Abstract
Peripheral T-cell lymphomas (PTCLs) are aggressive malignancies of mature T lymphocytes with 5-year overall survival rates of only ∼ 35%. Improvement in outcomes has been stymied by poor understanding of the genetics and molecular pathogenesis of PTCL, with a resulting paucity of molecular targets for therapy. We developed bioinformatic tools to identify chromosomal rearrangements using genome-wide, next-generation sequencing analysis of mate-pair DNA libraries and applied these tools to 16 PTCL patient tissue samples and 6 PTCL cell lines. Thirteen recurrent abnormalities were identified, of which 5 involved p53-related genes (TP53, TP63, CDKN2A, WWOX, and ANKRD11). Among these abnormalities were novel TP63 rearrangements encoding fusion proteins homologous to ΔNp63, a dominant-negative p63 isoform that inhibits the p53 pathway. TP63 rearrangements were seen in 11 (5.8%) of 190 PTCLs and were associated with inferior overall survival; they also were detected in 2 (1.2%) of 164 diffuse large B-cell lymphomas. As TP53 mutations are rare in PTCL compared with other malignancies, our findings suggest that a constellation of alternate genetic abnormalities may contribute to disruption of p53-associated tumor suppressor function in PTCL.
Introduction
Peripheral T-cell lymphomas (PTCLs) represent ∼ 12% of non-Hodgkin lymphomas and have 5-year overall survival rates of only ∼ 35% with conventional chemotherapy.1-4 Identifying new predictive biomarkers and therapeutic targets for PTCL could lead to improved outcomes, as they have in other hematologic malignancies. However, this has been challenging in PTCL because of marked clinical and pathologic heterogeneity, rarity of individual subtypes, and poor understanding of the genetics and molecular pathogenesis of PTCL.
Recurrent chromosomal rearrangements are critical in the molecular pathogenesis of nearly all types of hematologic neoplasms. The only well-characterized rearrangements in PTCLs included in the current World Health Organization (WHO) classification are those involving the anaplastic lymphoma kinase (ALK) gene, which occur in ∼ 6% of PTCLs.1,3 ALK rearrangements generate oncogenic ALK fusion proteins, the presence of which defines a specific WHO subtype, ALK-positive anaplastic large-cell lymphoma (ALCL). ALK also serves as a prognostic marker, as ALK-positive ALCLs have more favorable outcomes than ALCLs and other PTCLs that lack ALK rearrangements and ALK protein expression.2 Recently, small-molecule inhibitors of the ALK tyrosine kinase have emerged as a new therapeutic strategy, not only for lymphomas but also for solid tumors such as non–small-cell lung cancer.5 These advances highlight the potential impact of discovering additional recurrent chromosomal rearrangements in PTCLs, particularly those that lack ALK rearrangements.
Next-generation sequencing (NGS) offers exciting new opportunities for genetic discovery in cancer. We recently discovered a recurrent rearrangement in PTCL involving the DUSP22-IRF4 locus on 6p25.3 and used NGS of mate-pair genomic DNA libraries to characterize the breakpoints.6-8 The mate-pair approach is a highly sensitive means to identify rearrangements across the entire genome at a fraction of the cost of whole-genome sequencing. In this study, we enhanced our previous bioinformatic algorithms to be able to identify chromosomal rearrangements de novo. We identified 13 recurrent rearrangements in 21 PTCLs. Five of these rearrangements involved p53-related genes, including TP53, TP63, CDKN2A, WWOX, and ANKRD11. Particularly interesting were novel TP63 rearrangements that encoded fusion proteins containing an N-truncated p63 (ΔNp63) similar to native ΔNp63 isoforms, which are known to have oncogenic properties and to inhibit the p53 pathway by a dominant-negative mechanism.9 These rearrangements were seen in 11 (5.8%) of 190 PTCLs and were mutually exclusive with ALK rearrangements. TP63 rearrangements were associated with inferior overall survival among PTCLs. They also were detected in 2 (1.2%) of 164 diffuse large B-cell lymphomas (DLBCLs). Because TP53 mutations are much rarer in PTCLs than in other malignancies, our findings suggest that a constellation of alternate genetic abnormalities may contribute to disruption of p53-associated tumor suppressor function in PTCL.
Methods
Patients and tissue samples
Pathologic material from patients with PTCL was reviewed and diagnoses and classifications were confirmed using WHO criteria.3 Sequencing studies were performed on 16 patient tissue samples, including 4 PTCLs, not otherwise specified (NOS; TCL5, TCL29, TCL32, and TCL65); 1 ALK-positive ALCL (TCL3); 8 ALK-negative ALCLs (TCL1, TCL2, TCL11, TCL13, TCL14, TCL15, TCL16, and TCL56); and 3 primary cutaneous ALCLs (TCL6, TCL8, and TCL9). DNA was isolated from frozen tissue blocks containing > 80% PTCL cells by phenol-chloroform extraction and ethanol precipitation. RNA was extracted using PerfectPure (5 PRIME). Paraffin-based studies on formalin-fixed PTCL or DLBCL tissues were performed on blocks or tissue microarrays (TMAs) described previously.6,10-12 Genomic DNA from anonymized donors without a cancer history was obtained from the Mayo Clinic Biobank. The study was approved by the Mayo Clinic Institutional Review Board.
Cell lines
The ALK-positive ALCL cell lines SU-DHL-1 and SR-786 were obtained from DSMZ, while Karpas 299 was obtained from ATCC. MAC1 and MAC2A were derived from a patient with primary cutaneous ALCL and established by M.E.K. The ALK-negative ALCL cell line FE-PD was generously provided by Dr K. Pulford (Oxford, United Kingdom) with kind permission from Dr A. Del Mistro (Padova, Italy). Cells were maintained in RPMI 1640 (Invitrogen) with 10% fetal bovine serum (HyClone; except SR-786, 15%). DNA and RNA were isolated using the QIAamp DNA Blood Mini Kit and RNeasy Plus Mini Kit, respectively (QIAGEN).
Mate-pair library construction and sequencing
Sequencing was performed on the 16 PTCL patient tissue samples and the 6 cell lines described in the preceding 2 paragraphs. Two of the cell lines (MAC1 and MAC2A) were derived from the same patient and were considered together. Thus, a total of 21 PTCLs were analyzed. Mate-pair libraries were constructed and sequenced as previously described,7 with minor changes as described in supplemental Methods (available on the Blood Web site, see the Supplemental Materials link at the top of the online article).
Mate-pair data mapping and bioinformatic analysis
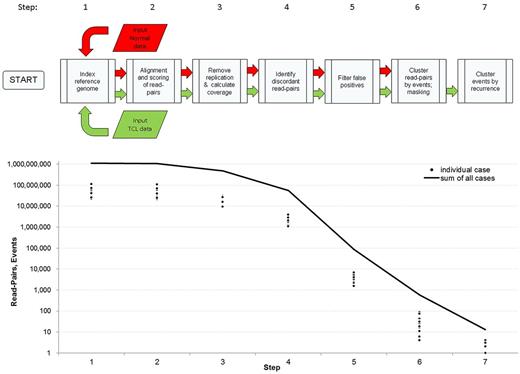
A set of algorithms was developed to detect unsuspected large chromosomal aberrations in PTCL with high specificity and sensitivity (Figure 1; supplemental Figures 1-6). The algorithms were designed specifically to handle NGS of unamplified DNA using mate-pair preanalytic protocols. The read-to-reference genome-mapping algorithm described previously7 was modified so that both mate-pair reads were evaluated together across the whole genome. The near simultaneous mapping of both reads eliminates a large number of false positives that would arise if read pairs were mapped independently, as the optimal read mappings often are far apart on the genome. Alternatively, multiple mapped positions can be stored and analyzed further at a later time, but that would require intermediate storage of large amounts of data and additional computing time.
Mapping of the mate-pair read pairs was facilitated by indexing the Hg19 reference genome using a binary-indexing routine that converted the genome nucleotide sequence to 32-bit binary numbers. These numbers served as addresses to the index array in random-access memory (RAM), where their corresponding genomic positions were stored if unique. If there were multiple positions with the same address, then a pointer to a linked list was stored in the index array (supplemental Figure 1). Evaluation and alignment of reads was performed with fast binary operations such as memory look-ups, XOR, and POPCNT. The algorithm could evaluate 10 000 read-pairs/s using 1 central processing unit. High mapping speeds were accomplished by indexing the whole reference genome in RAM, requiring computers with large RAM (∼ 30 Gb) and 64-bit architecture and operating system.
After mapping all the read pairs to the reference genome, postprocessing analysis was facilitated by cleaning and ordering the mapping output. A set of postmapping algorithms and plotting scripts were written in R including (1) replication removal, (2) bridged and base coverage analysis, (3) intrachromosomal detection; (4) translocation detection, (5) detection of recurrent abnormalities, (6) breakpoint junction plotting, and (7) count plotting. Replication removal was critical to reduce false positives, as the mate-pair protocol resulted in identical fragments because of amplification. The rate of replication varied across samples and was attributed to varied reduction of diversity during the early steps of the protocol (see “Mate-pair library construction and sequencing”). After removal of replicate read pairs, coverage calculations indicated the expected number of mate pairs covering a break point. The intrachromosomal and translocation detection scripts used homology scores to filter false positives and detect chromosomal abnormalities. Detection of abnormalities was performed by an R script that first used masking to remove false positives and then clustering techniques to find recurrent events. Finally, plotting scripts were used to visually inspect and choose events for further experimental validation. Descriptions of the scripts and the criteria used for masking and filtering are included as supplemental Methods.
Validation of mate-pair data and identification of chromosomal breakpoints
Primers specific for each abnormality were designed using Oligo Version 6.71 (Molecular Biology Insights; supplemental Table 1). PCR was performed on genomic DNA using the HotStar Taq DNA polymerase kit (QIAGEN) and a GeneAmp PCR System 9700 (Applied Biosystems). PCR-cycle parameters were 95°C × 15 minutes, 30 cycles of (95°C × 30 seconds; 60°C × 30 seconds; and 72°C × 5 minutes), followed by extension at 72°C × 12 minutes and 4°C × ∞. Amplicons were visualized on 0.8% agarose gels. Either the entire post-PCR product or DNA extracted from the gel using the QIAquick Gel Extraction kit (QIAGEN) was bidirectionally sequenced with the same primers used for amplification on a 3730xl DNA Analyzer (Applied Biosystems). Sequences were analyzed using Sequencher Version 4.8 software (GeneCodes). Sequence data were aligned to the human genome (February 2009 build; GRCh37/hg19) using BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat) to characterize the chromosomal breakpoints.
TP53 gene sequencing
Genomic DNA corresponding to exons 4 to 10 of TP53 was sequenced using the sequencing and amplification primers listed in supplemental Table 2. DNA was PCR amplified with GoTaq Hot Start Polymerase (Promega), checked for amplification and product size using the QIAxcel DNA Large Fragment kit (QIAGEN), treated with EsoSAP-IT (USB Corp), sequenced using the BigDye Terminator Version 1.1 Cycle Sequencing kit (Applied Biosystems) on an ABI 3130XL Genetic Analyzer (Applied Biosciences), and analyzed using Sequencher.
Multiple ligation-dependent probe amplification
A combination of multiple ligation-dependent probe amplification (MLPA) and Luminex xTAG (Luminex Molecular Diagnostics Inc) technologies was used for deletion detection as previously described.13 Briefly, DNA (200 ng) was denatured, followed by addition of M13-tailed probes incorporating sequences targeting exon 1 of ANKRD11 (ANKR1, 5′-agaggccgccctgagacggtgc-3′, 5′-gcgatggaccgagggcccca-3′; ANKR12, 5′-ctgcatgggcagcgcccgcg-3′,5′-ccccgccgctgagccgtcgc-3′) and control genes as described previously.13 Samples were denatured (95°C, 5 minutes) before probe hybridization (60°C, ≥ 16 hours). Ligase-65 (MRC Holland) was added for probe ligation (54°C, 15 minutes). The ligase was inactivated and the ligation mixes were added to premade PCR mixes containing universal M13 primers (forward primer 5′ biotin-labeled). PCR consisted of 23 cycles. PCR products were mixed with xTAG beads, 2000 of each bead per sample, denatured (96°C, 2 minutes) and hybridized (37°C, 1 hour). Hybridized products were incubated with 25 μL of a 1:125 mixture of streptavidin R-PE and 1× tetramethylammonium chloride buffer and applied to a Luminex 200 instrument. Two lasers with different wavelengths were used to detect the bead identity and the reporter molecule signal. For each probe, deviations of > 20% from a 1:1 ratio between signals from normal control samples and patient samples indicated possible deletions or insertions/duplications. Less than 4% of normal samples fell outside this range. Data were analyzed semiautomatically with Gene Marker (Softgenetics), using CNNTB (3p22.1), DMD (Xp21.1), HIRA (22q11.2), NPC-E17 (18q11.2), and TNFRSF7 (12p13.31) as reference/control genes.
Fluorescence in situ hybridization
A dual-fusion fluorescence in situ hybridization (FISH) probe to detect TBL1XR1/TP63 fusions and a breakapart FISH probe to detect TP63 rearrangements (regardless of partner locus) were designed and prepared similarly to previous probes developed in our laboratory.7 Briefly, bacterial artificial chromosomes (BACs) corresponding to the regions of interest were obtained from ResGen Invitrogen. DNA was isolated using the QIAGEN Plasmid Maxi Kit and labeled using the Vysis Nick Translation kit and either SpectrumGreen-dUTP or SpectrumOrange-dUTP (Vysis). Each BAC clone was tested individually by hybridization to normal metaphases. Vysis LSI TP53 SpectrumOrange/CEP 17 SpectrumGreen and Vysis LSI CDKN2A/SpectrumOrange/CEP 9 SpectrumGreen probes were obtained from Abbott Molecular.
Interphase FISH from paraffin material was performed on PTCLs, DLBCLs, or normal tonsil control sections as previously described.7 Briefly, 5-μm sections were deparaffinized in CitriSolve, dehydrated in ethanol, pretreated in 1M Tris/0.5M EDTA, and treated with NaCl protease. After ethanol dehydration, labeled DNA probe was applied and the slides were codenatured, hybridized overnight, and counterstained with 6-diamidino-2-phenylindole dihydrochloride. Metaphase FISH was performed from cell lines or archived bone marrow cell pellets when available as previously described. Slides were analyzed using standard fluorescence microscopy techniques.
RT-PCR and real-time quantitative PCR
The High Capacity Reverse Transcription kit (Applied Biosystems) was used to prepare cDNA from total RNA extracted from frozen PTCL tissue specimens. PCR for TBL1XR1 fusion transcripts 1 and 2 was performed using 1 μL of cDNA and PCR conditions as for genomic DNA (described in “Validation of mate-pair data and identification of chromosomal breakpoints”). Primers for fusion 1 were: forward, TGCGACAAACAAGATTGA; reverse, GCAAACCCATCATAGGA. Primers for fusion 2 were: forward, CAGTGGCTCTACACAGTTAG; reverse, TGGGTAGTCGGTGTTG. TaqMan quantitative PCR (QPCR) primer/probe sets were obtained from Applied Biosystems (TP63, Hs00978340_m1; TBL1XR1, Hs01037554_m1) or Integrated DNA Technologies (GAPDH: forward, GAAGGTGAAGGTCGGAGTC; reverse, GAAGATGGTGATGGGATTTC; probe, /56-FAM/CAAGCTTCCCGTTCTCAGCC/3IAbRQSp/). QPCR was performed in duplicate on a 7900HT Fast Real-Time PCR System (Applied Biosystems). Expression values of unknowns were derived from standard curves generated from serial dilutions of reference standards at known copy numbers for each gene and shown as copies of target transcript per copy of GAPDH.
For Western blotting, immunohistochemistry, detection of fusion transcripts, and statistics, see supplemental Methods.
Results
Discovery and validation of recurrent chromosomal rearrangements in PTCL
We identified 13 recurrent rearrangements in 21 PTCLs (Figure 1, Table 1, supplemental Table 3). Nine of these were intrachromosomal rearrangements; the type of rearrangement could be predicted in 8, based on the relative orientations (strands) of the reads. One of these was a novel inv(3)(q26q28) involving the TP53 homologue, TP63, and the nuclear corepressor (NCOR)–associated gene, TBL1XR1. Seven of the intrachromosomal rearrangements corresponded to deletions, with the deleted regions including TP53, genes encoding proteins involved in p53 stabilization (WWOX, CDKN2A), and the p53 coactivator gene ANKRD11.14-16 Thus, 5 of 13 recurrent rearrangements (38%) involved p53-related genes. The remaining 4 rearrangements were interchromosomal abnormalities, including the previously known ALK and DUSP22-IRF4 translocations.
Approach to detect chromosomal rearrangements from mate-pair genomic DNA sequencing data. (Top) schematic of bioinformatic algorithm. False-positive calls were minimized using filters based on quality of mapping, quality of nearby sequence in the reference genome, and a mask developed from noncancerous samples (see “Mate-pair data mapping and bioinformatic analysis”). Candidate abnormalities were supported by at least 4 mate pairs with reads mapping > 15 kb apart. Events were classified as recurrent abnormalities when they shared breakpoints within 1 Mb in 2 or more cases. (Bottom) numbers of read pairs or rearrangement events after executing each step of the algorithm, shown for individual cases and as the sum of all cases.
Approach to detect chromosomal rearrangements from mate-pair genomic DNA sequencing data. (Top) schematic of bioinformatic algorithm. False-positive calls were minimized using filters based on quality of mapping, quality of nearby sequence in the reference genome, and a mask developed from noncancerous samples (see “Mate-pair data mapping and bioinformatic analysis”). Candidate abnormalities were supported by at least 4 mate pairs with reads mapping > 15 kb apart. Events were classified as recurrent abnormalities when they shared breakpoints within 1 Mb in 2 or more cases. (Bottom) numbers of read pairs or rearrangement events after executing each step of the algorithm, shown for individual cases and as the sum of all cases.
Recurrent chromosomal rearrangements in PTCL predicted by mate-pair sequencing
| Breakpoint 1 . | Breakpoint 2 . | Rearrangement . | Samples with abnormality (of 21) . | Notations . | ||||
|---|---|---|---|---|---|---|---|---|
| Locus . | Gene . | Locus . | Gene . | Primary samples . | Cell lines . | n . | ||
| Xp22.31 | STS, HDHD1A | Xp22.31 | STS, HDHD1A | Deletion | TCL9 | FE-PD, SU-DHL-1, MAC1/2A* | 4 | Steroid sulfatase; pseudouridine-5′-monophosphatase |
| 2p23.2 | ALK | 5q35.1 | NPM1 | Translocation | TCL3 | SU-DHL-1, Karpas 299, SR-786 | 4 | Anaplastic lymphoma kinase; nucleophosmin |
| 3q26.32 | TBL1XR1 | 3q28 | TP63 | Inversion | TCL5, TCL29 | — | 2 | Transducin (beta)–like 1 X-linked receptor 1, tumor protein p63 |
| 4p15.1 | None | M† | Unknown | Unknown | TCL5, TCL8, TCL3, TCL56 | — | 4 | — |
| 6p25.3 | DUSP22 | 7q32.3 | FLJ43663 | Translocation | TCL1, TCL8, TCL11 | FE-PD | 4 | Dual-specificity phosphatase 22; near FRAH7 fragile site on 7q32.37 |
| 6q27 | None | 8q22.1 | Unknown | Unknown | TCL56 | Karpas 299 | 2 | — |
| 7q31.31 | None | 7q31.31 | None | Deletion | TCL13, TCL65 | — | 2 | No intervening known genes |
| 8q24.21 | Unknown | 8q24.21 | None | Unknown | TCL15 | MAC1/2A | 2 | — |
| 9p21.3 | MTAP | 9p21.3 | MTAP, CDKN2BAS | Deletion | TCL15, TCL29, TCL32 | — | 3 | Deleted region contains CDKN2A encoding p16INK4A and p19ARF (stabilizes p53)14 |
| 10q21.3 | CTNNA3 | 10q21.3 | CTNNA3 | Deletion | TCL6, TCL56 | — | 2 | Catenin alpha-3 |
| 16q23.1 | WWOX | 16q23.1 | WWOX | Deletion | TCL6, TCL29 | SU-DHL-1, Karpas 299, MAC1/2a̱* | 5 | WW domain-containing oxidoreductase (stabilizes p53)15 |
| 16q24.3 | ANKRD11 | 16q24.3 | SPG7 | Deletion | TCL1, TCL2, TCL9, TCL29 | FE-PD, MAC1/2A* | 6 | Ankyrin repeat domain-containing protein 11 (p53 co-activator)16 ; spastic paraplegia 7 |
| 17p13.1 | TMEM95, POLR2A | 17p13.1 | DNAH2, WRAP53 | Deletion | TCL3 | SR-786 | 2 | Deleted region contains TP53 |
| Breakpoint 1 . | Breakpoint 2 . | Rearrangement . | Samples with abnormality (of 21) . | Notations . | ||||
|---|---|---|---|---|---|---|---|---|
| Locus . | Gene . | Locus . | Gene . | Primary samples . | Cell lines . | n . | ||
| Xp22.31 | STS, HDHD1A | Xp22.31 | STS, HDHD1A | Deletion | TCL9 | FE-PD, SU-DHL-1, MAC1/2A* | 4 | Steroid sulfatase; pseudouridine-5′-monophosphatase |
| 2p23.2 | ALK | 5q35.1 | NPM1 | Translocation | TCL3 | SU-DHL-1, Karpas 299, SR-786 | 4 | Anaplastic lymphoma kinase; nucleophosmin |
| 3q26.32 | TBL1XR1 | 3q28 | TP63 | Inversion | TCL5, TCL29 | — | 2 | Transducin (beta)–like 1 X-linked receptor 1, tumor protein p63 |
| 4p15.1 | None | M† | Unknown | Unknown | TCL5, TCL8, TCL3, TCL56 | — | 4 | — |
| 6p25.3 | DUSP22 | 7q32.3 | FLJ43663 | Translocation | TCL1, TCL8, TCL11 | FE-PD | 4 | Dual-specificity phosphatase 22; near FRAH7 fragile site on 7q32.37 |
| 6q27 | None | 8q22.1 | Unknown | Unknown | TCL56 | Karpas 299 | 2 | — |
| 7q31.31 | None | 7q31.31 | None | Deletion | TCL13, TCL65 | — | 2 | No intervening known genes |
| 8q24.21 | Unknown | 8q24.21 | None | Unknown | TCL15 | MAC1/2A | 2 | — |
| 9p21.3 | MTAP | 9p21.3 | MTAP, CDKN2BAS | Deletion | TCL15, TCL29, TCL32 | — | 3 | Deleted region contains CDKN2A encoding p16INK4A and p19ARF (stabilizes p53)14 |
| 10q21.3 | CTNNA3 | 10q21.3 | CTNNA3 | Deletion | TCL6, TCL56 | — | 2 | Catenin alpha-3 |
| 16q23.1 | WWOX | 16q23.1 | WWOX | Deletion | TCL6, TCL29 | SU-DHL-1, Karpas 299, MAC1/2a̱* | 5 | WW domain-containing oxidoreductase (stabilizes p53)15 |
| 16q24.3 | ANKRD11 | 16q24.3 | SPG7 | Deletion | TCL1, TCL2, TCL9, TCL29 | FE-PD, MAC1/2A* | 6 | Ankyrin repeat domain-containing protein 11 (p53 co-activator)16 ; spastic paraplegia 7 |
| 17p13.1 | TMEM95, POLR2A | 17p13.1 | DNAH2, WRAP53 | Deletion | TCL3 | SR-786 | 2 | Deleted region contains TP53 |
Values shown in bold indicate rearrangement validated by PCR and breakpoint identified by Sanger sequencing.
PTCL indicates peripheral T-cell lymphoma; and –, no data.
Present in either cell line from a single patient.
M indicates mitochondrial genome; the mate pairs for breakpoint 2 may map to PCBD2 on 5q31.1, but homology to that locus did not meet alignment criteria.
We used several complementary approaches to validate rearrangements identified by mate-pair NGS. First, we designed tumor-specific primers from aberrant mate-pair reads and Sanger sequenced the precise breakpoints for 7 of the 13 recurrent rearrangements identified by NGS. This analysis confirmed all 7 of these rearrangements (bold values in Table 1; Figure 2A-B). To validate deletions and to further evaluate copy number variations (CNVs), we developed algorithms to count mate pairs mapping between the breakpoints (Figure 2C). Briefly, a window size was determined based on the overall genomic coverage, the genomic region of interest was divided into windows of this size, and the number of mate pairs mapping within each window was determined. In addition to validating deletions predicted from the mate-pair data, these analyses identified CNVs in these regions in additional cases without aberrant mate pairs (eg, cases with deletions extending to the telomere; supplemental Figure 7, supplemental Tables 4-6). Deletions also were examined using multiplex ligation-dependent probe amplification (MLPA; Figure 2D) or FISH (Figure 2E). Based on the results of these validation assays, at least 1 of the 5 p53-related rearrangements (ie, involving TP53, TP63, CDKN2A, WWOX, or ANKRD11) was present in 14 (67%) of 21 PTCLs (Figure 2F).
Recurrent abnormalities in p53-related genes in peripheral T-cell lymphomas. (A) PCR using tumor-specific primers (supplemental Table 1) designed from aberrant mate-pair sequences from SR-786 indicating juxtaposition of POLR2A and WRAP53, situated on either side of TP53. (B) Partial results of Sanger sequencing (nucleotides in red) of the SR-786 amplicon from panel A spanning the POLR2A/WRAP53 breakpoints. (C) Count plot showing chr17p based on mate-pair data from TCL3, showing TP53 deletion. Each circle represents a 52-kb window (W, based on genomic coverage; see “Mate-pair data mapping and bioinformatic analysis”); the position along the y-axis represents the number of mate pairs mapping within each window. The horizontal red line represents the number of mate pairs expected for a copy-neutral locus based on normalization across the entire genome. The horizontal blue line represents the aberrant mate pairs, which map ∼ 174 kb apart (instead of the expected ∼ 5 kb based on the size selection used for mate-pair library preparation). (D) MLPA (see “Methods”) for ANKRD11 in cases without (TCL5, left) and with (TCL29, right) homozygous deletions. Numerals represent ratios of patient samples (blue peaks) to normal female controls (red peaks) for 2 probes (ANKR1 and ANKR12) to exon 1 of ANKRD11. The remaining gene probes are reference controls. (E) Metaphase CDKN2A/B FISH image from TCL29. Arrows point to centromeres of chromosomes 9 (CEP9; green signals). There is homozygous deletion of CDKN2A/B (absent red signals). For comparison, an interphase nucleus with 2 copies of the CDKN2A/B gene region is seen at bottom right (arrowhead). (F) Summary of abnormalities in p53-related genes in 21 sequenced PTCLs, with status of translocations involving DUSP22-IRF4 or ALK. There were 16 patient samples (“TCL” identifiers) and 6 cell lines (SR-786, Karpas [K] 299, SU-DHL-1, FEPD, MAC1, and MAC2A). MAC1 and MAC2A were considered together as they were derived from the same patient.
Recurrent abnormalities in p53-related genes in peripheral T-cell lymphomas. (A) PCR using tumor-specific primers (supplemental Table 1) designed from aberrant mate-pair sequences from SR-786 indicating juxtaposition of POLR2A and WRAP53, situated on either side of TP53. (B) Partial results of Sanger sequencing (nucleotides in red) of the SR-786 amplicon from panel A spanning the POLR2A/WRAP53 breakpoints. (C) Count plot showing chr17p based on mate-pair data from TCL3, showing TP53 deletion. Each circle represents a 52-kb window (W, based on genomic coverage; see “Mate-pair data mapping and bioinformatic analysis”); the position along the y-axis represents the number of mate pairs mapping within each window. The horizontal red line represents the number of mate pairs expected for a copy-neutral locus based on normalization across the entire genome. The horizontal blue line represents the aberrant mate pairs, which map ∼ 174 kb apart (instead of the expected ∼ 5 kb based on the size selection used for mate-pair library preparation). (D) MLPA (see “Methods”) for ANKRD11 in cases without (TCL5, left) and with (TCL29, right) homozygous deletions. Numerals represent ratios of patient samples (blue peaks) to normal female controls (red peaks) for 2 probes (ANKR1 and ANKR12) to exon 1 of ANKRD11. The remaining gene probes are reference controls. (E) Metaphase CDKN2A/B FISH image from TCL29. Arrows point to centromeres of chromosomes 9 (CEP9; green signals). There is homozygous deletion of CDKN2A/B (absent red signals). For comparison, an interphase nucleus with 2 copies of the CDKN2A/B gene region is seen at bottom right (arrowhead). (F) Summary of abnormalities in p53-related genes in 21 sequenced PTCLs, with status of translocations involving DUSP22-IRF4 or ALK. There were 16 patient samples (“TCL” identifiers) and 6 cell lines (SR-786, Karpas [K] 299, SU-DHL-1, FEPD, MAC1, and MAC2A). MAC1 and MAC2A were considered together as they were derived from the same patient.
TP63 rearrangements in PTCL and DLBCL
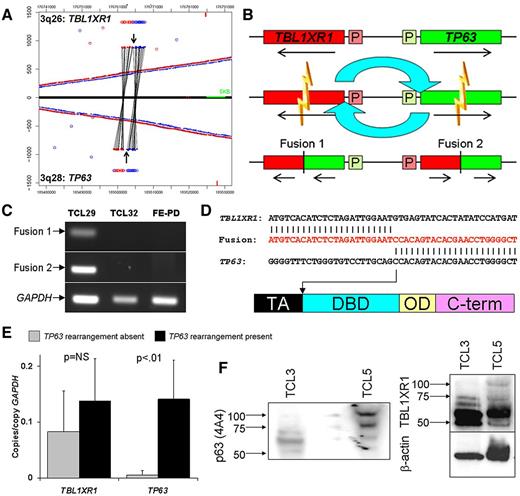
TP63 rearrangements were a novel finding in our study and, to our knowledge, represent the first rearrangement of any TP53 family member. In addition to the 2 cases with inv(3)(q26q28) leading to TBL1XR1/TP63 (Figure 3A), a third case had t(3;6)(q28;p22.3) corresponding to TP63/ATXN1 (supplemental Figures 8, 9D). We studied TBL1XR1/TP63 further at the transcript and protein levels. The TBL1XR1/TP63 breakpoints of the inv(3)(q26q28) leading to (Figure 3A) predicted 2 potential fusion transcripts (Figure 3B). RT-PCR and Sanger sequencing demonstrated that both were present in affected cases (Figure 3C-D). We focused on fusion 2, transcribed from the highly active TBL1XR1 promoter and containing exons 1-7 of TBL1XR1 fused in frame to exons 4-8 or 4-10 of TP63 (depending on C-terminal splicing, not shown).
Validation and characterization of TBL1XR1/TP63 fusion in peripheral T-cell lymphomas. (A) Junction plot of mate-pair data showing inv(3)(q26q28) involving TBL1XR1 and TP63. Arrows indicate breakpoints. (B) Schematic diagram of inv(3)(q26q28) with resultant gene fusions. P indicates promoter region. (C) RT-PCR for fusion transcripts 1 and 2 in cases with (TCL29) and without (TCL32 and FE-PD) inv(3)(q26q28). (D) Partial results of Sanger sequencing (nucleotides in red) of TBL1XR1/TP63 (fusion transcript 2) in TCL29, with TP63 breakpoint (arrow) showing loss of transactivation (TA) domain and retention of DNA-binding domain (DBD), similar to TP63 isoforms encoding dominant-negative ΔNp63. (E) QPCR in 16 PTCL tissue samples shows higher expression of p63 in cases with inv(3)(q26q28) (n = 3) than in cases without this rearrangement (n = 13); TBL1XR1 is expressed more uniformly. (F) Western blot of protein from PTCL tissue samples without (TCL3) and with (TCL5) TBL1XR1/ΔNp63; the fusion protein runs at ∼ 100 kDa.
Validation and characterization of TBL1XR1/TP63 fusion in peripheral T-cell lymphomas. (A) Junction plot of mate-pair data showing inv(3)(q26q28) involving TBL1XR1 and TP63. Arrows indicate breakpoints. (B) Schematic diagram of inv(3)(q26q28) with resultant gene fusions. P indicates promoter region. (C) RT-PCR for fusion transcripts 1 and 2 in cases with (TCL29) and without (TCL32 and FE-PD) inv(3)(q26q28). (D) Partial results of Sanger sequencing (nucleotides in red) of TBL1XR1/TP63 (fusion transcript 2) in TCL29, with TP63 breakpoint (arrow) showing loss of transactivation (TA) domain and retention of DNA-binding domain (DBD), similar to TP63 isoforms encoding dominant-negative ΔNp63. (E) QPCR in 16 PTCL tissue samples shows higher expression of p63 in cases with inv(3)(q26q28) (n = 3) than in cases without this rearrangement (n = 13); TBL1XR1 is expressed more uniformly. (F) Western blot of protein from PTCL tissue samples without (TCL3) and with (TCL5) TBL1XR1/ΔNp63; the fusion protein runs at ∼ 100 kDa.
TP63 isoforms are classified as TAp63 variants that contain a transactivation (TA) domain or as ΔNp63 variants that are truncated at their N termini and lack a fully active TA domain. Fusion 2 encoded the DNA binding and oligomerization domains common to all p63 isoforms, but, similar to ΔNp63 isoforms, lacked the TA domain (Figure 3D). QPCR targeting the DNA-binding domain demonstrated 28-fold higher expression in PTCLs with TP63 rearrangements (P < .01; Figure 3E). No significant difference in TBL1XR1 expression was detected between the 2 groups. The TBL1XR1/ΔNp63 fusion protein was recognized by both TBL1XR1 and p63 antibodies on Western blot, with a molecular mass of ∼ 100 kDa, similar to that predicted by the fusion sequence (Figure 3F).
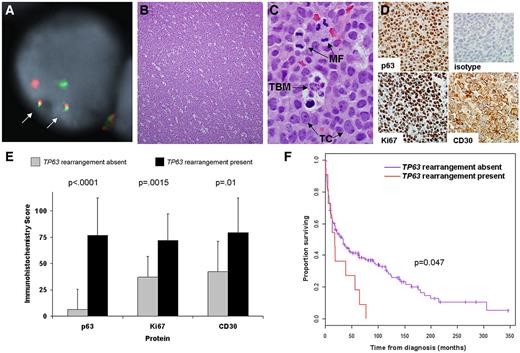
By FISH, 11 (5.8%) 190 PTCL tissue samples had TP63 rearrangements (Figure 4A, supplemental Figure 9). The distribution of these rearrangements by PTCL subtype is shown in Table 2. Rearrangements were seen in PTCL, NOS (9.4%); ALK-negative ALCL (12.5%); and primary cutaneous ALCL (10.5%). They were absent in other PTCL subtypes studied. The partner gene was TBL1XR1 in 7 (63.6%) of the 11 cases with TP63 rearrangements. TP63 rearrangements were present in the initial diagnostic biopsies from 7 patients; biopsies taken at relapse were tested in the remaining 4 rearranged cases as the diagnostic biopsies were not available. Histologically, 7 (63.6%) of the 11 rearranged cases demonstrated a diffuse growth pattern, with sheet-like growth of the tumor cells (Figure 4B). Three of the remaining cases had a patchy or nodular growth pattern, while the fourth had numerous admixed acute inflammatory cells. All 11 rearranged cases demonstrated apoptotic debris, which in 4 cases was accompanied by prominent tingible body macrophages (Figure 4C).
Clinicopathologic findings in peripheral T-cell lymphomas with TP63 rearrangements. (A) Dual-fusion (D-) and/or breakapart FISH probes were used to screen 190 PTCLs for TP63 rearrangements. In this case, D-FISH showed 2 abnormal fusion signals (arrows), corresponding to fusion of TBL1XR1 (green) to TP63 (red). The remaining green and red signals represent the nonrearranged copies of TBL1XR1 and TP63, respectively. (B) Most PTCLs with TP63 rearrangements showed a diffuse, sheet-like growth pattern (hematoxylin and eosin, 100× magnification; image acquired using an Olympus DP71 camera and Olympus BX51 microscope). (C) All rearranged cases demonstrated apoptotic debris, and 4 of 11 cases had prominent tingible body macrophages (TBM) phagocytozing cellular debris (1000×). Mitotic figures (MF) also were present. TC indicates tumor cells. (D) Immunohistochemistry of the tumor shown in panels A through C shows strong, uniform nuclear staining for p63 protein (400×). Virtually all tumor cells express the proliferation marker Ki67, as well as the lymphocyte activation marker, CD30. Top right panel indicates isotype control antibody. (E) Associations between TP63 rearrangement and results of immunohistochemical studies. (F) PTCL patients with TP63 rearrangements (n = 11) had significantly poorer overall survival than those without TP63 rearrangements (n = 179; median survival with and without rearrangements, 17.9 months vs 33.4 months, respectively).
Clinicopathologic findings in peripheral T-cell lymphomas with TP63 rearrangements. (A) Dual-fusion (D-) and/or breakapart FISH probes were used to screen 190 PTCLs for TP63 rearrangements. In this case, D-FISH showed 2 abnormal fusion signals (arrows), corresponding to fusion of TBL1XR1 (green) to TP63 (red). The remaining green and red signals represent the nonrearranged copies of TBL1XR1 and TP63, respectively. (B) Most PTCLs with TP63 rearrangements showed a diffuse, sheet-like growth pattern (hematoxylin and eosin, 100× magnification; image acquired using an Olympus DP71 camera and Olympus BX51 microscope). (C) All rearranged cases demonstrated apoptotic debris, and 4 of 11 cases had prominent tingible body macrophages (TBM) phagocytozing cellular debris (1000×). Mitotic figures (MF) also were present. TC indicates tumor cells. (D) Immunohistochemistry of the tumor shown in panels A through C shows strong, uniform nuclear staining for p63 protein (400×). Virtually all tumor cells express the proliferation marker Ki67, as well as the lymphocyte activation marker, CD30. Top right panel indicates isotype control antibody. (E) Associations between TP63 rearrangement and results of immunohistochemical studies. (F) PTCL patients with TP63 rearrangements (n = 11) had significantly poorer overall survival than those without TP63 rearrangements (n = 179; median survival with and without rearrangements, 17.9 months vs 33.4 months, respectively).
Distribution of TP63 rearrangements by PTCL subtype
| PTCL subtype . | Cases studied, n (%) . | TP63 rearrangement, n/tested (%) . | TBL1XR1 partner, n/tested (%) . |
|---|---|---|---|
| Angioimmunoblastic TCL | 38 (20.0) | 0/38 (0.0) | — (—) |
| PTCL, NOS | 53 (27.9) | 5/53 (9.4) | 3/5 (60.0) |
| ALCL, ALK positive | 21 (11.1) | 0/21 (0.0) | — (—) |
| ALCL, ALK negative | 32 (16.8) | 4/32 (12.5) | 3/4 (75.0) |
| ALCL, primary cutaneous | 19 (10.0) | 2/19 (10.5) | 1/2 (50.0) |
| Mycosis fungoides* | 7 (3.9) | 0/7 (0.0) | — (—) |
| Enteropathy-associated TCL | 4 (2.1) | 0/4 (0.0) | — (—) |
| Hepatosplenic TCL | 1 (0.5) | 0/1 (0.0) | — (—) |
| SCPTCL | 1 (0.5) | 0/1 (0.0) | — (—) |
| LGL | 5 (2.6) | 0/5 (0.0) | — (—) |
| NKTL | 9 (4.7) | 0/9 (0.0) | — (—) |
| Total | 190 (100.0) | 11/190 (5.8) | 7/11 (63.6) |
| PTCL subtype . | Cases studied, n (%) . | TP63 rearrangement, n/tested (%) . | TBL1XR1 partner, n/tested (%) . |
|---|---|---|---|
| Angioimmunoblastic TCL | 38 (20.0) | 0/38 (0.0) | — (—) |
| PTCL, NOS | 53 (27.9) | 5/53 (9.4) | 3/5 (60.0) |
| ALCL, ALK positive | 21 (11.1) | 0/21 (0.0) | — (—) |
| ALCL, ALK negative | 32 (16.8) | 4/32 (12.5) | 3/4 (75.0) |
| ALCL, primary cutaneous | 19 (10.0) | 2/19 (10.5) | 1/2 (50.0) |
| Mycosis fungoides* | 7 (3.9) | 0/7 (0.0) | — (—) |
| Enteropathy-associated TCL | 4 (2.1) | 0/4 (0.0) | — (—) |
| Hepatosplenic TCL | 1 (0.5) | 0/1 (0.0) | — (—) |
| SCPTCL | 1 (0.5) | 0/1 (0.0) | — (—) |
| LGL | 5 (2.6) | 0/5 (0.0) | — (—) |
| NKTL | 9 (4.7) | 0/9 (0.0) | — (—) |
| Total | 190 (100.0) | 11/190 (5.8) | 7/11 (63.6) |
TCL indicates T-cell lymphoma; PTCL, peripheral TCL; NOS, not otherwise specified; ALCL, anaplastic large-cell lymphoma; ALK, anaplastic lymphoma kinase; SCPTCL, subcutaneous panniculitis-like TCL; LGL, T-cell large granular lymphocytic leukemia; —, not applicable; and NKTL, extranodal NK/TCL, nasal type.
Including 2 cases with large-cell transformation.
We used immunohistochemistry to evaluate cases with and without TP63 rearrangements for expression of p63 (n = 188), the proliferation marker Ki67 (n = 162), and CD30 (an adverse prognostic factor in PTCL, NOS2 ; n = 170; Figure 4D-E). Mean immunohistochemistry scores (± SDs) for p63 in PTCLs with (n = 11) and without (n = 177) TP63 rearrangements were 76.8 ± 35.2 and 6.4 ± 19.1, respectively (P < .0001). The scores for the 11 rearranged cases were 0, 15, 80, 80, 90, 90, 90, 100, 100, 100, and 100. Interestingly, the cases with scores of 0 and 15 were PTCLs, NOS with non-TBL1XR1 partners. Mean scores for Ki67 in PTCLs with (n = 11) and without (n = 151) TP63 rearrangements were 72.0 ± 32.9 and 37.0 ± 28.8, respectively (P = .0015). Mean scores for CD30 in PTCLs with (n = 11) and without (n = 159) TP63 rearrangements were 79.6 ± 32.8 and 42.3 ± 46.8, respectively (P = .01). No case with TP63 rearrangement expressed ALK.
Patients with PTCLs carrying TP63 rearrangements had significantly inferior overall survival compared with patients without TP63 rearrangements (median survival: 17.9 months vs 33.4 months, respectively; P = .047; Figure 4F). Because p63 protein is expressed in approximately one-third of DLBCLs,17 we examined these tumors by FISH and found 2 (1.2%) of 164 DLBCLs had TP63 rearrangements (Figure 5A-D). The SnowShoes-FTD algorithm did not detect TP63 fusions in transcriptome sequencing data from carcinomas of the breast, lung, or head and neck, nor in transcriptome data from normal samples (Figure 5E).
TP63 rearrangements in DLBCLs. (A) DLBCL with TP63 rearrangement (hematoxylin and eosin, 400×). (B) Immunohistochemistry for the B-cell antigen, CD20 (400×). (C) Tumor cells showed strong nuclear expression of p63 protein (400×; inset, isotype control). (D) Dual-fusion FISH showed 2 fusion signals (arrows), corresponding to TBL1XR1/TP63 fusion. (E) TP63 rearrangements were seen in 1.2% of DLBCLs by FISH. The SnowShoes fusion detection algorithm did not identify TP63 fusions in transcriptome data from carcinomas (ca) of the breast, lung, or head and neck (H&N), nor in data from normal controls.
TP63 rearrangements in DLBCLs. (A) DLBCL with TP63 rearrangement (hematoxylin and eosin, 400×). (B) Immunohistochemistry for the B-cell antigen, CD20 (400×). (C) Tumor cells showed strong nuclear expression of p63 protein (400×; inset, isotype control). (D) Dual-fusion FISH showed 2 fusion signals (arrows), corresponding to TBL1XR1/TP63 fusion. (E) TP63 rearrangements were seen in 1.2% of DLBCLs by FISH. The SnowShoes fusion detection algorithm did not identify TP63 fusions in transcriptome data from carcinomas (ca) of the breast, lung, or head and neck (H&N), nor in data from normal controls.
Discussion
Our genome-wide analysis of PTCLs using the mate-pair approach identified structural rearrangements of at least 1 of 5 p53-related genes in 67% of PTCLs analyzed. We elected to focus on TP63 rearrangements based on the novelty of these events and the finding that they resulted in fusion proteins homologous to ΔNp63, a group of p63 isoforms with known oncogenic function. p63 is 1 of 3 members of the p53 family, which includes p53, p63, and p73. The p53 pathway is commonly deregulated in human neoplasia, most commonly by TP53 mutation, which may be seen in up to 50% of cancers.18 Surprisingly, despite in vitro and clinical evidence indicating the importance of p53 in PTCL, TP53 mutations are relatively rare in this disease.19-21 We found TP53 mutations in 4 of 21 PTCLs (and only 1 of 16 PTCL tissue samples; Figure 2F, supplemental Table 7). Furthermore, mutations of TP63 rarely are seen in any human cancer.22 Taken together, these findings suggest that structural abnormalities of TP63 and other p53-related genes might represent an alternate means of deregulating the p53 pathway in PTCLs.
ΔNp63 is overexpressed and associated with oncogenic function in various human malignancies, including carcinomas of the breast, lung, and head and neck.9,23-26 This is in contrast to the tumor suppressor activity of full-length TP63 splice variants retaining the N-terminal TA domain, which promote apoptosis, cell-cycle arrest, and cell senescence.27-29 The best-characterized mechanism for the oncogenic effects of ΔNp63 is its dominant-negative function,27,30,31 including the ability to sequester TA domain-containing p53 family members or compete for binding to transcriptional targets of these proteins.24,32-34 ΔNp63 also may function through direct transcriptional activity distinct from that of TAp63 or by associating with heterogeneous nuclear ribonucleoproteins to modulate pre-mRNA processing.35-37 Finally, a role for ΔNp63 in the regulation of stem cell dynamics recently has been proposed.38,39
TP63 rearrangements were found exclusively in 3 PTCL subtypes: PTCL, NOS; ALK-negative ALCL; and primary cutaneous ALCL. Of 5 PTCLs, NOS with TP63 rearrangements, 4 (80%) had CD30 staining in ≥ 80% of cells (the threshold used by the International Peripheral T-cell Lymphoma Project, which found 5-year overall survival rates of only 19% in CD30-positive PTCLs, NOS.2 ) In contrast, CD30 staining in ≥ 80% of cells was seen in only 14 (30.4%) of 46 of PTCLs, NOS tested without TP63 rearrangements. ALCLs express CD30 by definition.3 TP63 rearrangements were not seen in ALK-positive ALCL and thus are mutually exclusive with ALK rearrangements in the cases studied thus far. A 6p25.3 rearrangement was detected by FISH in 1 of 11 cases with TP63 rearrangements (a primary cutaneous ALCL), and thus these 2 events are not mutually exclusive.
ALK-negative ALCL has a prognosis poorer than ALK-positive ALCL but better than CD30-positive PTCL, NOS, while primary cutaneous ALCL typically has an excellent prognosis (5-year overall survival of 90%).2 Interestingly, both patients with primary cutaneous ALCLs bearing TP63 rearrangements had aggressive cutaneous disease refractory to multiple therapy regimens and died 56 and 64 months after diagnosis, respectively. It is possible that testing for TP63 rearrangements could identify patients with ALK-negative or primary cutaneous ALCL at higher risk for adverse outcomes. Because TP63 rearrangements were associated with p63 protein expression, we also investigated whether immunohistochemical staining for p63 was associated with prognosis; however, no such association was found. This lack of association likely is because of the presence of p63 protein expression in a subset of PTCLs without TP63 rearrangements, and the inability of immunohistochemistry to distinguish between p63 fusion proteins and wild-type p63.
It is not clear whether TP63 rearrangements are primary or secondary events during lymphomagenesis. Among B-cell non-Hodgkin lymphomas, TP53 mutations have been associated with both clinical progression and histologic transformation.40 However, TP63 rearrangements were seen in the initial diagnostic biopsy specimens in all 7 rearranged PTCLs for which the initial biopsy was available. Furthermore, well-defined criteria for transformation of most PTCLs have not been established. Thus, determining when during lymphomagenesis these rearrangements occur will require additional study. It also is apparent that TP63 rearrangements are not specific to PTCLs. Since the original submission of this manuscript, Scott et al also reported TBL1XR1/TP63 fusions in B-cell non-Hodgkin lymphomas (DLBCL and follicular lymphoma).41 We did not find evidence of TP63 fusions in carcinoma types associated with expression of ΔNp63, including breast, lung, and head and neck cancers. Because the incidence of TP53 mutations is lower in both B-cell non-Hodgkin lymphomas and PTCLs than in solid tumors,40 these observations raise the possibility that TP63 rearrangements are relatively specific for non-Hodgkin lymphomas and perhaps other tumors in which TP53 mutations are rare. Although the sample size was limited, the fact that TP63 rearrangements and TP53 mutations were mutually exclusive in our mate-pair sequencing study set provides some preliminary evidence for this hypothesis.
In addition to rearrangements directly affecting TP63 and TP53, we identified recurrent deletions of CDKN2A, ANKRD11, and WWOX in PTCLs. CDKN2A is a tumor suppressor gene encoding p16INK4A and p19ARF, the latter of which stabilizes p53 to promote senescence and apoptosis.14 CDKN2A is deleted in numerous cancers, including PTCLs.42,43 ANKRD11 encodes the tumor suppressor, ANR11, a p53 coactivator that can restore p53 transcriptional activity in the presence of TP53 mutations.16,44,45 WWOX encodes the tumor suppressor, WW domain-containing oxidoreductase, which binds to and stabilizes p53 during cellular stress, thus promoting apoptosis.15 WWOX loss of heterozygosity occurs in numerous malignancies, and may be associated with adverse clinical outcomes.46 Further corroborating our findings, TP53, ANKRD11, and WWOX were found to reside within recurrently deleted regions in a recent comparative genomic hybridization study of cutaneous ALCLs.47 A more comprehensive characterization of these p53-related deletion events in a larger sample set will be necessary to assess the interplay between these deletions and TP63 rearrangements. It is possible that the mechanisms of p53 pathway deregulation in PTCLs are more heterogeneous than in cancers in which TP53 mutations are common, and that the relative frequency of each genetic mechanism is lower. This might explain the fact that TP63 rearrangements were relatively infrequent in PTCLs (5.8%), although 67% of cases in the mate-pair study set had at least 1 of the 5 p53-related rearrangements identified. Although this study set reflects cell lines as well as tissue samples, including cell lines in our analysis, it is unlikely to affect the relevance of our findings. Of the p53-related abnormalities studied, only TP53 mutations (ie, not the rearrangements) were substantially more common in PTCL cell lines (3 of 5) than in patient tissue samples (1 of 16); it is unclear whether this reflects greater success developing cell lines from cases with TP53 mutations or acquisition of these mutations in vitro.
The current study represents the first report of genome-wide NGS of PTCLs. In contrast to our previous approach, which characterized a rearrangement in which 1 partner locus was known,7 the present study identified previously unknown events using an enhanced bioinformatic algorithm that could query the entire reference genome in RAM simultaneously. The finding of multiple structural abnormalities affecting p53-related genes may explain the apparent paradox of rare TP53 mutations in these tumors despite the apparent importance of inactivating the p53 pathway. TP63 rearrangements may represent a novel mechanism deregulating this pathway in lymphomas. ΔNp63 has been proposed as a therapeutic target and various means to down-regulate expression or increase degradation of ΔNp63 have been described, including drugs such as cisplatin and poly(ADP-ribose) polymerase inhibitors.23,27,48 Such approaches might be applicable to PTCLs and DLBCLs expressing ΔNp63 fusion proteins. Small molecules also might be designed to inhibit the interaction of ΔNp63 fusion proteins with other p53 family members. Finally, the finding that TP63 rearrangements are associated with specific microscopic, phenotypic, and clinical features among PTCLs illustrates the power of NGS to refine molecular classification systems that serve as the basis for individualized cancer therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank R. Sikkink for technical support and S. Kaufmann for valuable comments on the manuscript.
This work was supported by a Waterman Biomarker Discovery grant from the Mayo Clinic Center for Individualized Medicine, and an Experimental Pathology Development Award from the Mayo Clinic Department of Laboratory Medicine and Pathology. A.L.F. is a Damon Runyon Clinical Investigator supported by the Damon Runyon Cancer Research Foundation (CI-48-09).
Authorship
Contribution: G.V. designed bioinformatic algorithms, analyzed data, and wrote the manuscript; S.H.J. and Y.W.A. performed bioinformatic analysis; R.A.K., L.A.F., D.M., J.C.P. and I.V.K. performed experiments and analyzed data; R.P.K., D.S.V., S.K.G., S.M.A., and A.D. designed experiments and analyzed data; E.B., R.F., M.E.L., N.S.K., and N.Ö. analyzed data; B.W.E. designed and performed experiments; E.A.T. provided data; M.E.K. provided critical reagents; D.I.S. designed experiments and provided data; and A.L.F. designed and performed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests. Mayo Clinic has filed a provisional patent application for technology associated with this research.
Correspondence: Andrew L. Feldman, MD, Department of Laboratory Medicine and Pathology, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail:feldman.andrew@mayo.edu.

![Figure 2. Recurrent abnormalities in p53-related genes in peripheral T-cell lymphomas. (A) PCR using tumor-specific primers (supplemental Table 1) designed from aberrant mate-pair sequences from SR-786 indicating juxtaposition of POLR2A and WRAP53, situated on either side of TP53. (B) Partial results of Sanger sequencing (nucleotides in red) of the SR-786 amplicon from panel A spanning the POLR2A/WRAP53 breakpoints. (C) Count plot showing chr17p based on mate-pair data from TCL3, showing TP53 deletion. Each circle represents a 52-kb window (W, based on genomic coverage; see “Mate-pair data mapping and bioinformatic analysis”); the position along the y-axis represents the number of mate pairs mapping within each window. The horizontal red line represents the number of mate pairs expected for a copy-neutral locus based on normalization across the entire genome. The horizontal blue line represents the aberrant mate pairs, which map ∼ 174 kb apart (instead of the expected ∼ 5 kb based on the size selection used for mate-pair library preparation). (D) MLPA (see “Methods”) for ANKRD11 in cases without (TCL5, left) and with (TCL29, right) homozygous deletions. Numerals represent ratios of patient samples (blue peaks) to normal female controls (red peaks) for 2 probes (ANKR1 and ANKR12) to exon 1 of ANKRD11. The remaining gene probes are reference controls. (E) Metaphase CDKN2A/B FISH image from TCL29. Arrows point to centromeres of chromosomes 9 (CEP9; green signals). There is homozygous deletion of CDKN2A/B (absent red signals). For comparison, an interphase nucleus with 2 copies of the CDKN2A/B gene region is seen at bottom right (arrowhead). (F) Summary of abnormalities in p53-related genes in 21 sequenced PTCLs, with status of translocations involving DUSP22-IRF4 or ALK. There were 16 patient samples (“TCL” identifiers) and 6 cell lines (SR-786, Karpas [K] 299, SU-DHL-1, FEPD, MAC1, and MAC2A). MAC1 and MAC2A were considered together as they were derived from the same patient.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/11/10.1182_blood-2012-03-419937/4/m_zh89991296500002.jpeg?Expires=1765900705&Signature=g3QsneOvCTzLUR61k40oH57Xd0LQYkt9l5mmzvkWjmmxDqHGIHAKqnSQ03QbTPZLBsd83w8ZCTfnF~536so19C5eV9VtRD8MPueKU5eNXd51QLT20gYVeEyIgPW77jieBzOc6njndauQnrVnbgVlYOEYMO76bhvnBHsmNCP2pxGSZGa~xEs2bZEM7cEJp63Oustd9dIWxkSVgJM2R76f9iCnoISTU0noSrLLOPG~rG8ejg-tC1jys2smb~snHJGjEMf-f8SgPZWQli4WN-LKLe6uPUxMjfyASJB33olzPbBh8DFzfyZ26xjmbQgrxfIhl3O0d9CM8Z~-c19a2sXPQQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal