ cytotoxic T lymphocytes (CTLs) in controlling HIV-1 infection. The antiviral activity of CTLs is mediated by cytolysis of infected cells on TCR recognition of viral epitopes that are presented by HLA-I molecules on the surface of infected cells.1 CTLs can mediate cytolysis of infected cells early after infection and therefore can reduce viral replication.1,2
HIV-1 Nef is a 27-kDa myristoylated protein with a central role in immunopathogenesis. Its down-regulation of surface HLA-I molecules on infected cells3,4 may facilitate viral persistence by the evasion of CTLs. In vitro studies have demonstrated that Nef-mediated HLA-I down-regulation impairs the antiviral efficiency of HIV-1–specific CTLs5-7 and that CTLs drive the selective pressure to maintain this function.8 Analogously, in vivo studies using the macaque SIV model have shown that Nef-mediated Mamu down-regulation impairs CTL antiviral responses, which exert strong selective pressure to maintain this Nef function.9,10 Moreover, the ability of Nef to down-regulate HLA-I in vivo is correlated with the breadth of the HIV-1–specific CTL response,11 further confirming the role of Nef in evasion of CTL antiviral activity.
The impact of HIV-1 Nef on CTL antiviral activity is epitope specific. HLA-I C–restricted CTLs are unaffected because Nef down-regulates cell surface HLA-I A and HLA-I B but spares HLA-I C molecules.7 Moreover, HLA-I A– and HLA-I B–restricted CTLs can vary in their susceptibility7 or even resist interference by Nef,6 illustrating the epitope-dependent variability of Nef antagonism of CTL antiviral activity.
A proposed factor determining the impact of Nef on CTL antiviral activity is the timing of epitope presentation versus Nef-mediated HLA-I down-regulation. Nef is one of proteins that is expressed the earliest after cell infection,12 and HLA-I down-regulation lags by comparison.13 It has been hypothesized that CTLs targeting epitopes presented before HLA-I down-regulation may preempt the antagonistic Nef effect.
Indirect evidence supporting this hypothesis was provided by van Baalen et al14 and Ali et al,15 who demonstrated separately that accelerating epitope expression can increase the antiviral efficacy of CTL clones. More direct evidence came from Sacha et al, who showed that SIV Gag and Pol epitopes from proteins carried by incoming virions can be presented before down-regulation of Mamu molecules by Nef and that CTLs targeting these epitopes can eliminate virus-infected cells before viral protein translation.16,17 These previous studies suggested a crucial role for epitope presentation timing in determining the degree of Nef impact on CTL antiviral activity. However, the kinetic relationship of HIV-1 epitope presentation versus Nef-mediated HLA-I down-regulation is poorly understood. Moreover, whether other factors implicated in the efficacy and shaping of the CTL response (eg, HLA-I restriction, functional avidity, and viral protein targeting)18-20 affect CTL interaction with Nef is not known. Given the contribution of Nef-mediated immune evasion to HIV-1 persistence in vivo, defining factors determining the ability of Nef to interfere with CTL antiviral activity would shed light on the requirements for optimizing or eliciting efficacious HIV-1–specific CTL responses.
Methods
HIV-1–permissive cells
CD4+ T1 cells21 (expressing HLA A*02 and B*40) and the T1/primary CD4+ T-lymphocyte hybridoma 1CC4.14 (expressing A*02, B*15, B*40, and B*57)7 were maintained in RPMI 1640 medium supplemented with 10% FCS, l-glutamine, HEPES, and penicillin-streptomycin (R10) as described previously.2 Primary CD4+ T lymphocytes (expressing A*02 and B*57) were expanded from PBMCs with a CD3/CD8-bispecific mAb22 confirmed to be > 95% CD3+/CD4+ and maintained in R10 supplemented with 50 U/mL of recombinant human IL-222 (R10-50) from the National Institutes of Health (NIH) AIDS Reagent Repository. Primary CD4+T lymphocytes were stimulated with 1 μg/mL of phytohemagglutinin for 3 days before viral infection.22
Viruses
Replication-competent whole HIV-1.
HIV-1 NL4-3.123 viruses containing wild-type or a methionine-to-alanine mutation at position 20 of Nef (Nef-M20A) were produced and titered as described previously.24 For experiments assessing the kinetics of Nef-mediated HLA-I down-regulation, the virus contained the murine CD24/heat stable antigen (HSA) reporter gene in the vpr locus.24
Single-round infectious HIV-1.
The NL4-3.1 genome was altered to contain mutations D368R in gp120,25 L26R in gp41,26 a truncation of 26 amino acids of the gp41 cytoplasmic domain,27 and an additional deletion in the V3 loop (300-329), and insertion of the gene for HSA in the vpr locus.24 This replication-defective genome was cotransfected into 293T cells with a vesicular stomatitis virus G glycoprotein (VSV-G) envelope expression vector, and supernatant was harvested after 2 days to produce a stock of single-round infectious HIV-1 pseudotyped with VSV-G envelope (NL4-3-ΔEnv/VSV-G-Env). The virus stock was concentrated by ultracentrifugation and titered by flow cytometry after infection of T1 cells.
HIV-1–specific CTL clones
CTL clones were derived previously from the PBMCs of HIV-1–infected patients by limiting-dilution cloning and maintained by periodic restimulation with anti-CD3 Ab and irradiated allogeneic PBMCs in R10-50 as described previously.1
Assessment of Nef impact on the antiviral activities of CTL clones
The Nef impact assay was established to allow standardized comparisons of Nef effects on CTL suppression of HIV-1 replication by comparing wild-type and mutant Nef-M20A viruses.7 Briefly, cells were acutely infected with NL4-3.1 containing wild-type Nef or Nef-M20A and cocultured with or without the CTL clone at an effector-to-target ratio of 0.25:1 in triplicate wells. The inhibition efficiency was calculated as: (log10 p24 without CTL − log10 p24 with CTL)/(log10 p24 without CTL). The Nef-effect ratio was then calculated as: (inhibition efficiency of HIV-1 with wild-type Nef)/(inhibition efficiency of HIV-1 with Nef-M20A), for which 1 indicates no impact of Nef (same efficiency) and 0 indicates complete ablation of CTL antiviral activity (efficiency of 0 for HIV-1 with wild-type Nef).
Kinetics of HIV-1–specific CTL killing of virus-infected cells
Infected cell killing by CTL clones was assessed by a modified 51Cr-release assay.2,19 Target cells (5 × 105) were incubated with virus stock containing 600 fg (or the indicated amount) of Gag p24 per target cell for 2 hours in the presence of 25 μCi 51Cr and 4 μg/mL of polybrene/100 μL, and washed 3 times with R10. For cells treated with 500μM tenofovir and 500μM zidovudine (NIH AIDS Research and Reference Reagent Program), the drugs were added to the cells at least 2 hours before infection and maintained throughout the experiment. The target cells were resuspended in R10-50 at a concentration of 6.6 × 104 cells/mL and cocultured with or without the specific CTL clone at an effector-to-target ratio of 3:1 (3 × 104 CTL and 104 target cells per well in 96-well U-bottom plates) in 250 μL of R10-50 in triplicate wells. At the indicated times, 30 μL of supernatant was harvested from each well for measurement of 51Cr release by microscintillation counting (Lumaplate [Perkin Elmer] and Microbeta [Wallac]). Controls included target cells with no added CTLs (spontaneous release) and target cells lysed with 2.5% Triton X-100 (maximal release). At each time point, the released 51Cr was calculated by multiplying the 51Cr concentration in cpm/μL by the remaining volume of the medium and then adding back the total 51Cr counts removed from each well from earlier time points. Specific lysis was calculated as: specific lysis = (total experimental 51Cr release − total spontaneous 51Cr release)/(total maximal 51Cr release − total spontaneous 51Cr release). An uninfected control was also included in the assay to determine any nonspecific killing by CTLs. To define the onset of infected cell killing, the logarithmic regression curve was fitted to the average infected cell lysis over time (with R2 > 0.9) to estimate the time corresponding to 10% specific lysis (K10; the choice of 10% is arbitrary based on the limit of reliable detection). This low threshold was chosen to minimize variability due to efficiency (rather than timing) of killing.
CTL functional avidity measurements
Functional avidity of CTL clones was determined by standard peptide titration 51Cr-release assays.1,2,19,23 Briefly, the 51Cr-labeled target cells were preincubated with serial dilutions of the cognate peptide before the 51Cr-release assay. Functional avidity was measured as the sensitizing dose (concentration) of peptide yielding 50% of maximal CTL killing (SD50).
Measuring kinetics of Nef-mediated HLA-I down-regulation
Target cells were infected at excess multiplicity of infection of 12 with recombinant NL4-3.1 expressing wild-type Nef or Nef-M20A for 4 hours at 37°C, washed twice, resuspended, and plated at 5 × 105 cells/well in a 24-well plate. In some experiments, infection was performed using NL4-3 ΔEnv/VSV-G virus stock at 600 fg of Gag p24 per cell. Primary CD4+ T lymphocytes were stimulated with 1 μg/mL of phytohemagglutinin for 3 days and infected with the indicated viruses at 600 fg of Gag p24/cell. To assess HLA-I down-regulation, the acutely infected cells were costained for the cell-surface reporter HSA (mAb M1/69-PE; eBiosciences), HLA-A*02 (mAb bb7.2–Alexa Fluor 488; AbD Serotec), and HLA-B*57 (mAb B17-Biotin [One Lambda] with secondary staining using streptavidin-PE/Cy5.5 [eBiosciences]) at the indicated times. In addition, CD4 expression (mAb OKT4-APC; eBiosciences) was monitored on acutely infected primary CD4+ T lymphocytes. Flow cytometry was performed on a FACScan or FACSCalibur flow cytometer (BD Biosciences) and analyzed using FlowJo Version 7.6.4 software (TreeStar). At the indicated time points, infected cells (positive for HSA expression) were gated for analysis of A*02 and B*57 expression as shown by mean fluorescence intensity (MFI). Productively infected primary CD4+T lymphocytes were identified by gating on low-CD416 and high-HSA reporter-expressing cells.24 The relative expression of HLA-I on cells infected with wild-type Nef virus was then calculated as a fraction of MFI compared with Nef-M20A virus after subtraction of background MFI (observed using an isotype control). Nef-mediated down-regulation of HLA-I on 1CC4.14 cells and primary CD4+ T lymphocytes was assessed in 3 and 6 independent experiments, respectively.
Mathematical modeling of the kinetics of Nef-mediated HLA-I down-regulation kinetics
HLA-I down-regulation kinetics were fitted using a modified 3 parameter Gompertz function: [f(t) = 1 −β1a−a−β2(t−β3), where a is any positive constant, and β1, β2, and β3 are unknown parameters]. The Gompertz curve has 2 asymptotes: 1 as t → −∞ and 1 − β1 as t → ∞. The left asymptote, 1 as t → −∞, was fitted to the down-regulation time points where the wild-type relative HLA-I expression remained 1 or above. The right asymptote, 1 − β1 as t → ∞, was fitted to the relative HLA-I expression that had reached a plateau, which represented the maximum HLA-I down-regulation observed in infected cells. The Gompertz curve is also asymmetrical, with a steeper decline from the left asymptote compared with the approach to the right asymptote, which is an appropriate model for fitting HLA-I down-regulation by Nef, in which down-regulation occurs abruptly and then reaches a plateau. We chose a = 10 so that the value of the function at β3 is f(β3) = 1 − β1/10, making β3 the point at which HLA expression has decreased 10% between asymptotes (the choice of a is arbitrary). To compare the fits for A*02 and B*57, we calculated the natural parameterization and estimates of the parameter and its variance. We also compared the slopes of the fitted curves at the estimated value of β3: f′(β3). This required an approximation (using the Delta method) to estimate the variance of the slope. Assuming independence of the estimates, we used the Wald test to compare for each experiment the following: (1) β3, (2) f′(β3), and (3) 1 − β1, the right asymptote. All statistical calculations were done using the procedure nl in Stata Version 10 software.
Statistical analyses
The Kruskal-Wallis and Dunn posthoc analysis tests were used to compare the functional avidities of CTL responses across different viral proteins, and for the multiple comparison analysis of Nef effects on viral inhibition across different groups of CTL clones; a 2-tailed Student t test was performed for comparison of Nef effects on viral inhibition or infected cell killing kinetics (K10) between 2 groups of CTL clones, using Microsoft Excel 2008. The Pearson test was used to assess the relationship between the functional avidities or infected cell killing kinetic (K10) and Nef effects; P values < .05 were considered significant.
Results
Gag-specific CTL antiviral activity is overall less susceptible to Nef-mediated HLA-I down-regulation
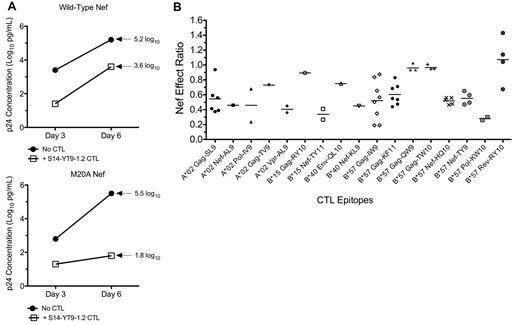
Using a previously described assay7 (Figure 1A), we assessed the impact of Nef on the antiviral activities of CTL clones targeting 17 different epitopes. These epitopes were derived from both structural and accessory proteins, and were presented by various HLA-I types (Table 1). Although most CTLs were affected by wild-type Nef, individual clones differed in their susceptibility to Nef (Figure 1B). To determine whether the impact of Nef was determined by the targeted viral protein, Nef-effect ratios were compared across epitopes located in different HIV-1 proteins. Gag epitopes in general had higher Nef-effect ratios (median, 0.73; range, 0.56-0.93), indicating low CTL susceptibility to Nef-mediated HLA-I down-regulation. Nef and Pol epitopes trended toward having lower ratios than Gag (Pol median, 0.29; range, 0.24-0.34; P = .04), suggesting greater CTL susceptibility to Nef-mediated HLA-I down-regulation (Figure 2A). In general, Gag epitopes had a higher Nef-effect ratio than all non-Gag–derived epitopes combined (P = .03; Figure 2B), suggesting that Gag-specific CTLs are overall less susceptible to Nef.
Impact of Nef on the antiviral activity of HIV-1–specific CTLs. The susceptibility of HIV-1–specific CTLs to Nef-mediated HLA-I down-regulation was measured using a previously described viral suppression assay. (A) CTL clone S14-YT9-1.2 (B*57 restricted and Nef specific) was tested for inhibition of NL4-3.1 virus containing wild-type Nef or Nef-M20A (unable to down-regulate HLA-I) in parallel. Replication was assessed by measuring supernatant Gag p24 antigen (log10 pg/mL) and plotted over time. Inhibition of wild-type virus at day 6 was 1.6 log10 units (5.2-3.6 = 1.6) and inhibition efficiency was 0.3 (1.6/5.2 = 0.31). Inhibition of M20A-Nef virus at day 6 was 3.7 log10 units (5.5-1.8 = 3.7) and inhibition efficiency was 0.7 (3.7/5.5 = 0.67). Therefore, the Nef-effect ratio was 0.42 (0.31/0.67 = 0.46). A ratio of 0 would therefore indicate complete evasion mediated by Nef and a ratio of 1 would indicate no effect of Nef. (B) The Nef effect on viral inhibition is plotted for a panel of HIV-1–specific CTL clones. Each data point represents the average Nef-effect ratio of a CTL clone across multiple independent experiments; the horizontal bar represents the mean.
