Abstract
The clinical course of patients with chronic lymphocytic leukemia (CLL) is heterogeneous. Several prognostic factors have been identified that can stratify patients into groups that differ in their relative tendency for disease progression and/or survival. Here, we pursued a subnetwork-based analysis of gene expression profiles to discriminate between groups of patients with disparate risks for CLL progression. From an initial cohort of 130 patients, we identified 38 prognostic subnetworks that could predict the relative risk for disease progression requiring therapy from the time of sample collection, more accurately than established markers. The prognostic power of these subnetworks then was validated on 2 other cohorts of patients. We noted reduced divergence in gene expression between leukemia cells of CLL patients classified at diagnosis with aggressive versus indolent disease over time. The predictive subnetworks vary in levels of expression over time but exhibit increased similarity at later time points before therapy, suggesting that degenerate pathways apparently converge into common pathways that are associated with disease progression. As such, these results have implications for understanding cancer evolution and for the development of novel treatment strategies for patients with CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by accumulation of monoclonal B cells in the blood, marrow, and secondary lymphoid tissues. The clinical course of patients with CLL is highly variable. Some patients are free of symptoms for many years, during which time treatment is typically not necessary. For others the disease is relatively aggressive and requires therapy soon after diagnosis. Because standard therapies are associated with potential morbidity and are not considered curative, current recommendations are to withhold treatment until the patient manifests disease-related complications or clear evidence of disease progression.1
Several prognostic markers have been defined that can identify patients with poor prognosis at early stages of the disease. For example, patients segregate into 2 major subgroups based on whether their leukemia cells express immunoglobulin heavy chain variable region (IGHV) genes that have incurred somatic mutations.2 Patients with CLL cells that express IGHV lacking mutations generally have more aggressive disease than patients with CLL cells that express IGHV genes that have incurred somatic mutations.3,4 Similarly, patients that have CLL B cells that express high levels of CD38 or the ζ-associated protein of 70 kDa (ZAP-70) progress on average more rapidly than those with CLL cells that have low or undetectable levels of these proteins.4,5
For many cancers, an increasing number of prognostic markers have been identified through analysis of genome-wide expression profiles.6 Marker sets are selected by scoring each individual gene for how well its expression level discriminates between different classes of disease. Several microarray studies have reported sets of genes that are useful as surrogate markers for known prognostic factors in CLL, such as the IGHV mutational status.7-9 Other studies have instead correlated gene expression levels directly with median time of patient survival or progression-free survival.10,11
Despite their promise, expression-based biomarkers continue to face serious challenges because of their variable accuracy for predicting patient outcomes.12 Furthermore, the marker sets identified by different research groups often share few genes in common. Two landmark studies, Rosenwald and colleagues7 and Klein and colleagues,8 each identified approximately 100 genes that were expressed differentially by CLL cells that use mutated versus unmutated IGHV genes. However, only 4 marker genes were identified in common between these studies. This discrepancy may reflect the subtle nature of the changes in expression of the relatively few genes governing disease progression compared with that of the downstream effectors, which can vary considerably from patient to patient.13
As an alternative approach for identifying disease markers, several groups have integrated gene expression measurements over sets of genes that encode proteins known to interact within protein subnetworks or pathway databases.14-21 Such prognostic profiles are not listings of individual genes or proteins, but the aggregate expression of subnetworks of genes or proteins within a vast interaction network. These subnetworks can identify gene expression differences between different populations of patients that account for their diverse clinical behavior and—unlike conventional analysis—the roles of these genes in disease are interpretable in the context of networks and pathways.
Here, we pursued a subnetwork-based analysis of gene expression profiles to discriminate between groups of patients with disparate risks for CLL progression. The clinical characterization of patients, blood sample preparation, and microarray processing all follow the unified protocol implemented by the Microarray Innovations in LEukemia (MILE) program,22,23 which proposed standards for microarray-based assays in the diagnosis and subclassification of leukemia. Unlike conventional prognostic algorithms using known factors or gene markers, we make no assumptions about the time of oncogenesis. From these analyses, we identified subnetwork signatures that changed over time, converging on a high-risk profile associated with progressive disease requiring therapy.
Methods
Gene expression profiling of peripheral blood from CLL patients
Leukemia cells were isolated from blood samples of CLL patients enrolled in the MILE study who had not received prior therapy for CLL, as per the MILE protocol.22 Expression data were gathered from samples found to have a CLL cell population with greater than 90% CD5+CD19+, as accessed via flow cytometry. Total RNA was isolated and hybridized to Affymetrix HG-U133+2 GeneChips. Of the total of 20 606 genes represented on the microarray, a total of 15 348 had expression levels that were reliably detected in at least 8 patients (5% of the University of California, San Diego cohort [UC San Diego cohort]). For details on microarray hybridization and data processing, see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus24 and are accessible through GEO Series accession no. GSE39671 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39671).
Scoring, searching, and pruning subnetworks
A subnetwork is defined as a gene set that induces a single connected component in the protein interaction network. Given a particular subnetwork M, let a represent its vector of activity scores over the patients, and let T represent the corresponding vector of treatment-free survival (time from sample collection to treatment [SC→TX]). To derive a, expression values gij are normalized to z-transformed scores zij which for each gene i have μ = 0 and σ = 1 over all samples j (supplemental Figure 3). The individual zij of each member gene in the subnetwork are averaged into a combined z score, which is designated the activity aj. The predictive score S(M) is an estimation of the statistical significance of a as the sole predictor variable on a patient's treatment need in a Cox proportional hazard model on T:H(t)/H0(t) = eka, where H(t) is the hazard function at time t and H0(t) is the baseline hazard for an individual when the value of a equals zero. S(M) is defined as −log P value of a χ2 test on the above model of the hazard over a null model of only the baseline hazard. Given the predictive score function S, a greedy search is performed to identify subnetworks within the protein interaction network for which the scores are locally maximal. Candidate subnetworks are seeded with a single protein and iteratively expanded, with every protein serving as a seed in a separate search. At each iteration, the search considers addition of a protein from the neighbors of proteins in the current subnetwork. The addition that yields the maximal score increase is adopted. After each addition, the search considers deletion of each protein from the current subnetwork (except those proteins essential to subnetwork connectivity), and deletions that yield higher score are accepted. The search ends when no addition or deletion increases the score over a specified improvement rate r. The parameter r is chosen as 0.1 to avoid overfitting to the expression data used. To assess the significance of the identified subnetworks, 3 tests of significance are performed. In this study, significant subnetworks are selected that satisfy all 3 tests with P1 < .05, P2 < .05, and P3 < .00005. For details on estimation of a null distribution of S by permuting the network and expression data as well as mergence of overlapped subnetworks, see supplemental Methods.
Prognosis evaluation
Given a set of subnetwork markers, patient samples in a training set are clustered into 2 subgroups by a 2-means clustering method based on similarity in activity. The 2 clusters of the training samples are labeled as low- or high-risk groups according to their treatment-free survival curves in a Kaplan-Meier analysis. A nearest shrunken centroid classifier25 is then trained on the subnetwork activity matrix (significant subnetworks vs patient samples) with the risk labels learned from the clustering analysis. For a new patient of unknown prognosis, the expression profile is first transformed into a subnetwork activity profile in the same way as for the training samples. The nearest shrunken centroid classifier assigns the new activity profile to 1 of the 2 risk groups whose shrunken mean activity of subnetworks over training samples is more similar to the activity of the new sample. For gene markers, similar risk stratification and outcome prediction procedures are performed on the original gene expression matrix.
Subnetwork and gene markers were evaluated using 2 approaches: (1) cross-validation within the UC San Diego cohort and (2) an independent validation using this cohort as training data and the European cohort and the cohort in Friedman and colleagues.26 In the cross-validation, one-fifth of the UC San Diego samples were designated as “test” data and withheld during risk-group assignment and classifier training. Subnetwork markers and top gene markers were identified using only the training data. Each of the 5 patient subsets in the UC San Diego cohort was evaluated in turn as the test set, while training on the other 4 sets. The risk-group predictions among the 5 test sets were pooled to plot 2 treatment-free survival curves in a Kaplan-Meier analysis. In the second validation approach, subnetwork markers or top gene markers were selected using the whole dataset. Patients in the independent cohorts were assigned to 1 of the 2 risk groups by the classifier learned from the UC San Diego cohort. In both validation schemes, a log-rank test was used to estimate the significance level of the difference between the survival curves in a Kaplan-Meier analysis.
For details on real-time PCR for serial gene expression and flow cytometry and immunoblot analyses for protein expression, see supplemental Methods.
Results
Gene expression profiling of peripheral blood from CLL patients
We profiled genome-wide mRNA expression of leukemia cell samples of 130 CLL patients registered at the University of California San Diego Moores Cancer Center (referred to as the UC San Diego cohort; see “Gene expression profiling of peripheral blood from CLL patients”). Lymphocytes were isolated from the blood samples of patients who had not received treatment before sample acquisition. We also examined an independent cohort of 17 patients from European sites (Rome and Munich) in the MILE study (referred to as the European cohort) who had leukemia cell gene expression profiles that were obtained using the same protocol as that used with the UC San Diego cohort.
Protein subnetworks stratify CLL patients into different risk groups
Figure 1 shows the overall process of subnetwork-based disease prognostics, which involves the integration of expression levels of genes encoding proteins known to interact with one another (Figure 1A), and then clustering the samples into subgroups based on their relative subnetwork activities (Figure 1B). Kaplan-Meier survival analysis then is used to assign low-risk and high-risk labels to each subgroup (Figure 1C). To define human protein interaction subnetworks, we assembled a pooled dataset comprising 45 526 experimentally validated interactions among 9800 human proteins, integrated from yeast 2-hybrid experiments27,28 and assessed from the literature for both protein-protein and protein-DNA binding29-34 (see supplemental Methods for details). Of the 15 348 genes reliably detected in CLL, a total of 7589 encoded proteins were covered in this compiled interaction network. We integrated the expression values of each gene encoding a protein in the interaction network, allowing us to consider sets of genes whose aggregate expression profiles defined a subnetwork “activity” score (see “Scoring, searching, and pruning subnetworks”).
Schematic overview of subnetwork identification and definition of risk groups. (A) The expression profile of each gene is projected onto its corresponding protein in a protein-protein interaction subnetwork. A greedy search is performed to find subnetworks for which the activities are associated with the time from sample collection to treatment (SC→TX). Significant subnetworks are selected based on null distributions estimated from permuted data. Subnetworks are used to identify disease genes, and the subnetwork activity is used to characterize the signatures of different risk groups. (B) K-means clustering segregates patients by their distinct subnetwork activity patterns. (C) Patient clusters are assigned high versus low risk based on median treatment-free probabilities in a Kaplan-Meier analysis.
Schematic overview of subnetwork identification and definition of risk groups. (A) The expression profile of each gene is projected onto its corresponding protein in a protein-protein interaction subnetwork. A greedy search is performed to find subnetworks for which the activities are associated with the time from sample collection to treatment (SC→TX). Significant subnetworks are selected based on null distributions estimated from permuted data. Subnetworks are used to identify disease genes, and the subnetwork activity is used to characterize the signatures of different risk groups. (B) K-means clustering segregates patients by their distinct subnetwork activity patterns. (C) Patient clusters are assigned high versus low risk based on median treatment-free probabilities in a Kaplan-Meier analysis.
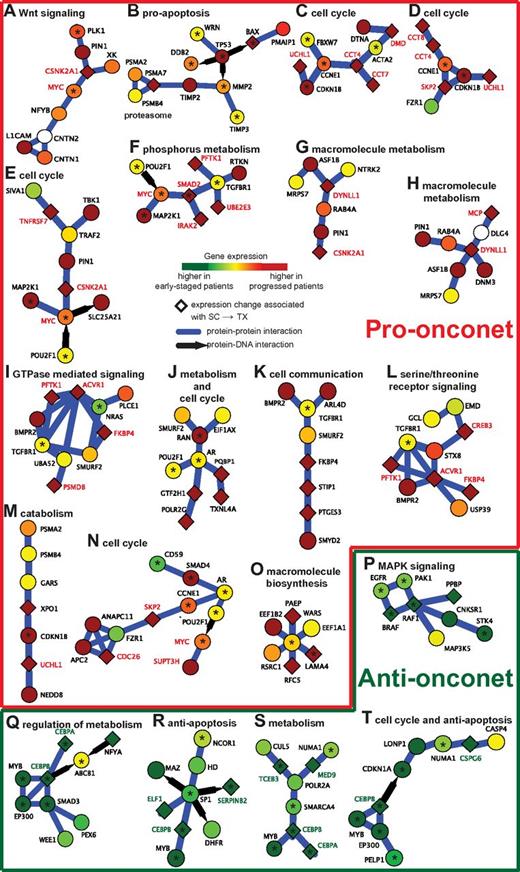
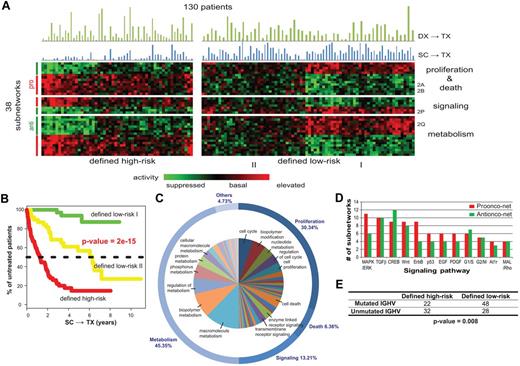
Using this framework, we searched for subnetworks whose activity scores across the 130 patients in the UC San Diego cohort were associated with the treatment-free survival from the time of sample collection (abbreviated as SC→TX). We identified 38 prognostic subnetworks that satisfied 3 separate tests for statistical significance, covering a total of 230 genes (see supplemental Methods). The prognostic subnetworks included proteins involved in WNT signaling35 (Figure 2A), sensitivity to apoptosis36 (Figure 2B), cell division (Figure 2C-E,J,N,T), cell-cell communication (Figure 2K), receptor signaling (Figure 2L,P), resistance to apoptosis (Figure 2R,T), or cell metabolism37 (Figure 2J,Q,S), all of which are known or potential factors in CLL pathogenesis. Clustering of the patients by subnetwork activity resulted in 1 cluster of 54 patients for which the median treatment-free survival was low and a second cluster of 76 patients for which the median SC→TX was substantially longer (Figure 3A). Interestingly, we found that the low-risk group could be divided further into 2 clear subgroups, designated low-risk I and II, with very different subnetwork activity profiles (Figure 3A). The low-risk I patients, whose subnetwork profiles were almost perfectly anticorrelated with those of the high-risk patients, were also associated with longest treatment-free survival SC→TX (Figure 3B).
Example subnetworks of CLL disease progression enriched for the hallmarks of cancer. (A-O) Pro-onconets. (P-T) Anti-onconets. Nodes and links represent human proteins and protein physical interactions, respectively. Blue links indicate protein-protein interactions; black arrows indicate protein-DNA binding. The color of each node scales with the change in gene expression in patients of shorter treatment-free survival intervals versus longer: red represents up-regulation in patients of shorter intervals whereas green represents down-regulation. The predominant cellular functions are indicated next to each subnetwork. Known cancer susceptibility genes are highlighted by a black asterisk. Names of genes marked in red/green are further probed for serial expression in an additional patient cohort (red indicates genes expressed in pro-onconets and green indicates those expressed in anti-onconets).
Example subnetworks of CLL disease progression enriched for the hallmarks of cancer. (A-O) Pro-onconets. (P-T) Anti-onconets. Nodes and links represent human proteins and protein physical interactions, respectively. Blue links indicate protein-protein interactions; black arrows indicate protein-DNA binding. The color of each node scales with the change in gene expression in patients of shorter treatment-free survival intervals versus longer: red represents up-regulation in patients of shorter intervals whereas green represents down-regulation. The predominant cellular functions are indicated next to each subnetwork. Known cancer susceptibility genes are highlighted by a black asterisk. Names of genes marked in red/green are further probed for serial expression in an additional patient cohort (red indicates genes expressed in pro-onconets and green indicates those expressed in anti-onconets).
Subnetwork signatures of CLL disease progression. (A) Activity of the 38 significant subnetworks (rows) across the 130 patients (columns). The color of each block scales with the activity level of a subnetwork in a particular patient. Patients are clustered into high/low-risk groups, and subnetworks are clustered into 3 functional categories (proliferation and death, signaling, and metabolism). Blue bars above the heatmap show the intervals of SC→TX for each sample while green bars chart the intervals of DX→TX. (B) Kaplan-Meier analysis yields treatment-free probabilities with regard to the 3 risk groups defined by subnetwork activity patterns. (C) Distribution of the predominant cellular functions associated with the 38 subnetworks. Related functions are clustered into categories named on the outer circle. The marked functions in the inner circle are associated with at least 2% of the subnetworks. See supplemental Figure 4 for all enriched functions. (D) Top enriched signaling cascades. Bars show numbers of the 38 subnetworks, which have member genes involved in each pathway. (E) Comparison of patient stratification by subnetwork prognosis versus IGHV mutation status.
Subnetwork signatures of CLL disease progression. (A) Activity of the 38 significant subnetworks (rows) across the 130 patients (columns). The color of each block scales with the activity level of a subnetwork in a particular patient. Patients are clustered into high/low-risk groups, and subnetworks are clustered into 3 functional categories (proliferation and death, signaling, and metabolism). Blue bars above the heatmap show the intervals of SC→TX for each sample while green bars chart the intervals of DX→TX. (B) Kaplan-Meier analysis yields treatment-free probabilities with regard to the 3 risk groups defined by subnetwork activity patterns. (C) Distribution of the predominant cellular functions associated with the 38 subnetworks. Related functions are clustered into categories named on the outer circle. The marked functions in the inner circle are associated with at least 2% of the subnetworks. See supplemental Figure 4 for all enriched functions. (D) Top enriched signaling cascades. Bars show numbers of the 38 subnetworks, which have member genes involved in each pathway. (E) Comparison of patient stratification by subnetwork prognosis versus IGHV mutation status.
Twenty-two of the 38 significant subnetworks had increased activity in the defined high-risk group (referred to as pro-onconets; eg, Figure 2A-O), whereas the other 16 had decreased activity (referred to as anti-onconets; eg, Figure 2P-T). Among the protein functions significantly enriched within the 38 subnetworks, the majority related to cell metabolism (45.4%), cell survival/proliferation/death (36.7%), and cell-signal transduction (13.2%, Figure 3C). Several key signaling proteins implicated in CLL, such as those encoded by MAPK/ERK, TGFβ, CREB, or WNT, were involved in regulation of multiple subnetworks (Figure 3D, supplemental Methods).
Predicting the time of therapy from the date of sample collection
We next explored the power of the subnetwork markers to predict the risk for requiring imminent therapy. For this purpose, a patient's average gene expression level was calculated for each of the 38 subnetworks; the list of 38 average levels was designated as the patient's subnetwork profile. This profile was predicted as “high risk” if it correlated with the average subnetwork profiles of the high-risk group better than those of the low-risk group. Conversely, the patient subnetwork profile was predicted as “low risk” if it better correlated with the average subnetwork profiles of the low-risk group.
Cross-validation within the UC San Diego cohort (see “Prognosis evaluation”) showed excellent predictive performance (P = 3.5 × 10−6; black lines in Figure 4A). A similar cross-validation procedure was applied using individual gene expression markers instead of subnetworks. Although these gene-based markers also held prognostic value (P = 5.24 × 10−4; gray lines in Figure 4A), they were significantly less robust than the network-based approach in predicting risk for disease requiring therapy. Both prognostics compared favorably with either the IGHV mutation status (P = .01) or those reported in previous microarray studies (the red bars in Figure 5D).
Use of expression levels of genes versus subnetworks to stratify patient samples. (A) Five-fold cross-validation on the 130 patients evaluated at UC San Diego. Survival analyses on SC→TX are shown for both the low- (dashed lines) and high- (solid lines) risk groups predicted by subnetwork signatures (black lines) or by gene signatures (gray lines). (B-C) Survival curves on SC→TX for (B) the 17 European patients or for (C) the patient cohort in Friedman et al.26 The 2 risk groups are predicted by 2 sets of markers developed on the UC San Diego cohort, including the 38 subnetworks (black lines) and the top 230 genes (gray lines).
Use of expression levels of genes versus subnetworks to stratify patient samples. (A) Five-fold cross-validation on the 130 patients evaluated at UC San Diego. Survival analyses on SC→TX are shown for both the low- (dashed lines) and high- (solid lines) risk groups predicted by subnetwork signatures (black lines) or by gene signatures (gray lines). (B-C) Survival curves on SC→TX for (B) the 17 European patients or for (C) the patient cohort in Friedman et al.26 The 2 risk groups are predicted by 2 sets of markers developed on the UC San Diego cohort, including the 38 subnetworks (black lines) and the top 230 genes (gray lines).
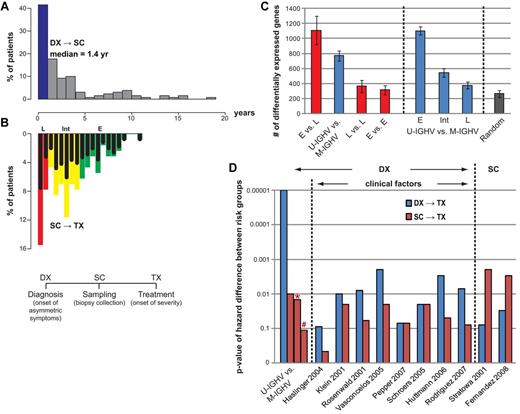
Disparity in gene expression between pretreatment samples collected at various times after diagnosis. (A) Histograms depicting the proportion of patients in the UC San Diego cohort who had sample collection (SC) at various years after diagnosis (DX), as indicated below the graph. The blue bar indicates the proportion of patients who had SC less than 1 year after DX. (B) Inverse histograms depicting the proportion of patients in the UC San Diego cohort who had SC at 1 or more years before therapy, as indicated in the scale above the graph. The samples are considered representative of patients with early-phase disease (“E”) if they were collected more than 4 years before therapy (green bars), intermediate phase (“I”) if collected 4 or less, but 1 or more, years before therapy (yellow bars), or late phase (“L”) if collected less than 1 year before therapy (red bars). The black bars in each colored bar depict the proportion of samples collected in that respective year before therapy that had CLL cells with mutated IGHV. (C) Gene expression differences between different phases of the disease (leftmost panel) and between IGHV subgroups at different phases (middle panel). Bars chart the mean number of differentially expressed genes from 5 trials of 2-tail t tests on 12 versus 12 samples with P value cutoffs at .05. Permutation tests on the same sample sets were performed to assess the numbers of false positives (rightmost panel). (D) Treatment-free survival analyses of all 130 UC San Diego patients using published marker sets. Bars chart the P value of the difference between the low- and high-risk groups, defined by each marker set reported previously. Each marker set is evaluated on both DX→TX (blue bars) and SC→TX (red bars). Bars with * or # denote P value of the difference between SC→TX for samples segregated via IGHV mutation status when the time from DX→SC was less than 1 year (*) or more than year (#), respectively.
Disparity in gene expression between pretreatment samples collected at various times after diagnosis. (A) Histograms depicting the proportion of patients in the UC San Diego cohort who had sample collection (SC) at various years after diagnosis (DX), as indicated below the graph. The blue bar indicates the proportion of patients who had SC less than 1 year after DX. (B) Inverse histograms depicting the proportion of patients in the UC San Diego cohort who had SC at 1 or more years before therapy, as indicated in the scale above the graph. The samples are considered representative of patients with early-phase disease (“E”) if they were collected more than 4 years before therapy (green bars), intermediate phase (“I”) if collected 4 or less, but 1 or more, years before therapy (yellow bars), or late phase (“L”) if collected less than 1 year before therapy (red bars). The black bars in each colored bar depict the proportion of samples collected in that respective year before therapy that had CLL cells with mutated IGHV. (C) Gene expression differences between different phases of the disease (leftmost panel) and between IGHV subgroups at different phases (middle panel). Bars chart the mean number of differentially expressed genes from 5 trials of 2-tail t tests on 12 versus 12 samples with P value cutoffs at .05. Permutation tests on the same sample sets were performed to assess the numbers of false positives (rightmost panel). (D) Treatment-free survival analyses of all 130 UC San Diego patients using published marker sets. Bars chart the P value of the difference between the low- and high-risk groups, defined by each marker set reported previously. Each marker set is evaluated on both DX→TX (blue bars) and SC→TX (red bars). Bars with * or # denote P value of the difference between SC→TX for samples segregated via IGHV mutation status when the time from DX→SC was less than 1 year (*) or more than year (#), respectively.
Although cross-validation is a useful starting point, it can inflate estimates of accuracy because both the training and testing phases are performed on the same cohort of patients. Therefore, we also examined the data collected independently on samples collected in the European cohort. The activity signatures of the 38 subnetworks identified from the UC San Diego cohort were able to deliver a robust prognosis on the European cohort (P = .027, Figure 4B). However, the gene expression markers identified from the UC San Diego cohort failed to correctly identify patients in this cohort who were at high risk for requiring therapy (P = .714, Figure 4B). Use of the IGHV mutation status also failed to segregate these patients (P = .681 in supplemental Figure 1A). Strikingly, these gene expression markers actually mis-segregated the high-risk patients into a subgroup that had a longer treatment-free survival than that of the other patients (Figure 4B). Furthermore, none of the 10 previously published gene marker sets could stratify patients in this European cohort into subgroups that differed significantly in their intervals of SC→TX.
As yet another independent test of prediction accuracy, we examined an external dataset drawn from a previous study outside of the MILE program.26 The subnetwork signature could stratify patients in this independent patient cohort (P = .035 in Figure 4C). However, neither the individual gene expression markers nor the IGHV mutation status (supplemental Figure 1B) were indicative of SC→TX of patients in this cohort.
Treatment-free survival from the time of sample collection provides an alternative measure of patient status and cannot be reliably predicted by gene markers from previous microarray studies
We found that the low- and high-risk groups defined on SC→TX of the UC San Diego cohort had a strong association with IGHV status. Patients with CLL cells that used mutated IGHV genes (with < 98% germline sequence homology) comprised 63% of the patients in the low-risk patient group (longer SC→TX), but only 40% of the high-risk group (association P = .008 using a Fisher exact test, Figure 3E). On the other hand, over one-third of the patients in each group were categorized differently by the subnetwork profiles than by IGHV mutation status. However, the risk groups did not show a significant difference in the length of the time from diagnosis (DX) to therapy (TX), a commonly used indicator of disease aggressiveness (abbreviated as DX→TX; see the green bars in Figure 3A).
We next sought to evaluate whether sets of marker genes proposed by previous studies were prognostic of SC→TX. On the UC San Diego cohort, 5 of the 8 CLL marker sets published previously for their prediction power on DX→TX were able to segregate patients into 2 risk groups with an acceptable difference in their median times of DX→TX (P ≤ .01 in 5-fold cross-validation in Figure 5D). However, none of these sets reached the same statistical significance in predicting DX→TX as did the IGHV mutation status. Moreover, none showed prognostic power on SC→TX. On the other hand, the 2 gene sets, both of which were from studies that took time of SC into consideration, are prognostic of SC→TX, but not of DX→TX (the rightmost 2 sets of bars in Figure 5D), further suggesting the dynamic difference in these 2 time measures.
Transcriptional activity converges between patients of different IGHV status as disease advances
As in most CLL studies, the time from diagnosis (DX) to sample collection (SC; abbreviated as DX→SC) varied significantly among the UC San Diego cohort of 130 patients, which were assayed at various times after diagnosis, but before therapy (Figure 5A). Approximately 40% of these patients were sampled within 1 year of diagnosis, whereas 16.9% of the patients had samples collected 5 years or more after diagnosis. As expected, the patients in this cohort experienced heterogeneity in the interval of DX→TX, as well as in the interval of SC→TX (Figure 5B). The samples collected from patients at an earlier disease phase (“E” in Figure 5B; defined as having SC→TX > 4 years) displayed different transcriptional activity than those of patients with an imminent need of treatment (“L” in Figure 5B; defined as having SC→TX < 1 year; the leftmost bar in Figure 5C). Such differences in gene expression could not be fully explained by differences in the IGHV mutation status of the patients (the second bar from the left in Figure 5C). This might reflect the fact that some patients with CLL cells that used mutated IGHV genes provided samples for these analyses many years after diagnosis, but shortly before requiring treatment (the “L” black bars in Figure 5B).
Next, we compared the expression profiles of CLL cells that used unmutated versus mutated IGHV genes and that were collected at similar disease phases (similar time lengths of SC→TX). The comparison showed that the level of differential gene expression between the 2 subgroups became lower as SC approached TX (Figure 5C middle panel), suggesting that transcriptional differences between CLL cells of different IGHV mutation status converged with disease progression. Interestingly, the number of gene differences between CLL cells that used unmutated versus mutated IGHV genes at a later disease phase was not significantly larger than that observed in comparisons between random samples (Figure 5C rightmost bar).
As expected, patients with leukemia cells that used unmutated IGHV genes had a shorter median time from DX→TX than did patients with CLL cells that used mutated IGHV genes (P = 10−5; Figure 5D leftmost blue bar). However, the IGHV mutation status was not predictive of SC→TX for patients whose SC was obtained more than a year after DX (P = .16, Figure 5D red bar marked by # sign), reflecting perhaps the fact that IGHV mutation status is a fixed parameter that does not change over time. As such, even patients with CLL cells that use mutated IGHV genes ultimately may progress to requiring therapy, even though they continue to have the so-called “good” prognostic feature.
Convergence of dynamic CLL subnetwork transcriptome with disease progression
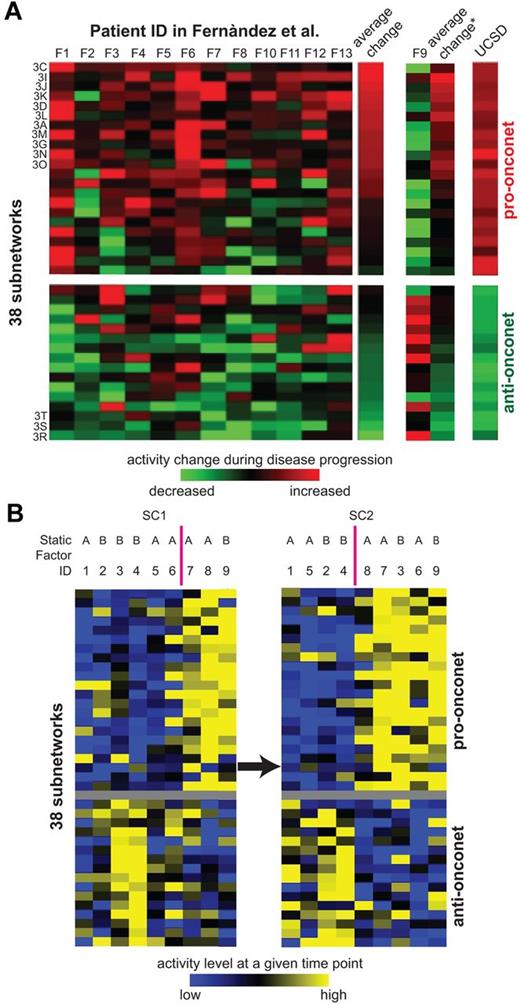
Thus far, patients were sampled at only 1 time point. To investigate the correlation between dynamic subnetwork activities and CLL progression, we next selected an additional 9 patients of various progression paces and sampled their leukemia cells serially at 2 different time points (SC1 and SC2) after DX, but before TX (Figure 6A). Some patients had an aggressive activity pattern on the onconets (high on pro-onconets and low on anti-onconets relatively soon after diagnosis (patients 7, 8, and 9 at SC1) whereas the others showed the reverse pattern, reflecting the heterogeneous nature of CLL disease progression. As disease progressed, 2 more patients (patients 3 and 6 at SC2) obtained the aggressive activity pattern on the onconets, suggesting over time the activity patterns can change in any one patient to converge on what appears to be that associated with more aggressive disease. As discussed in Figure 5C, the activity convergence could not be explained by the static type of clinical factors, such as mutation status of IGHV and ZAP70 expression.
Serial expression of example subnetwork genes and the subnetwork signature along disease progression. (A) Subnetwork activities in serial samples of 9 additional patients registered at UC San Diego. Rows and columns represent subnetworks and patients, respectively. SC1 marks the earlier sample while SC2 represents the later sample of the same patient. Both SC1 and SC2 are before TX. The color of each block scales with the activity level of a subnetwork in a particular patient at either SC1 or SC2 relative to the average activity of all the studied samples. The static factor status of patients is listed above the columns: “A” indicates unmated IGHV and ZAP70 positive; and “B”, mutated IGHV and ZAP70 negative. (B) Subnetwork activity changes in serial samples of 13 patients from Fernandez and colleagues.10 Rows and columns represent subnetworks and patients, respectively. The color of each block scales with the activity change in a subnetwork from the initial versus subsequent sample of each particular patient. The heatmap of patient F9 is separately displayed because of its contrasting pattern versus the other 12 patients. The average change column illustrates the averaged activity change in a subnetwork across patients: the column with an asterik (*) represents the average of all the 13 patients, whereas the column labeled “average change” without the asterisk excludes the data from patient 9. The rightmost column denotes the prognosis power of the 38 subnetworks on UC San Diego samples (the coefficient of each subnetwork as the predictor in a univariate Cox hazard model on SC→TX). The subnetworks that have significant differences between initial and subsequent samples from each patient in Fernandez and colleagues10 (P < .05 from a 1-tailed t test) are indicated by the figure panels in which they are displayed (“3C”, “3I”, etc).
Serial expression of example subnetwork genes and the subnetwork signature along disease progression. (A) Subnetwork activities in serial samples of 9 additional patients registered at UC San Diego. Rows and columns represent subnetworks and patients, respectively. SC1 marks the earlier sample while SC2 represents the later sample of the same patient. Both SC1 and SC2 are before TX. The color of each block scales with the activity level of a subnetwork in a particular patient at either SC1 or SC2 relative to the average activity of all the studied samples. The static factor status of patients is listed above the columns: “A” indicates unmated IGHV and ZAP70 positive; and “B”, mutated IGHV and ZAP70 negative. (B) Subnetwork activity changes in serial samples of 13 patients from Fernandez and colleagues.10 Rows and columns represent subnetworks and patients, respectively. The color of each block scales with the activity change in a subnetwork from the initial versus subsequent sample of each particular patient. The heatmap of patient F9 is separately displayed because of its contrasting pattern versus the other 12 patients. The average change column illustrates the averaged activity change in a subnetwork across patients: the column with an asterik (*) represents the average of all the 13 patients, whereas the column labeled “average change” without the asterisk excludes the data from patient 9. The rightmost column denotes the prognosis power of the 38 subnetworks on UC San Diego samples (the coefficient of each subnetwork as the predictor in a univariate Cox hazard model on SC→TX). The subnetworks that have significant differences between initial and subsequent samples from each patient in Fernandez and colleagues10 (P < .05 from a 1-tailed t test) are indicated by the figure panels in which they are displayed (“3C”, “3I”, etc).
We further examined the overall activity changes of all the 38 subnetworks in a prior study examining for changes in gene expression of CLL cells collected from patients at different times before therapy10 (Figure 6B). In this study, 13 patients were profiled at each of 2 time points, 1 obtained at diagnosis and the other just before therapy. On average, more than half of the pro-onconets increased in activity between the time of diagnosis and the time of therapy. Conversely, the anti-onconets decreased in activity over the course of the disease. Remarkably, among the 22 pro-onconets, 11 showed significant activity induction before therapy (P ≤ .05 from a paired t test in Figure 2A,C-D,G,I-L,N-O); 3 of the 16 anti-onconets were significantly repressed before treatment (Figure 2R-T).
To verify the relationship between the onconet activity changes and disease progression, we selected another 30 patients and performed serial expression of 27 genes by quantitative RT-PCR as an orthogonal validation tool. We measured expression changes of genes in a panel of 27 genes in the onconets significantly associated with disease progression in the data from the study by Fernandez and colleagues10 (Figure 6A). Genes were selected based on 2 criteria: (1) their inclusion in the predictive subnetworks (pro- or anti-onconets) related to cell cycle (Figure 2C-D), regulation of c-MYC (Figure 2E-F,N), G-protein signaling (Figure 2I,L), macromolecule metabolism (Figure 2G-H,S), or resistance to apoptosis (Figure 2R,T), and (2) their differential gene expression observed in the UC San Diego cohort (more suitable to be quantified by RT-PCR).
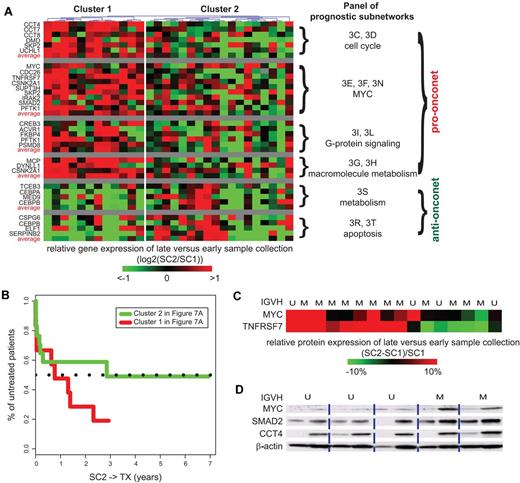
Based on the changes on gene expression in the onconets, the patients could be segregated into 2 groups (Figure 7A). Cluster 1, which had samples that increased expression levels of the probed genes in the pro-onconets over time and decreased expression levels for the anti-onconets, resembles the transcriptome changes of the high-risk patients seen on subnetwork analysis of microarray data. As suggested by the transcriptome changes in the onconets, patients in cluster 1 indeed had a higher likelihood to be in need of treatment compared with the rest of the patients (Figure 7B).
Serial gene and protein expression of example subnetwork genes during disease progression. (A) Gene expression changes in serial samples of 30 additional patients registered at UC San Diego. Rows and columns represent genes and patients, respectively. The color of each block scales with the log2-transformed ratio of a gene in the earlier sample (SC1) compared with the later sample (SC2) of a particular patient. Both SC1 and SC2 are before TX. The “average” rows illustrate the averaged expression change of genes in similar subnetworks across patients. Genes participating in similar subnetworks are clustered together and the figures of the corresponding subnetworks are indexed next to each cluster. Patients are clustered based on their changes on gene expression by a hierarchical clustering dendrogram. (B) Survival analyses on SC2→TX are shown for both cluster 1 (red line) and cluster 2 (green line) segregated by gene expression changes in panel A. (C) Heatmap of protein expression changes of MYC and TNFRSF7 measured by flow cytometry in serial samples of 16 patients registered at UC San Diego. Colors represent the percentage of change in median florescence intensity of a protein in the later sample compared with the earlier sample of a particular patient. (D) Immunoblotting of MYC, SMAD2, and CCT4 in serial samples of 5 patients.
Serial gene and protein expression of example subnetwork genes during disease progression. (A) Gene expression changes in serial samples of 30 additional patients registered at UC San Diego. Rows and columns represent genes and patients, respectively. The color of each block scales with the log2-transformed ratio of a gene in the earlier sample (SC1) compared with the later sample (SC2) of a particular patient. Both SC1 and SC2 are before TX. The “average” rows illustrate the averaged expression change of genes in similar subnetworks across patients. Genes participating in similar subnetworks are clustered together and the figures of the corresponding subnetworks are indexed next to each cluster. Patients are clustered based on their changes on gene expression by a hierarchical clustering dendrogram. (B) Survival analyses on SC2→TX are shown for both cluster 1 (red line) and cluster 2 (green line) segregated by gene expression changes in panel A. (C) Heatmap of protein expression changes of MYC and TNFRSF7 measured by flow cytometry in serial samples of 16 patients registered at UC San Diego. Colors represent the percentage of change in median florescence intensity of a protein in the later sample compared with the earlier sample of a particular patient. (D) Immunoblotting of MYC, SMAD2, and CCT4 in serial samples of 5 patients.
To determine whether the activity changes inferred from transcription have a functional effect on CLL progression, we selected a MYC-associated subnetwork involved in cell-cycle regulation (Figure 2E) and examined for changes in protein expression of some of the genes encoded in that subnetwork over time in 16 CLL patients (a subset of patients in Figure 7A; see supplemental Methods). Several patients had samples with elevated expression of c-MYC that increased over time (Figure 7C-D). Elevated expression levels also were observed in the samples of such patients in the c-MYC interacting partner encoded by SMAD2 (Figure 7D). Another protein TNFRSF7 (Figure 7C) included in this subnetwork also showed increasing expression levels over time. Besides the MYC subnetwork, the increase in expression of subnetworks encoding proteins that promote progression though the cell cycle, exemplified by CCT4 (Figure 7D), further suggested that our transcriptome-based subnetworks have functional implications for disease progression.
Discussion
In this study, we examined for gene expression differences that could segregate patients who were at different risks for requiring therapy relatively soon after sample collection. Many of the prognosis indicators used for segregating CLL patients into different risk categories for disease progression defined subgroups that differed in the median times from diagnosis to initial therapy. However, many patients are asymptomatic at diagnosis, but are detected through incidental laboratory findings. Some patients who receive infrequent medical evaluations may have undetected CLL for years before diagnosis, potentially shortening the interval between diagnosis and initial therapy. Taking such uncertainties and needs into consideration, we sought to identify gene expression subnetworks that could distinguish patients who soon would require therapy from those who would have continued indolent disease after sample collection. Unlike many prior microarray studies, which segregated patients using established prognostic markers, we focused on defining markers associated with treatment-free survival. Support for using this approach came from our analyses of gene expression differences between samples collected from patients at different phases of the disease. Samples from patients segregated using established fixed prognostic parameters, such as IGHV mutation status, had the most divergent gene expression profiles when collected within 1 year of diagnosis, whereas samples collected from patients years after diagnosis had gene expression profiles that apparently converged on that of samples from patients with high-risk disease. This observation suggests that the leukemia might evolve over time into one that has characteristics of disease requiring therapy, albeit at different rates, which may depend on factors that differentially segregate with fixed parameters such IGHV mutation status.
To interrogate for gene expression signatures that might be associated with disease progression, we used subnetwork-aided gene expression analysis, which had several advantages over previous single-gene expression analyses in identifying signatures associated with CLL disease progression. First, the subnetwork-based prognosis appeared more robust. When applied to 2 independent validation datasets, the subnetwork-based approach could more reliably stratify patients at different risks for requiring therapy than the expression signatures of individual marker genes selected without network information. By summarizing multiple gene variables into a network of a holistic view, the network-based prognosis also reduced potential noise when the sample size was small. Although 1 of the 2 validation cohorts used to confirm the subnetwork prognosis—the European cohort—is of a small sample size, the data were collected in the multi-institutional MILE study, which was the same as that used to collect the data for the training set (the UC San Diego cohort). The MILE study implemented the same clinical and experimental protocols at each site. These consistencies in patient management and sample processing minimize artifacts resulting from techniques, arrays, or machines, making the performance on the data from the validation set highlight the true biologic and clinical values of the subnetwork prognosis analyses. The strong performance of the subnetwork prognosis on the validation set of data collected from a center outside the MILE study26 further strengthens our confidence in the prognostic values of the subnetwork signatures. Moreover, the subnetworks identified in this study were significantly superior to prognostic algorithms developed from analyses of expression levels of single genes in stratifying patients of other cohorts.
Another advantage to using the subnetworks approach is that this method provides models of molecular mechanisms, which might contribute to CLL disease progression. Indeed, the subnetwork transcriptomes that distinguish the CLL cells of patients who will or will not require immanent therapy imply that CLL cells associated with more aggressive disease have higher rates of metabolism and cell division, but lower resistance to apoptosis, than CLL cells associated with more indolent disease. For example, the MAPK/ERK signaling cascade has 20 member genes found in our subnetworks. Activation of ERK functions in cellular proliferation and differentiation.38 Aberrations in the MAPK/ERK cascade have been implicated in a high proportion of human cancers and deregulation in this cascade has been implicated in the generation of mitogenic signals in essentially all hematologic malignancies.39 The observations of the 5 MYC-participating subnetworks and the 14 CREB target genes included in the subnetworks also suggest the impact of MAPK/ERK signaling on CLL disease progression, given that activation of MAPK can lead to phosphorylation of MYC and CREB. Recent studies in both mice and patients reveal the potential role of MYC in aggressive disease.40,41
Another prominent signaling protein is TGFβ, which induces apoptosis in numerous cell types.42 It acts as an antiproliferative factor at early phases of oncogenesis; however, later it might enhance tumor progression. The participation of TGFβ in several pro-onconets implies its promoting role in tumor progression, consistent with the observation that in vitro addition of TGFβ does not increase spontaneous apoptosis of B cells in CLL patients,43 but rather serves as an endogenous growth inhibitor.44 The same numbers of pro-onconets and anti-onconets in our subnetwork signature include genes involved in TGFβ signaling (Figure 3D), supporting the potential dual role of TGFβ in CLL development and progression. Furthermore, the subnetwork signature from expression analysis also recovers several genes found to have somatic nonsilent mutations in recent genomic sequencing studies, including FBXW7 (Figure 2C) in the NOTCH1 signaling pathway,45 the WNT (Figure 2A) signaling pathway,46 TP53 (Figure 2B) for DNA damage and cell-cycle control,47 and SF3B1 or XPO1 (Figure 2M), involved in RNA processing or nuclear export of proteins and mRNAs, respectively.48
Although genes with known cancer mutations, such as MYC, TGFβ, and TP53, are typically not detected through analysis of differential expression, they play a central role in the protein network by interconnecting many expression-responsive genes. We observed that many known cancer genes were connected with each other through subnetworks. In all, approximately 27% of the genes in CLL subnetworks (62 of 230 genes total) were known to be associated with cancer (hypergeometric P = 2 × 10−15 in supplemental Figure 2, see supplemental Methods). This fraction was very high compared with conventional expression analysis, for which 16.5% of genes (38 of top 230 genes) were known to be associated with cancer. This higher enrichment was not due solely to the bias of using literature-curated subnetworks (compared with random subnetworks in supplemental Figure 2). As one explanation for why subnetwork analysis performs better, we found that the majority of the cancer genes identified by subnetwork analysis (49 of 62) did not exhibit an altered expression pattern as the disease progressed (P > .01 from an univariate Cox hazard model on SC→TX). Rather, they were included in the subnetworks because of their connectivity—ie, they were required to interconnect many expression-responsive genes (Figure 2).
It should be recognized that transcriptome-based analyses cannot distinguish subnetworks that have different activation states that are not reflected at the level of transcription. This might be the case for subnetworks involving the T-cell leukemia 1 (TCL1) proto-oncogene. Consistent with previous reports,49,50 the expression of TCL1 correlated modestly with disease progression (P = .01 from an univariate Cox hazard model) in the larger UC San Diego cohort. However, TCL1 did not show up in any of the onconets, probably because the expression of genes encoding the proteins interacting with TCL1, including 3 AKT kinases, did not change with disease progression (P > .5 from an univariate Cox hazard model). Instead, the activity of this and other such subnetworks may be governed at a posttranscription level.
The success of correlating treatment-free survival from the date of sample collection with CLL subnetwork transcriptome suggests the association between inner cell states and the disease phase, suggesting that there might be changes in the leukemia cell population over time. Alternatively, there might be emergence of subclones from the tissue microenvironment that express different subnetworks,41 providing for greater growth and/or survival characteristics that allows them to overtake leukemia cell subpopulations that express subnetworks associated with indolent disease.
The idea of cancer as an evolutionary process is not new,51 but little attention has been drawn on the applications of understanding and predicting neoplastic progression. The association observed here between treatment-free survival and the subnetwork transcriptome supports the notion that transcriptional activity of these subnetworks contributes to, or results from, the dynamic evolution of leukemic cells. With proper normalization on the diverse clinical courses between patients, we found considerable differences in gene expression between CLL cells that use mutated versus unmutated IGHV genes at diagnosis that diminishes as the disease progresses to the point of requiring therapy. That the transcriptome difference fades when the 2 subgroups progress, albeit at different rates, supports the idea of cancer evolution.
Putting these together, we rechallenge the “2 distinct disease” hypothesis and speculate that (1) the CLL disease transcriptome evolves over time to reach a state associated with disease requiring treatment, (2) leukemia cells that use unmutated IGHV genes have a higher risk for rapid evolution to develop the transcriptome associated with disease requiring treatment, and (3) the transcriptome of leukemia cells that use mutated IGHV genes transforms gradually to a subnetwork transcriptome similar to that of leukemia cells that use unmutated IGHV genes before therapy. Regardless of their IGHV mutation status, our serial patient samples (as well as those in a previous longitudinal CLL study), demonstrate elevated expression of the pro-onconets and declining expression of the anti-onconets in the identified subnetwork signature over time, further suggesting that degenerate pathways may converge into common pathways that govern disease progression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
H.-Y.C. and T.I. were supported by grants from the National Science Foundation (NSF425926), National Institutes of Health (ES14811), Pfizer and Agilent Labs. H.-Y.C. was additionally supported by a Trainee Research Award from the American Society of Hematology. L.R. and T.J.K. were supported by National Institutes of Health grants for the CLL Research Consortium (P01-CA081534) and a Merit Award to T.J.K. (R37-CA049870).
National Institutes of Health
Authorship
Contribution: H.-Y.C., L.R., M.S., and K.L. performed research; H.-Y.C., L.R., T.I., and T.J.K. wrote and edited the manuscript; H.-Y.C., L.R., T.I., and T.J.K. analyzed the data; and L.R., A.K., T.H., R.F., and T.J.K. contributed vital new reagents and/or analytical tools.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Trey Ideker, University of California San Diego, SKAGGS, Rm 4244, 9500 Gilman Dr, La Jolla, CA 92093-0688; e-mail: tideker@ucsd.edu; or Thomas J. Kipps, University of California San Diego, 3855 Health Sciences Dr, Moores Cancer Center, Room 4307, MC #0820, La Jolla, CA 92093-0820; e-mail: tkipps@ucsd.edu.