Impact of Nef on the antiviral activity of HIV-1–specific CTLs. The susceptibility of HIV-1–specific CTLs to Nef-mediated HLA-I down-regulation was measured using a previously described viral suppression assay. (A) CTL clone S14-YT9-1.2 (B*57 restricted and Nef specific) was tested for inhibition of NL4-3.1 virus containing wild-type Nef or Nef-M20A (unable to down-regulate HLA-I) in parallel. Replication was assessed by measuring supernatant Gag p24 antigen (log10 pg/mL) and plotted over time. Inhibition of wild-type virus at day 6 was 1.6 log10 units (5.2-3.6 = 1.6) and inhibition efficiency was 0.3 (1.6/5.2 = 0.31). Inhibition of M20A-Nef virus at day 6 was 3.7 log10 units (5.5-1.8 = 3.7) and inhibition efficiency was 0.7 (3.7/5.5 = 0.67). Therefore, the Nef-effect ratio was 0.42 (0.31/0.67 = 0.46). A ratio of 0 would therefore indicate complete evasion mediated by Nef and a ratio of 1 would indicate no effect of Nef. (B) The Nef effect on viral inhibition is plotted for a panel of HIV-1–specific CTL clones. Each data point represents the average Nef-effect ratio of a CTL clone across multiple independent experiments; the horizontal bar represents the mean.
CTL clones tested for Nef-effect
| HLA . | Epitope (location) . | Clone (n)* . | Mean Nef-effect ratio (SD) . | Mean functional avidity (SD)† . |
|---|---|---|---|---|
| A*02 | SLYNTVATL (Gag 77-85) | S1-SL9-1.8 (11) | 0.54 (0.19) | 2.3 (0.6) |
| S1-SL9-1.7 (1) | ||||
| S1-SL9-3.23 (18) | ||||
| S36-SL9-1.9 (10) | ||||
| S36-SL9-10.18 (3) | ||||
| S31-SL9-10.11 (4) | ||||
| TLNAWVKVV (Gag 151-159) | S82-TV9-10.28 (2) | 0.73 (0.09) | 5.0 | |
| AAVDLSHFL (Nef 83-91) | S58-AL9-10.18 (2) | 0.46 | ND | |
| ILKEPVHGV (Pol 446-472) | 68A62-IV9 (8) | 0.46 (0.31) | 4.9 | |
| S31-IV9-10.4 (1) | ||||
| AIIRILQQL (Vpr 59-67) | S36-AL9-1.1 (15) | 0.41 (0.06) | ND | |
| S36-AL9-10.10 (1) | ||||
| B*15 | RLRPGGKKKY (Gag 20-29) | MO471-RY10-1.1 (5) | 0.89 (0.24) | ND |
| TQGYFPDWQNY (Nef 117-127) | 42 871-TY11-10.4 (5) | 0.34 (0.10) | 4.6 (0.2) | |
| 42 871-TY11-10.37 (2) | ||||
| B*40 | QELKNSAVNL (Env 805-814) | S82-QL10-1.6 (2) | 0.75 (0.12) | ND |
| KEKGGLEGL (Nef 92-100) | S16-KL9-4.1 (1) | 0.45 | ND | |
| B*57 | ISPRTLNAW (Gag 147-155) | S11-IW9-10.73 (5) | 0.52 (0.27) | 4.8 (0.3) |
| S11-IW9-3.5 (1) | ||||
| S11-IW9-10.68 (2) | ||||
| S11-IW9-10.65 (1) | ||||
| S14-IW9-3.14 (1) | ||||
| S14-IW9-3.21 (3) | ||||
| S14-IW9-10.15 (6) | ||||
| S36-IW9-10.37 (2) | ||||
| KAFSPEVIPMF (Gag 162-172) | S14-KF11-10.2 (10) | 0.60 (0.14) | 4.3 (0.2) | |
| S14-KF11-10.12 (2) | ||||
| S14-KF11-10.36 (2) | ||||
| S14-KF11-10.47 (5) | ||||
| S14-KF11-3.22 (9) | ||||
| S14-KF11-1.3 (5) | ||||
| S14-KF11-10.6 (1) | ||||
| TSTLQEQIGW (Gag 240-249) | S11-TW10-3.24 (2) | 0.97 (0.04) | 4.7 (0.7) | |
| S11-TW10-10.38 (7) | ||||
| S11-TW10-10.47 (3) | ||||
| QASQEVKNW (Gag 308-316) | S34-QW9-1.1 (1) | 0.96 (0.05) | 5.2 (0.8) | |
| S16-QW9-1.10 (1) | ||||
| S34-QW9-10.53 (1) | ||||
| HTQGYFPDWQ (Nef 116-125) | 42 871-HQ10-3.6 (4) | 0.52 (0.05) | 4.7 (1.1) | |
| S14-HQ10-1.1 (2) | ||||
| S14-HQ10-1.3 (3) | ||||
| S11-HQ10-10.10 (1) | ||||
| S11-HQ10-10.31 (2) | ||||
| YFPDWQNYT (Nef 120-128) | S14-YT9-1.2 (2) | 0.55 (0.09) | 5.9 | |
| S14-YT9-10.4 (2) | ||||
| S14-YT9-10.8 (1) | ||||
| S36-YT9-3.36 (2) | ||||
| KIATESIVIW (Pol 529-538) | S34-KW10-10.38 (6) | 0.28 (0.03) | 3.9 | |
| S34-KW10-10.55 (1) | ||||
| RTVRLIKLLY (Rev 14-23) | S36-RY10-3.20 (2) | 1.00 (0.29) | 4.9 | |
| S36-RY10-3.3 (4) | ||||
| S36-RY10-3.4 (2) | ||||
| S36-RY10-3.30 (1) |
| HLA . | Epitope (location) . | Clone (n)* . | Mean Nef-effect ratio (SD) . | Mean functional avidity (SD)† . |
|---|---|---|---|---|
| A*02 | SLYNTVATL (Gag 77-85) | S1-SL9-1.8 (11) | 0.54 (0.19) | 2.3 (0.6) |
| S1-SL9-1.7 (1) | ||||
| S1-SL9-3.23 (18) | ||||
| S36-SL9-1.9 (10) | ||||
| S36-SL9-10.18 (3) | ||||
| S31-SL9-10.11 (4) | ||||
| TLNAWVKVV (Gag 151-159) | S82-TV9-10.28 (2) | 0.73 (0.09) | 5.0 | |
| AAVDLSHFL (Nef 83-91) | S58-AL9-10.18 (2) | 0.46 | ND | |
| ILKEPVHGV (Pol 446-472) | 68A62-IV9 (8) | 0.46 (0.31) | 4.9 | |
| S31-IV9-10.4 (1) | ||||
| AIIRILQQL (Vpr 59-67) | S36-AL9-1.1 (15) | 0.41 (0.06) | ND | |
| S36-AL9-10.10 (1) | ||||
| B*15 | RLRPGGKKKY (Gag 20-29) | MO471-RY10-1.1 (5) | 0.89 (0.24) | ND |
| TQGYFPDWQNY (Nef 117-127) | 42 871-TY11-10.4 (5) | 0.34 (0.10) | 4.6 (0.2) | |
| 42 871-TY11-10.37 (2) | ||||
| B*40 | QELKNSAVNL (Env 805-814) | S82-QL10-1.6 (2) | 0.75 (0.12) | ND |
| KEKGGLEGL (Nef 92-100) | S16-KL9-4.1 (1) | 0.45 | ND | |
| B*57 | ISPRTLNAW (Gag 147-155) | S11-IW9-10.73 (5) | 0.52 (0.27) | 4.8 (0.3) |
| S11-IW9-3.5 (1) | ||||
| S11-IW9-10.68 (2) | ||||
| S11-IW9-10.65 (1) | ||||
| S14-IW9-3.14 (1) | ||||
| S14-IW9-3.21 (3) | ||||
| S14-IW9-10.15 (6) | ||||
| S36-IW9-10.37 (2) | ||||
| KAFSPEVIPMF (Gag 162-172) | S14-KF11-10.2 (10) | 0.60 (0.14) | 4.3 (0.2) | |
| S14-KF11-10.12 (2) | ||||
| S14-KF11-10.36 (2) | ||||
| S14-KF11-10.47 (5) | ||||
| S14-KF11-3.22 (9) | ||||
| S14-KF11-1.3 (5) | ||||
| S14-KF11-10.6 (1) | ||||
| TSTLQEQIGW (Gag 240-249) | S11-TW10-3.24 (2) | 0.97 (0.04) | 4.7 (0.7) | |
| S11-TW10-10.38 (7) | ||||
| S11-TW10-10.47 (3) | ||||
| QASQEVKNW (Gag 308-316) | S34-QW9-1.1 (1) | 0.96 (0.05) | 5.2 (0.8) | |
| S16-QW9-1.10 (1) | ||||
| S34-QW9-10.53 (1) | ||||
| HTQGYFPDWQ (Nef 116-125) | 42 871-HQ10-3.6 (4) | 0.52 (0.05) | 4.7 (1.1) | |
| S14-HQ10-1.1 (2) | ||||
| S14-HQ10-1.3 (3) | ||||
| S11-HQ10-10.10 (1) | ||||
| S11-HQ10-10.31 (2) | ||||
| YFPDWQNYT (Nef 120-128) | S14-YT9-1.2 (2) | 0.55 (0.09) | 5.9 | |
| S14-YT9-10.4 (2) | ||||
| S14-YT9-10.8 (1) | ||||
| S36-YT9-3.36 (2) | ||||
| KIATESIVIW (Pol 529-538) | S34-KW10-10.38 (6) | 0.28 (0.03) | 3.9 | |
| S34-KW10-10.55 (1) | ||||
| RTVRLIKLLY (Rev 14-23) | S36-RY10-3.20 (2) | 1.00 (0.29) | 4.9 | |
| S36-RY10-3.3 (4) | ||||
| S36-RY10-3.4 (2) | ||||
| S36-RY10-3.30 (1) |
ND indicates not determined.
Number of independent experiments performed.
Functional avidity in log10 picograms/mL peptide.
Gag-specific CTLs overall are less susceptible to Nef. The effect of Nef on CTL antiviral activity was compared according to the epitope source protein. Gag epitopes in general had higher Nef-effect ratios than epitopes located in Pol and Nef. (A) The data plotted according to proteins. Each dot represents the Nef-effect ratio of an epitope; the horizontal bar represents the mean. Statistical significance was assessed using the Kruskal-Wallis test followed by the Dunn post test for multiple comparisons. (B) Gag epitopes compared with all other epitopes from non-Gag proteins. A 2-tailed Student t test was used to compare the 2 groups.
Gag-specific CTLs overall are less susceptible to Nef. The effect of Nef on CTL antiviral activity was compared according to the epitope source protein. Gag epitopes in general had higher Nef-effect ratios than epitopes located in Pol and Nef. (A) The data plotted according to proteins. Each dot represents the Nef-effect ratio of an epitope; the horizontal bar represents the mean. Statistical significance was assessed using the Kruskal-Wallis test followed by the Dunn post test for multiple comparisons. (B) Gag epitopes compared with all other epitopes from non-Gag proteins. A 2-tailed Student t test was used to compare the 2 groups.
The influence of Nef on CTL antiviral activity is not determined by HLA-I restriction
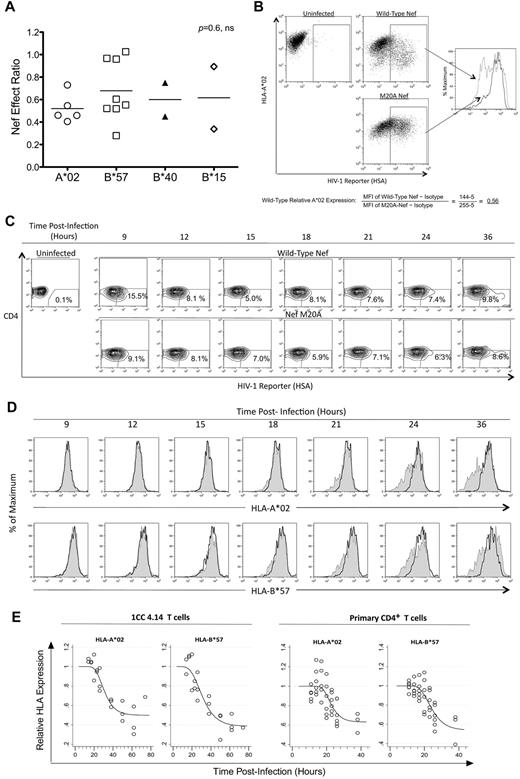
Different HLA-I molecules have been associated with better immune control of HIV-1 infection, particularly HLA-B*57, which is overrepresented in persons with low viremia and slow disease progression.28 Therefore, we investigated whether CTL susceptibility to Nef varied according to the presenting HLA-I molecule (Figure 3). CTLs targeting epitopes presented by various neutral HLA-I alleles and the protective B*57 allele were compared for their susceptibilities to Nef, revealing no significant difference (P = .57; Figure 3A). Furthermore, direct comparison of Nef-mediated HLA-I down-regulation of A*02 versus B*57 (Figure 3B-C) revealed similar magnitudes and kinetics of down-regulation (Figure 3D-E). These results suggest that the differential Nef effects on HIV-1–specific CTLs are unrelated to the presenting HLA-I type and that the in vivo protective effect of B*57-restricted responses in HIV-1 infection is not due to inefficient down-regulation of B*57 by Nef.
The effect of Nef on HIV-1–specific CTL antiviral activity is unrelated to HLA-I restriction of the epitope. The impact of Nef was compared across presenting HLA-I types. (A) Nef-effect ratios plotted for the indicated HLA-I types. Each dot represents one epitope; the horizontal bar represents the mean. There was no significant difference between groups (Kruskal-Wallis test). (B) A flow cytometric approach was used to measure Nef-mediated down-regulation of HLA-I A*02 and HLA-I B*57 on acutely HIV-1–infected cells. A representative experiment shows the analysis of HLA-A*02 down-regulation after gating on infected cells (positive for the HSA reporter) and determination of MFI of A*02 staining. The relative expression of A*02 on cells infected with wild-type Nef virus (gray dotted histogram) was then calculated as a fraction of MFI compared with Nef-M20A virus (black solid histogram) after subtraction of background MFI (from an isotype control). (C) Expression of cell-surface CD4 and the HSA reporter over time is demonstrated after acute infection of primary CD4+ T lymphocytes from an HIV-1–infected donor with both A*02 and B*57 (subject number 00036, a slow progressor not on treatment). (D) The infected (HSAhigh+ and CD4dim/−, percentages shown) primary CD4+ T lymphocytes were gated and analyzed for A*02 (top panel) and B*57 (bottom panel) expression. HLA-I expression is plotted for HIV-1 with wild-type Nef (gray shaded histograms) versus Nef-M20A (black histograms). (E) Gompertz plots of Nef-mediated down-regulation of A*02 versus B*57 are shown for the laboratory T-cell line 1CC4.14 (top panel) or primary CD4+ T lymphocytes* (bottom panel). In the top panel, the estimates for time to 10% A*02 versus B*57 down-regulation by Nef are 22.1 and 19.6 hours after infection, respectively. The slopes of the fitted curves at 10% down-regulation of A*02 versus B*57 are −0.016 and −0.015, respectively. The maximum levels of down-regulation of A*02 versus B*57 are estimated to be 50% and 40%, respectively. These parameters are not significantly different for A*02 versus B*57. In the bottom panel, the estimates for time to 10% A*02 versus B*57 down-regulation by Nef are 18.1 and 18.0 hours after infection, respectively. The slopes of the fitted curves at 10% down-regulation of A*02 versus B*57 are −0.023 and −0.021, respectively. The maximum levels of down-regulation of A*02 versus B*57 are estimated to be 60% and 50%, respectively. These parameters are not significantly different for A*02 versus B*57. The efficiency of infection was higher (approximately 40%-50% vs the 10% shown in Figure 3C) and Nef-mediated HLA-I down-regulation occurred earlier (approximately 12 hours after infection) in virus-infected primary CD4+ T lymphocytes of HIV-1–uninfected donors (data not shown).
The effect of Nef on HIV-1–specific CTL antiviral activity is unrelated to HLA-I restriction of the epitope. The impact of Nef was compared across presenting HLA-I types. (A) Nef-effect ratios plotted for the indicated HLA-I types. Each dot represents one epitope; the horizontal bar represents the mean. There was no significant difference between groups (Kruskal-Wallis test). (B) A flow cytometric approach was used to measure Nef-mediated down-regulation of HLA-I A*02 and HLA-I B*57 on acutely HIV-1–infected cells. A representative experiment shows the analysis of HLA-A*02 down-regulation after gating on infected cells (positive for the HSA reporter) and determination of MFI of A*02 staining. The relative expression of A*02 on cells infected with wild-type Nef virus (gray dotted histogram) was then calculated as a fraction of MFI compared with Nef-M20A virus (black solid histogram) after subtraction of background MFI (from an isotype control). (C) Expression of cell-surface CD4 and the HSA reporter over time is demonstrated after acute infection of primary CD4+ T lymphocytes from an HIV-1–infected donor with both A*02 and B*57 (subject number 00036, a slow progressor not on treatment). (D) The infected (HSAhigh+ and CD4dim/−, percentages shown) primary CD4+ T lymphocytes were gated and analyzed for A*02 (top panel) and B*57 (bottom panel) expression. HLA-I expression is plotted for HIV-1 with wild-type Nef (gray shaded histograms) versus Nef-M20A (black histograms). (E) Gompertz plots of Nef-mediated down-regulation of A*02 versus B*57 are shown for the laboratory T-cell line 1CC4.14 (top panel) or primary CD4+ T lymphocytes* (bottom panel). In the top panel, the estimates for time to 10% A*02 versus B*57 down-regulation by Nef are 22.1 and 19.6 hours after infection, respectively. The slopes of the fitted curves at 10% down-regulation of A*02 versus B*57 are −0.016 and −0.015, respectively. The maximum levels of down-regulation of A*02 versus B*57 are estimated to be 50% and 40%, respectively. These parameters are not significantly different for A*02 versus B*57. In the bottom panel, the estimates for time to 10% A*02 versus B*57 down-regulation by Nef are 18.1 and 18.0 hours after infection, respectively. The slopes of the fitted curves at 10% down-regulation of A*02 versus B*57 are −0.023 and −0.021, respectively. The maximum levels of down-regulation of A*02 versus B*57 are estimated to be 60% and 50%, respectively. These parameters are not significantly different for A*02 versus B*57. The efficiency of infection was higher (approximately 40%-50% vs the 10% shown in Figure 3C) and Nef-mediated HLA-I down-regulation occurred earlier (approximately 12 hours after infection) in virus-infected primary CD4+ T lymphocytes of HIV-1–uninfected donors (data not shown).
Functional avidity is not correlated with the degree of Nef interference with HIV-1–specific CTL antiviral efficacy
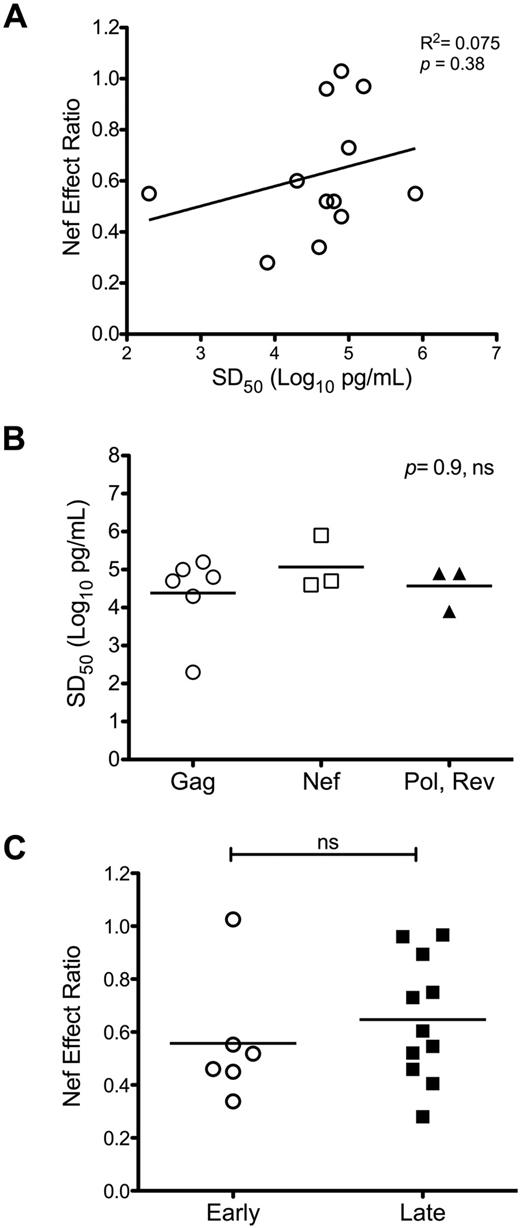
Previously reported data suggest that functional avidity is a key determinant of CTL antiviral efficacy.19,29,30 To explore the possibility that higher avidity allows better resistance to Nef, we investigated the relationship between the functional avidity (SD50) and the Nef-effect ratios of CTLs. When SD50 values were plotted against Nef-effect ratios, no significant correlation was apparent (Figure 4A). Because Gag-specific CTL responses appeared to be less susceptible to Nef-mediated HLA-I down-regulation compared with those directed against all non-Gag epitopes overall, we also investigated whether Gag-specific CTLs exhibited higher functional avidity than those targeting non-Gag epitopes. The SD50 values were not significantly different between CTLs targeting epitopes located in Gag versus other proteins (Figure 4B). These data suggest that differential Nef effects on HIV-1–specific CTL antiviral activity are not determined by differences in functional avidity.
Impact of Nef on HIV-1–specific CTL antiviral activity is not influenced by CTL functional avidity or Rev dependence of the epitope source protein. Nef-effect ratios for the tested CTL clones were compared according to functional avidity and Rev-dependence status. (A) Nef-effect ratio plotted against functional avidity (SD50) for each tested CTL clone; there was no significant correlation by the Pearson test. (B) Functional avidities of CTLs targeting Gag-derived epitopes compared with those directed against epitopes located in non-Gag proteins; the horizontal bars represent the means. There was no significant difference between groups by the Kruskal-Wallis test for multiple comparisons. (C) Nef-effect ratios plotted for CTLs recognizing epitopes from “early” (Rev-independent, Nef and Rev) versus “late” (Rev-dependent, Gag, Pol, Vpr, and Env) proteins. There was no significant difference between groups by a 2-tailed Student t test.
Impact of Nef on HIV-1–specific CTL antiviral activity is not influenced by CTL functional avidity or Rev dependence of the epitope source protein. Nef-effect ratios for the tested CTL clones were compared according to functional avidity and Rev-dependence status. (A) Nef-effect ratio plotted against functional avidity (SD50) for each tested CTL clone; there was no significant correlation by the Pearson test. (B) Functional avidities of CTLs targeting Gag-derived epitopes compared with those directed against epitopes located in non-Gag proteins; the horizontal bars represent the means. There was no significant difference between groups by the Kruskal-Wallis test for multiple comparisons. (C) Nef-effect ratios plotted for CTLs recognizing epitopes from “early” (Rev-independent, Nef and Rev) versus “late” (Rev-dependent, Gag, Pol, Vpr, and Env) proteins. There was no significant difference between groups by a 2-tailed Student t test.
The influence of Nef on CTL antiviral activity is not determined by early versus late transcription of the genes coding for the epitope source proteins
It has been hypothesized that CTLs recognizing epitopes derived from the early expressed HIV-1 proteins Rev, Tat, and Nef (translated from Rev-independent RNA transcripts) might be less susceptible to Nef because of a kinetic advantage in relation to HLA-I down-regulation.14,15 Addressing this hypothesis, we compared the effect of Nef on viral inhibition between CTLs targeting epitopes derived from the “late” expressed proteins Gag, Pol, Env, and Vpr (translated from Rev-dependent RNA transcripts) with those targeting epitopes derived from Nef and Rev (translated from Rev-independent RNA transcripts). The distributions of Nef-effect ratios, however, were not significantly different between the 2 groups (Figure 4C). Despite the “early” expression of Nef, CTLs directed against Nef-derived epitopes remained susceptible to Nef-mediated HLA-I down-regulation (Figure 2A), which is consistent with prior data.7 In contrast, the antiviral activity of CTLs targeting a Rev epitope were unaffected functionally by Nef (Figure 1B), suggesting that some “early” protein epitopes could have a kinetic advantage; although only one Rev epitope was assessed here, it has been shown previously that Rev-specific CTLs can be sensitive to Nef.7 Furthermore, the resistance of the tested Rev-specific CTLs to Nef-mediated HLA-I down-regulation appeared to be an outlier compared with the Nef-specific CTLs; in fact, the latter alone were statistically more susceptible to Nef-mediated HLA-I down-regulation than the group of CTLs targeting “late” proteins. However, removal of the Gag-specific CTLs from the “late” group resulted in loss of this significance. These findings indicate that “early” versus “late” protein epitopes do not necessarily correspond to early versus late CTL triggering in relation to Nef-mediated HLA-I down-regulation.
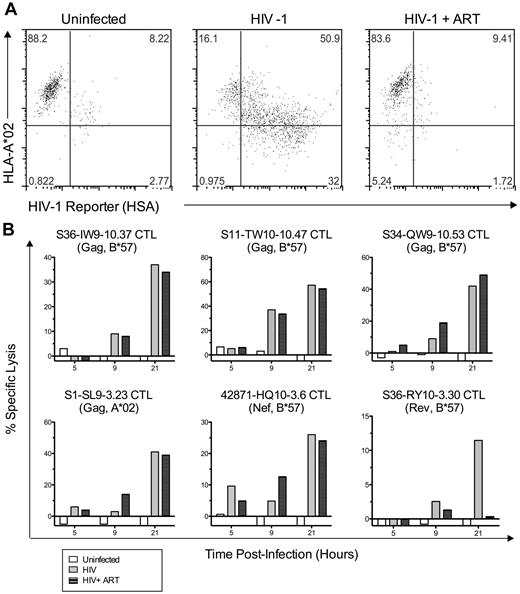
CTL resistance to Nef-mediated HLA-I down-regulation can be mediated by very early killing of HIV-1–infected cells
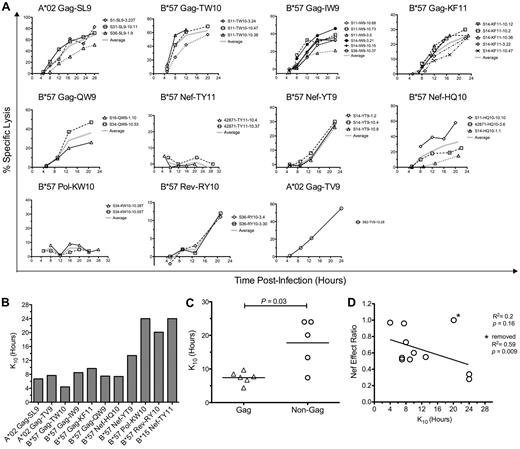
Despite the observations described in the previous section, the temporal relationship between HIV-1 epitope presentation and Nef-mediated HLA-I down-regulation clearly can influence the antiviral efficacy of HIV-1–specific CTLs, as shown previously in controlled experiments.14,15 Therefore, we assessed the kinetics of infected cell clearance by HIV-1–specific CTLs to investigate these kinetic relationships in greater detail directly (Figure 5). The onset of infected cell killing (using an arbitrary 10% threshold, K10) was estimated by fitting logarithmic regression curves to the observed specific lysis of acutely infected cells over time (Figure 5A). Across epitopes, K10 values ranged from as early as 4.4 hours to > 24 hours after infection (Figure 5B). Because Gag-specific CTLs were overall less susceptible to Nef than those targeting all non-Gag epitopes combined (Figure 2), we compared the killing kinetics of Gag-specific CTLs versus non-Gag–specific CTLs, finding that Gag-derived epitopes as a whole exhibited significantly lower K10 values than non-Gag epitopes (P = .03; Figure 5C), suggesting faster generation of Gag versus non-Gag epitopes (although this was not exclusive to Gag because one Nef epitope exhibited similarly early killing). Comparison of K10 values with Nef-effect ratios between epitopes suggested an inverse relationship (Figure 5D), although this was not statistically significant for this small number of epitopes (R2 = 0.2 and P = .16 improved to R2 = 0.58 and P = .009 with the removal of one outlier). These data indicate that the generally greater resistance of Gag-specific CTL antiviral activity to Nef was due to earlier killing of cells after infection, but that this was not specific to Gag.
CTL resistance to Nef is associated with early killing of HIV-1–infected cells. The timing of CTL killing of 1CC4.14 cells after acute infection with VSV-G Env-pseudotyped NL4-3-ΔEnv was assessed by serial 51Cr measurements over 24 hours. (A) Specific lysis plotted over time by epitope. Results with CTL clones are plotted with symbols and the average across clones is plotted with a broad gray line. (B) Estimates of time to reach 10% specific lysis (K10) are indicated. The estimates were obtained by fitting average lysis curves for each epitope with logarithmic regression. Note that CTLs targeting the B*15-restricted Nef TY11 and the B*57-restricted Pol KW10 epitopes were unable to recognize virus-infected cells within the first 24 hours of infection, and therefore each epitope was assigned a conservative K10 value of 24 hours. The inability of these CTL clones to recognize virus-infected targets was not because of inactivity of the clones, because they efficiently killed cells infected with HIV-1 containing the Nef M20A mutation (data not shown). (C) K10 values of Gag-specific versus non-Gag–specific CTLs are compared. Each point represents the K10 value for an epitope; the horizontal bar represents the mean across epitopes. Statistical significance was evaluated with a 2-tailed Student t test. (D) K10 values are plotted against Nef-effect ratios for all epitopes. Statistical significance was tested with a Pearson test. Note that these results were obtained using HIV-1 containing wild-type Nef; nearly identical values were obtained using virus with Nef-M20A, although the efficiencies of infected cell killing were slightly higher (data not shown). Asterisk indicates results after removing an outlier, the B*57-restricted Rev RY10 epitope.
CTL resistance to Nef is associated with early killing of HIV-1–infected cells. The timing of CTL killing of 1CC4.14 cells after acute infection with VSV-G Env-pseudotyped NL4-3-ΔEnv was assessed by serial 51Cr measurements over 24 hours. (A) Specific lysis plotted over time by epitope. Results with CTL clones are plotted with symbols and the average across clones is plotted with a broad gray line. (B) Estimates of time to reach 10% specific lysis (K10) are indicated. The estimates were obtained by fitting average lysis curves for each epitope with logarithmic regression. Note that CTLs targeting the B*15-restricted Nef TY11 and the B*57-restricted Pol KW10 epitopes were unable to recognize virus-infected cells within the first 24 hours of infection, and therefore each epitope was assigned a conservative K10 value of 24 hours. The inability of these CTL clones to recognize virus-infected targets was not because of inactivity of the clones, because they efficiently killed cells infected with HIV-1 containing the Nef M20A mutation (data not shown). (C) K10 values of Gag-specific versus non-Gag–specific CTLs are compared. Each point represents the K10 value for an epitope; the horizontal bar represents the mean across epitopes. Statistical significance was evaluated with a 2-tailed Student t test. (D) K10 values are plotted against Nef-effect ratios for all epitopes. Statistical significance was tested with a Pearson test. Note that these results were obtained using HIV-1 containing wild-type Nef; nearly identical values were obtained using virus with Nef-M20A, although the efficiencies of infected cell killing were slightly higher (data not shown). Asterisk indicates results after removing an outlier, the B*57-restricted Rev RY10 epitope.
Some Gag- and Nef-specific CTLs do not require presentation of de novo–synthesized viral proteins for infected cell killing
To determine the mechanism of early infected cell killing by Gag-specific CTLs, we treated acutely infected cells with the reverse transcriptase inhibitors tenofovir and zidovudine (ART) at concentrations that blocked viral replication and therefore prevented viral protein expression (Figure 6A). Treatment had minimal effects on the kinetics and magnitude of infected cell killing by several Gag-specific CTLs (Figure 6B). A Nef-specific CTL clone also appeared to be unaffected by drug treatment. In contrast, infected cell killing by a Rev-specific CTL clone was abolished by the treatment, indicating that this phenomenon is epitope specific. These data were consistent with previous findings that virion-derived proteins can be processed and presented to CTLs before de novo protein production by acutely SIV-infected cells,16,17 and suggest that our observation of early killing and Nef resistance is explained by this phenomenon.
Some Gag- and Nef-specific CTLs cells do not require de novo viral protein synthesis for early killing of HIV-1–infected cells. 1CC4.14 cells were infected with 600 fg of Gag p24/cell of VSV-G Env-pseudotyped NL4-3-ΔEnv-HSA and assessed for killing by CTLs (51Cr release) in the presence and absence of ART. (A) Uninfected or infected cells with or without ART were examined by flow cytometry for cell-surface expression of HLA A*02 and HSA reporter at 48 hours after infection, confirming that ART blocked the de novo viral protein expression seen in untreated cells (HSA expression and A*02 down-regulation by Nef). (B) Specific lysis of uninfected or infected target cells (with or without ART) was assessed by 51Cr-release assay over time. The data shown are representative of 2 independent experiments.
Some Gag- and Nef-specific CTLs cells do not require de novo viral protein synthesis for early killing of HIV-1–infected cells. 1CC4.14 cells were infected with 600 fg of Gag p24/cell of VSV-G Env-pseudotyped NL4-3-ΔEnv-HSA and assessed for killing by CTLs (51Cr release) in the presence and absence of ART. (A) Uninfected or infected cells with or without ART were examined by flow cytometry for cell-surface expression of HLA A*02 and HSA reporter at 48 hours after infection, confirming that ART blocked the de novo viral protein expression seen in untreated cells (HSA expression and A*02 down-regulation by Nef). (B) Specific lysis of uninfected or infected target cells (with or without ART) was assessed by 51Cr-release assay over time. The data shown are representative of 2 independent experiments.
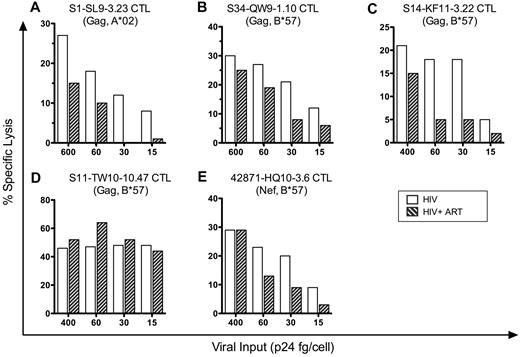
Early CTL killing of infected cells through incoming virion-derived epitopes depends on the viral inoculum, and the required inoculum varies by epitope
To investigate the amounts of virion-derived proteins that are required to trigger early CTL antiviral activity, we performed a dose-response analysis in the presence and absence of drug treatment. Target cells were infected with various concentrations of pseudotyped HIV-1 stocks corresponding to various ratios of Gag p24 protein concentration per cell (ranging from 15-600 fg of p24/cell), cocultured with HIV-1–specific CTLs with or without ART, and assessed for lysis at 12 hours after infection (Figure 7). Each CTL clone that had exhibited early killing was unaffected by ART at a viral inoculum of 400-600 fg of p24/cell, but differences were seen between clones with decreasing virus inputs of 60 fg of p24/cell or lower. Most clones showed a reduction in activity with decreasing inocula and were unable to kill infected cells at 15 fg of p24/cell, although one clone retained efficient killing activity at 15 fg of p24/cell (Figure 7D). ART treatment of the infected target cells had no effect on killing at a viral inoculum of 400 fg of p24/cell or higher, but variably affected the efficiency of killing at lower levels of viral input for different clones, suggesting various contributions of de novo–synthesized proteins to killing by 12 hours. The clone that killed infected cells efficiently at an input of 15 fg of p24/cell also maintained killing in the presence of ART at that inoculum (Figure 7D), suggesting more efficient recognition of incoming virion-derived epitopes. These data indicate that HIV-1–specific CTLs have various capacity to clear infected cells through recognition of incoming virion-derived epitopes.
Early killing of HIV-1–infected cells by CTLs is viral inoculum dependent. 1CC4.14 cells were infected with various amounts of VSV-G Env-pseudotyped NL4-3-ΔEnv and assessed for killing by CTLS (51Cr release) in the presence or absence of ART over time at 12 hours after infection. The data are representative of 3 independent experiments, except those shown in panel C, which are representative of 2 independent experiments.
Early killing of HIV-1–infected cells by CTLs is viral inoculum dependent. 1CC4.14 cells were infected with various amounts of VSV-G Env-pseudotyped NL4-3-ΔEnv and assessed for killing by CTLS (51Cr release) in the presence or absence of ART over time at 12 hours after infection. The data are representative of 3 independent experiments, except those shown in panel C, which are representative of 2 independent experiments.
Discussion
Nef-mediated HLA-I down-regulation impairs the effectiveness of CTL antiviral activity against HIV-1 in an epitope-specific manner,6,7 but the determining factors have thus far been unclear. In the present study, we investigated a panel of 17 viral CTL epitopes varying in sensitivity to Nef and assessed the effects of factors including HLA-I restriction, functional avidity, protein source, and kinetics of CTL recognition. The latter factor is the major determinant of Nef impact, which is consistent with other studies suggesting a role for antigen-presentation kinetics and CTL antiviral efficiency in general.14-17
Although it has been suggested that CTL targeting of the “early” proteins Tat, Rev, and Nef (translated from fully spliced transcripts that are Rev independent for nuclear exit) yields superior antiviral activity,14,15 our present data demonstrate that “early” versus “late” protein epitope source does not dictate early versus late CTL recognition of infected cells. A Rev- and a Nef-specific CTL clone recognized infected cells relatively late compared with Gag-specific CTLs, although another Nef-specific CTL clone did mediate early recognition. Therefore, it is likely that epitope processing from incoming virion proteins and/or other epitope factors (eg, efficiency of processing) override the role of protein-expression kinetics.
Nef-mediated interference of CTL antiviral activity also appears to be unaffected by HLA-I restriction. Given that some HLA-I types have strong associations with the degree of immune control of HIV-128 and that Nef is a critical protein for HIV-1 virulence,31 various down-regulation of different HLA-I types by Nef would be a potential unifying mechanism. However, 2 observations in the present study make this unlikely. First, the functional impact of Nef on CTL antiviral activity did not differ consistently between CTLs targeting epitopes restricted by B*57 versus those restricted by A*02, B*15, or B*40. Second, the magnitude and timing of B*57 down-regulation by HIV-1 Nef was similar to that of A*02. Therefore, it is likely that the influence of HLA-I type on immune control is not related to Nef function, but rather to other factors determined by targeting, such as epitope sequence constraints for escape.32,33
Similarly, functional avidity was not correlated with either the degree of Nef impact or the kinetics of antiviral responses (data not shown). Although it has been suggested that higher avidity CTLs can eliminate infected cells more rapidly and have higher antiviral efficacy30 and that CTLs targeting Gag or restricted by B*57 are superior because of higher functional avidity,34 we did not find significant differences in functional avidity of CTLs targeting different proteins or restricted by different HLA-I types. This finding was consistent with prior studies from our group demonstrating no correlation between functional avidity and antiviral efficacy35 and no greater antiviral activity for additional avidity beyond a required threshold for killing infected cells.19 Epitope targeting independent of such factors appears to be the key determinant of Nef impact on CTLs.
There has been considerable interest in previous observations that the magnitude and breadth of Gag targeting by the CTL response is correlated with better immune control in vivo.20,36,37 Whereas the Gag-specific CTL response appears to have superior antiviral activity compared with the Env-specific CTL response when tested ex vivo,22 the mechanism is unclear and this finding could be related to better matching of in vivo viral sequences to the test strain or to differences in CTL phenotype and function, rather than targeting per se. Our present data suggest a potential mechanism that is directly related to targeting. In our experimental conditions, most Gag-specific CTLs demonstrated early killing of acutely infected cells, which could translate directly to better clearance of infected cells and evasion of Nef-mediated HLA-I down-regulation by Gag-specific CTLs on average.
In agreement with similar findings in the SIV-macaque experimental system,16 in the present study, we found that CTLs against virion-contained proteins other than Gag also can mediate early killing of infected cells. Our data further suggest that the efficiency of triggering of this early killing varies by epitope: it is likely that Gag-specific CTLs exhibit efficient earlier killing on average than CTLs targeting other incoming virion-derived epitopes, but that this property is not uniformly dictated by the source protein. This is consistent with the finding that whereas Gag-specific CTL targeting is associated with significantly better immune control, this is a loose correlation that is not predictive on the individual level.
The contribution of incoming virion-derived epitopes to CTL antiviral activity in vivo has been debatable due to questions about the physiologic relevance of the viral inoculum used for in vitro studies. Our data examine the dose response of virus inoculum for early killing, demonstrating that at least 30 fg of p24/cell of viral inoculum is required to induce early cytolysis of infected target cells by the tested Gag- and Nef-specific CTLs that mediated early killing. Because each picogram of p24 corresponds to approximately 104 virions,38 this corresponds to approximately 300 virions/cell. A study examining virus-infected splenic T lymphocytes in 2 persons with HIV-1 infection revealed that 80% of infected cells harbored more than 2 distinct proviruses and approximately 20% contained 5-8 proviruses.39 Whereas these numbers suggest that target cells in vivo are infected by fewer virions than required for early killing in our experimental system, it is conceivable that a multiplicity of viral entry sufficient for early killing is achievable in vivo. Productively infected memory CD4+ T lymphocytes are estimated to produce from 103 to 5 × 104 virions,40 and thus an adjacent cell could be exposed to hundreds or even thousands of virions. It is also thought that a minority of released virions are replication competent41 and therefore an infected cell may take up many more virions than the number of successfully integrated proviruses. Therefore, in tissues in which activated CD4+ T lymphocytes are very tightly clustered and cell-to-cell viral spread is likely,42 such as the gastrointestinal tract, it is conceivable that infected cells could take up enough virions for early killing. This concept is further supported by recently published data showing that cell-to-cell spread of HIV-1 results functionally in a markedly higher multiplicity of infection.43 Whether this phenomenon is relevant in vivo, however, remains to be determined.
Among the evaluated Gag-specific CTLs, those recognizing the B*57-restricted TW10 (Gag 240-249) epitope are particularly efficient in killing infected cells before de novo viral protein production. TW10-specific CTLs required an inoculum of approximately 6-15 fg of p24/cell of input virus (approximately 60-150 virions/cell), whereas the other CTLs required greater than 30 fg of p24/cell input virus (approximately 300 virions/cell). Targeting of this immunodominant epitope in acutely infected persons has been associated with better immune control,44 and it has been demonstrated that this epitope has a particularly long intracellular half-life that contributes to high levels of presentation.45 This supports our observation regarding the lower inoculum required by TW10-specific CTLs to recognize epitopes derived from incoming virions, and our present findings may further suggest a mechanism for the strong immunodominance of this epitope.
Early infected cell cytolysis through recognition of virion-derived epitopes is not unique to HIV-1 Gag targeting. Studies of SIV-specific CTLs and SIV-infected cells have described this phenomenon for CTLs targeting non-Gag proteins, including epitopes in Pol-derived,17 Vpr-derived,46 and Rev-derived epitopes,46 but not Nef.17 Although we did not observe early infected cell killing by the Pol- or Rev-specific CTLs tested, a Nef-specific CTL clone targeting the B*57-restricted HQ10 epitope (Nef 116-125) exhibited recognition of virion-derived epitopes. Myristoylated Nef associates with the infected cell's plasma membrane and is packaged into virions during the process of budding; however, this process is inefficient and the virions contain approximately 10% reverse transcriptase and 0.5% Gag.47 The observation of early killing by HQ10-specific CTLs indicates that Nef is carried in sufficient amounts to allow recognition of virion-derived epitopes at viral inocula similar to what is required for several Gag-specific CTLs. Furthermore, our observation of Pol-targeted CTLs that did not mediate early killing suggests that epitope-specific variability in efficiency of epitope processing and presentation determines the likelihood for CTL recognition of epitopes from incoming virions, emphasizing that the kinetics of epitope presentation and recognition by CTLs are multifactorial and not specific to the targeted viral protein.
One caveat to the results of the present study are the use of VSV-G Env-pseudotyped HIV-1 to evaluate CTL recognition of epitopes derived from incoming virions, which targets a different cellular compartment than wild-type HIV-1. Whereas native HIV-1 Env induces viral internalization into the cytoplasm through membrane fusion, VSV-G Env induces endocytosis into clathrin-coated pits for early endosomal uptake.48 Endosomal proteins are processed and presented via the HLA-II antigen pathway, whereas cytoplasmic proteins undergo proteasomal processing and enter the HLA-I pathway used for CTL recognition. Although cross-presentation from the HLA-II to the HLA-I pathway can occur in professional APCs,49 this process is highly inefficient in T lymphocytes.50 This suggests that our use of VSV Env-pseudotyped HIV-1 likely conservatively underestimates the sensitivity of this phenomenon.
In summary, the results of the present study provide evidence for a direct link between the kinetics of CTL recognition of acutely infected cells and susceptibility to Nef-mediated immune evasion. CTLs that can recognize epitopes processed from proteins carried by incoming virions before de novo viral protein production temporally bypass the effect of Nef. This capability varies according to individual epitopes and is not determined solely by the source protein, although high copy numbers of Gag in virions makes Gag epitopes the most common triggers of early killing. This may be one factor contributing to the overall beneficial effect of Gag targeting by CTLs derived from proteins carried in virions, but this phenomenon is not unique to Gag and can also occur for epitopes from other proteins carried in virions, such as Pol and Nef.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the persons from whose blood they derived the CTL clones used in this study.
This work was supported by NIH grant AI043203 to O.O.Y. NIH research grant AI028697 to the UCLA AIDS Institute Center for AIDS Research provided infrastructure support for this project. Recombinant human IL-2, tenofovir, and zidovudine were provided by the NIH AIDS Reference and Reagent Program.
National Institutes of Health
Authorship
Contribution: D.Y.C. designed the experimental approaches, performed the experiments, analyzed the data, and wrote the manuscript; A.B. and H.L.N. performed the experiments and analyzed the data; W.G.C. analyzed the data and wrote the manuscript; and O.O.Y. conceived the project, designed the experimental approaches, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Otto O. Yang, Division of Infectious Diseases, 37-121 CHS, UCLA Medical Center, 10833 Le Conte Ave, Los Angeles, CA 90095; e-mail: oyang@mednet.ucla.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal