Abstract
Recurrent somatic mutation of SRSF2, one of the RNA splicing machinery genes, has been identified in a substantial proportion of patients with myelodysplastic syndrome (MDS). However, the clinical and biologic characteristics of MDS with this mutation remain to be addressed. In this study, 34 (14.6%) of the 233 MDS patients were found to have SRSF2 mutation. SRSF2 mutation was closely associated with male sex (P = .001) and older age (P < .001). It occurred concurrently with at least 1 additional mutation in 29 patients (85.3%) and was closely associated with RUNX1, IDH2, and ASXL1 mutations (P = .004, P < .001, and P < .001, respectively). Patients with SRSF2 mutation had an inferior overall survival (P = .010), especially in the lower risk patients. Further exploration showed that the prognostic impact of SRSF2 mutation might be attributed to its close association with old age. Sequential analyses in 173 samples from 66 patients showed that all SRSF2-mutated patients retained their original mutations, whereas none of the SRSF2-wild patients acquired a novel mutation during disease evolution. In conclusion, SRSF2 mutation is associated with distinct clinical and biologic features in MDS patients. It is stable during the clinical course and may play little role in disease progression.
Introduction
Pre-mRNA needs splicing process to become mature mRNA for further translation into proteins. The splicing of pre-mRNA, a highly regulated cascade of processes, is important for gene expression and genetic diversity; more than 90% of human genes undergo alternative splicing to make various protein isoforms for different biologic functions.1,2 SRSF2, located at 17q25.1, encodes serine/arginine-rich splicing factor 2 that belongs to the serine/arginine-rich protein family, important for splice-site selection, spliceosome assembly, and both constitutive and alternative splicing.3 Recently, through next-generation whole exome sequencing, recurrent somatic mutations involving the RNA splicing machinery were identified in a substantial proportion of patients with myelodysplastic syndrome (MDS),4-7 a myeloid hematopoietic disorder characterized by ineffective hematopoiesis, cytopenias, and risk of transformation to acute leukemia.8,9 Among these mutations, the clinical relevance of 2 prevalent mutations, U2AF354,6,10 and SF3B1,4,5,7,11-14 have been explored. Mutations in SRSF2 also have been reported in MDS patients,4,7,10,15,16 but studies concerning the clinical correlations of the mutations are limited. In addition, the interaction of SRSF2 mutation with other genetic alternations in MDS patients and its stability during disease progression remain to be determined.
The pathogenesis of MDS has not been clearly identified; genomic damage with accumulation of genetic aberrations,8,17 deregulated or autoreactive immune responses,18,19 and abnormal bone marrow (BM) microenvironment20,21 might all contribute to the development and progression of this preleukemic disease. Because the regulation of RNA splicing is important for normal cell function, genetic alternation in SRSF2 may play an important role in the pathogenesis of MDS.4 Here, we aimed to define the clinical correlations of SRSF2 mutation in patients with MDS and examine the interactions of this mutation with other genetic alternations. In addition, sequential analysis of SRSF2 mutation during the clinical follow-ups was performed to evaluate the stability of this mutation in disease progression. To the best of our knowledge, this is the first report to show a close association of SRSF2 mutation with RUNX1, IDH2, and ASXL1 mutations in MDS and the stability of this mutation during the clinical follow-ups.
Methods
Patients
Two hundred and thirty-three adult patients with de novo MDS diagnosed according to French-American-British (FAB) criteria at the National Taiwan University Hospital who had adequate cryopreserved BM cells were recruited into the study. Among them, the disease of 171 patients fulfilled the criteria of MDS according to World Health Organization (WHO) 2008 classification.22 All patients signed informed consents for sample collection in accordance with the Declaration of Helsinki. This study was approved by the Institutional Review Board of the National Taiwan University Hospital.
Mutation analyses
Mononuclear cells obtained from BM aspirates were isolated by Ficoll–Hypaque gradient centrifugation and cryopreserved on the same day of sample collection. Genomic DNA was extracted from 5 to 10 × 106 BM cells using the DNAzol kit (Invitrogen) according to the manufacturer's instructions. Detection of mutations in SRSF2 exon2 on BM cells was done by polymerase chain reaction (PCR) followed by direct sequencing. Approximately 50 ng of DNA was used for each PCR reaction. The primer pairs were the same as those designed by Yoshida et al.4 The PCR conditions were as follows: 95°C for 2 minutes; followed by 40 cycles of 95°C for 30 seconds, 62°C for 30 seconds, and 72°C for 30 seconds; and finally, 72°C for 2 minutes. The mutations were confirmed by at least 2 repeated analyses in this study. Detection of mutations in other genes, including IDH1,23 IDH2,24 JAK2,25 MLL/PTD,26 RUNX1,27 FLT3/ITD,28 WT1,29 N-RAS, K-RAS,30 ASXL1,31 DNMT3,32 and EZH2,33 was performed as described previously. Sequential studies during clinical courses were performed on 173 samples from 66 patients.
Cytogenetics
BM cells were harvested directly or after 1 to 3 days of unstimulated culture, and the metaphase chromosomes were banded by the G-banding method as described previously.34
Statistics
The χ2 test or Fisher exact test was performed to calculate the significance of association between SRSF2 mutation and other parameters of discrete variables, including sex, FAB subtypes, WHO classification, karyotypes, International Prognosis Scoring System (IPSS) scores, and mutations of other genes. Student t test or Mann-Whitney test was used to compare continuous variables and medians of distributions, such as age and hemograms. Kaplan-Meier estimation was used to plot survival curves, and log-rank tests were used to calculate the difference of overall survival (OS) between groups. Cox proportional hazard regression analysis was used to dissect the individual impacts of prognostic factors of OS. All tests were 2-tailed, and a P value of less than .05 was considered statistically significant. All statistical analyses were performed with SPSS 18 software (SPSS).
Results
Mutation patterns of SRSF2
SRSF2 mutation was detected in 34 (14.6%) of the 233 patients (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Twenty-five patients had missense mutations, including P95H in 13 patients, P95L in 9, P95R in 2, and P95A in 1. Eight patients had P95_R102del(c.284_307del), a 24-base pair deletion resulting in an 8-amino acid deletion starting from proline 95. One patient had a novel mutation: P95_D97del (c.283_291del), a 9-base pair deletion that resulted in a 3-amino acid deletion starting from Pro95 (supplemental Figure 1). Among the 171 patients with MDS according to WHO classification, 20 (11.7%) showed SRSF2 mutation. The mutations in all SRSF2-mutated patients were heterozygous.
Correlations of SRSF2 mutations with clinical features
Among 233 patients, 72 were females and 161, males, with a median age of 66 years (range, 18-95 years). The correlations of SRSF2 mutation with the patients' clinical features are summarized in Table 1. SRSF2 mutation was closely associated with male sex (19.9% in males vs 2.8% in females, P = .001) and older age (median, 74.5 years for SRSF2-mutated patients vs 63.0 years for SRSF2-wild patients, P < .001). With the exception of 1 patient, the mutation occurred exclusively in patients older than 50 years (Table 1). The same were also true if the analyses were performed on the 171 MDS patients based on WHO classification (P = .007 for sexes and P < .001 for age, respectively; data not shown). The mutation was more frequently detected in patients with chronic myelomonocytic leukemia (CMML) than in other FAB subtypes (P = .025; Table 1). However, the incidence of SRSF2 mutation was similar between patients with lower risk MDS (refractory anemia [RA] and RA with ring sideroblasts [RARS], 10.2%) and higher risk MDS (RA with excess blasts and RA with excess blasts in transformation, 13.7%). No significant differences were seen in the hemograms and serum lactate dehydrogenase levels between patients with and without SRSF2 mutation. There were no obvious associations between SRSF2 mutation and the cytogenetic groups, IPSS risk groups, or WHO subgroups (Table 1).
Clinical manifestation and laboratory features in MDS patients with and without SRSF2 mutation
| Variable . | SRSF2 mutation (−) (n = 199, 85.4%) . | SRSF2 mutation (+) (n = 34, 14.6%) . | Total (n = 233) . | P . |
|---|---|---|---|---|
| Sex* | .001 | |||
| Male | 129 (80.1) | 32 (19.9) | 161 | |
| Female | 70 (97.2) | 2 (2.8) | 72 | |
| Age, y† | 63.0 (18-91) | 74.5 (49-95) | 66.0 | < .001 |
| > 50* | 151 (82.1) | 33 (17.9) | 184 | .005 |
| ≦ 50* | 48 (98.0) | 1 (2.0) | 49 | |
| Lab data† | ||||
| WBC, /μL | 4205 (440-3 55300) | 4705 (1 250-38 180) | 4215 | .649 |
| Hb, g/dL | 8.3 (3.4-14.4) | 8.45 (4.0-14.6) | 8.4 | .236 |
| Platelet, × 1000/μL | 83.0 (2.0-607.0) | 86.0 (7.0-535.0) | 83.0 | .929 |
| LDH, U/L | 491.5 (145-6807) | 526.5 (230-1261) | 501.5 | .721 |
| FAB subtype* | .025 | |||
| RA/RARS | 88 (89.8) | 10 (10.2) | 98 | |
| RAEB/RAEBT | 88 (86.3) | 14 (13.7) | 102 | |
| CMML | 23 (69.7) | 10 (30.3) | 33 | |
| WHO subtype (n = 171)*‡ | .766 | |||
| RA/RCMD/RCUD§ | 87 (89.7) | 10 (10.3) | 97 | |
| RAEB1 | 31 (86.1) | 5 (13.9) | 36 | |
| RAEB2 | 33 (86.8) | 5 (13.2) | 38 | |
| Karyotype risk (n = 214)‖ | .128 | |||
| Good | 100 (84.0) | 19 (16.0) | 119 | |
| Intermediate | 38 (79.2) | 10 (20.8) | 48 | |
| Poor | 44 (93.6) | 3 (6.4) | 47 | |
| IPSS (n = 214)¶ | .995 | |||
| Low | 26 (86.7) | 4 (13.3) | 30 | |
| INT-1 | 72 (84.7) | 13 (15.3) | 85 | |
| INT-2 | 50 (84.7) | 9 (15.3) | 59 | |
| High | 34 (85.0) | 6 (15.0) | 40 |
| Variable . | SRSF2 mutation (−) (n = 199, 85.4%) . | SRSF2 mutation (+) (n = 34, 14.6%) . | Total (n = 233) . | P . |
|---|---|---|---|---|
| Sex* | .001 | |||
| Male | 129 (80.1) | 32 (19.9) | 161 | |
| Female | 70 (97.2) | 2 (2.8) | 72 | |
| Age, y† | 63.0 (18-91) | 74.5 (49-95) | 66.0 | < .001 |
| > 50* | 151 (82.1) | 33 (17.9) | 184 | .005 |
| ≦ 50* | 48 (98.0) | 1 (2.0) | 49 | |
| Lab data† | ||||
| WBC, /μL | 4205 (440-3 55300) | 4705 (1 250-38 180) | 4215 | .649 |
| Hb, g/dL | 8.3 (3.4-14.4) | 8.45 (4.0-14.6) | 8.4 | .236 |
| Platelet, × 1000/μL | 83.0 (2.0-607.0) | 86.0 (7.0-535.0) | 83.0 | .929 |
| LDH, U/L | 491.5 (145-6807) | 526.5 (230-1261) | 501.5 | .721 |
| FAB subtype* | .025 | |||
| RA/RARS | 88 (89.8) | 10 (10.2) | 98 | |
| RAEB/RAEBT | 88 (86.3) | 14 (13.7) | 102 | |
| CMML | 23 (69.7) | 10 (30.3) | 33 | |
| WHO subtype (n = 171)*‡ | .766 | |||
| RA/RCMD/RCUD§ | 87 (89.7) | 10 (10.3) | 97 | |
| RAEB1 | 31 (86.1) | 5 (13.9) | 36 | |
| RAEB2 | 33 (86.8) | 5 (13.2) | 38 | |
| Karyotype risk (n = 214)‖ | .128 | |||
| Good | 100 (84.0) | 19 (16.0) | 119 | |
| Intermediate | 38 (79.2) | 10 (20.8) | 48 | |
| Poor | 44 (93.6) | 3 (6.4) | 47 | |
| IPSS (n = 214)¶ | .995 | |||
| Low | 26 (86.7) | 4 (13.3) | 30 | |
| INT-1 | 72 (84.7) | 13 (15.3) | 85 | |
| INT-2 | 50 (84.7) | 9 (15.3) | 59 | |
| High | 34 (85.0) | 6 (15.0) | 40 |
WBC indicates white blood cells; Hb, hemoglobin; LDH, lactate dehydrogenase; RAEBT, refractory anemia with excess blasts in transformation; RCUD, refractory cytopenia with unilineage dysplasia; and RCMD, refractory cytopenia with multilineage dysplasia.
Number of patients (%).
Median (range).
One patient with MDS, unclassifiable, by WHO classification was excluded from the statistical analysis.
With or without ring sideroblasts.
Good, normal karyotype, isolated -Y, del(5q) or del(20q); poor, complex (≥ 3 abnormalities) or chromosome 7 anomalies; and intermediate, other abnormalities.
International prognosis scoring system: low, 0; intermediate (INT)–1, 0.5-1; INT-2, 1.5-2; and high, ≥ 2.5.
Association of SRSF2 mutation with other mutations
The association of SRSF2 mutation with other gene mutations also was explored (Table 2). Among the 34 patients with SRSF2 mutation, 29 had at least 1 additional mutation concurrently: 19 had 1, 5 each had 2 and 3, respectively. The patients with SRSF2 mutation had significantly higher incidences of RUNX1, ASXL1, and IDH2 mutations than those without the mutation (29.4% vs 10.1%, P = .004; 47.1% vs 19.6%, P < .001; and 20.6% vs 2.5%, P < .001, respectively; Table 2). The same were also true if the analyses were performed in the 171 MDS patients based on WHO classification (P = .001, P < .001, and P = .022, respectively; supplemental Table 1).
Comparison of concurrent alterations of other genes between MDS patients with and without the SRSF2 mutation
| Genes (no. of patients tested) . | No. (%) of patients with mutation of other gene* . | P . | |
|---|---|---|---|
| SRSF2-wild patients . | SRSF2-mutated patients . | ||
| JAK2 (n = 209) | 2 (1.1) | 1 (3.2) | .384 |
| MLL/PTD (n = 216) | 4 (2.2) | 2 (5.9) | .240 |
| K-RAS (n = 231) | 3 (1.5) | 0 (0) | 1.000 |
| N-RAS (n = 232) | 8 (4.0) | 1 (2.9) | 1.000 |
| RUNX1 (n = 233) | 20 (10.1) | 10 (29.4) | .004 |
| FLT3/ITD (n = 233) | 6 (3.0) | 0 (0) | .596 |
| WT1 (n = 228) | 1 (0.5) | 0 (0) | 1.000 |
| IDH1 (n = 233) | 3 (1.5) | 0 (0) | 1.000 |
| IDH2 (n = 233) | 5 (2.5) | 7 (20.6) | < .001 |
| ASXL1 (n = 233) | 39 (19.6) | 16 (47.1) | < .001 |
| DNMT3 (n = 230) | 11 (5.6) | 2 (5.9) | 1.000 |
| EZH2 (n = 217) | 19 (10.4) | 5 (14.7) | .549 |
| Genes (no. of patients tested) . | No. (%) of patients with mutation of other gene* . | P . | |
|---|---|---|---|
| SRSF2-wild patients . | SRSF2-mutated patients . | ||
| JAK2 (n = 209) | 2 (1.1) | 1 (3.2) | .384 |
| MLL/PTD (n = 216) | 4 (2.2) | 2 (5.9) | .240 |
| K-RAS (n = 231) | 3 (1.5) | 0 (0) | 1.000 |
| N-RAS (n = 232) | 8 (4.0) | 1 (2.9) | 1.000 |
| RUNX1 (n = 233) | 20 (10.1) | 10 (29.4) | .004 |
| FLT3/ITD (n = 233) | 6 (3.0) | 0 (0) | .596 |
| WT1 (n = 228) | 1 (0.5) | 0 (0) | 1.000 |
| IDH1 (n = 233) | 3 (1.5) | 0 (0) | 1.000 |
| IDH2 (n = 233) | 5 (2.5) | 7 (20.6) | < .001 |
| ASXL1 (n = 233) | 39 (19.6) | 16 (47.1) | < .001 |
| DNMT3 (n = 230) | 11 (5.6) | 2 (5.9) | 1.000 |
| EZH2 (n = 217) | 19 (10.4) | 5 (14.7) | .549 |
Number (%) of patients showing the specific gene mutation listed in the far left column among the SRSF2-wild or SRSF2-mutated group.
Prognostic impact of SRSF2 mutation on OS
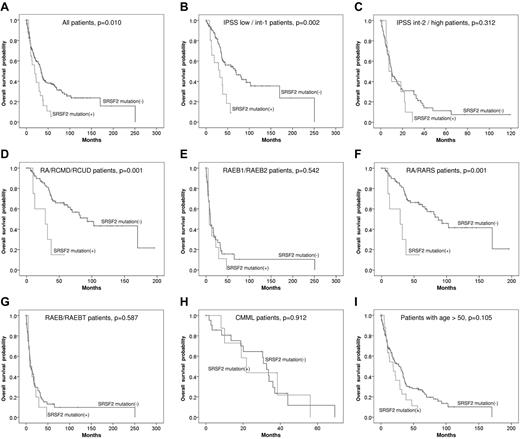
With a median follow-up period of 60.2 months, patients with SRSF2 mutation had an inferior OS than those without this mutation (median, 33.9 months vs 19.0 months, P = .010; Figure 1A). Subgroup analysis showed that the poor impact of SRSF2 mutation on OS was only demonstrated in the patients of lower risk groups defined either by IPSS (low and intermediate-1, 69.3 months vs 32.0 months, P = .002; Figure 1B), by World Health Organization classification (RA, RARS, refractory cytopenia with unilineage dysplasia, and refractory cytopenia with multilineage dysplasia, with or without ring sideroblasts, 93.0 months vs 28.7 months, P = .001; Figure 1D), or by FAB classification (RA and RARS, 87.4 months vs 28.7 months, P = .001; Figure 1F). No survival differences were found between the patients with and without SRSF2 mutation in the higher risk groups (Figure 1C,E,G). Among CMML patients, there was no OS difference between SRSF2-mutated and SRSF2-wild patients, either (Figure 1H). To evaluate whether the inferior survival associated with SRSF2 mutations was attributed to the age effect, we performed survival analysis restricted to the patients older than 50 years; in this group of patients, SRSF2 mutation did not influence the OS (Figure 1I). A multivariate Cox regression analysis using variables including SRSF2 mutation and other known prognostic clinical factors on OS also showed that, after adjustments, the impact of SRSF2 mutation per se on OS vanished (Table 3). Given that the prognostic impact of SRSF2 mutation was greatest in lower risk patients, a Cox regression test limiting to patients with low or intermediate-1 risk disease was done; SRSF2 mutation was similarly not an independent prognostic factor for OS in this group of patients (supplemental Table 2). These results suggest the interactive effects of SRSF2 mutation and age on the patients' OS.
Kaplan-Meier curves. Kaplan-Meier curves of OS stratified by SRSF2 mutation for all patients (A), lower and higher risk IPSS subgroups (B-C), lower and higher risk WHO subgroups (D-E), lower and higher risk FAB subgroups (F-G), CMML patients (H), and the patients older than 50 years (I).
Kaplan-Meier curves. Kaplan-Meier curves of OS stratified by SRSF2 mutation for all patients (A), lower and higher risk IPSS subgroups (B-C), lower and higher risk WHO subgroups (D-E), lower and higher risk FAB subgroups (F-G), CMML patients (H), and the patients older than 50 years (I).
Cox regression analysis for the overall survival in MDS patients
| Factor . | Hazard ratio . | 95% CI . | P . | |
|---|---|---|---|---|
| SRSF2 | Mutation (+) | 1.41 | 0.87∼2.30 | .166 |
| Mutation (−) | 1.0 (reference) | |||
| IPSS | INT-2/high | 2.33 | 1.40∼3.90 | .001 |
| Low/INT-1 | 1.0 (reference) | |||
| FAB subtype | CMML | 0.79 | 0.46∼1.36 | .387 |
| RA/RARS | 0.40 | 0.23∼0.72 | .002 | |
| RAEB/RAEBT | 1.0 (reference) | |||
| Age, y | > 50 | 3.36 | 1.91∼5.89 | < .001 |
| ≦ 50 | 1.0 (reference) | |||
| Factor . | Hazard ratio . | 95% CI . | P . | |
|---|---|---|---|---|
| SRSF2 | Mutation (+) | 1.41 | 0.87∼2.30 | .166 |
| Mutation (−) | 1.0 (reference) | |||
| IPSS | INT-2/high | 2.33 | 1.40∼3.90 | .001 |
| Low/INT-1 | 1.0 (reference) | |||
| FAB subtype | CMML | 0.79 | 0.46∼1.36 | .387 |
| RA/RARS | 0.40 | 0.23∼0.72 | .002 | |
| RAEB/RAEBT | 1.0 (reference) | |||
| Age, y | > 50 | 3.36 | 1.91∼5.89 | < .001 |
| ≦ 50 | 1.0 (reference) | |||
Mutation statuses of SRSF2 during disease evolution
SRSF2 mutations were studied sequentially in 173 samples from 66 patients, including 8 patients with SRSF2 mutation and 58 patients without mutation at diagnosis. During the clinical follow-ups, 6 of the 8 SRSF-mutated patients showed disease progression to higher risk subtypes, including 5 with acute myeloid leukemia (AML) transformation, whereas 31 of the 58 SRSF2-wild patients showed disease progression, including 23 with AML transformation. Among the 8 patients with SRSF2 mutation, all retained the original mutations in the subsequent samples (Table 4). In contrast, none of the 58 patients who had no SRSF2 mutation initially acquired a novel mutation during follow-ups, suggesting that this mutation might play little role in MDS progression. Four patients (50%) with SRSF2 mutation acquired novel mutations of other genes during follow-ups (Table 4), whereas 11 patients (29%) without SRSF2 mutation did so (P = .071; data not shown).
Sequential studies in the 8 MDS patients with SRSF2 mutation
| Patient no. . | Time after the initial BM sampling . | Diagnosis . | Karyotype . | SRSF2 mutation . | Other mutations . |
|---|---|---|---|---|---|
| 1 | (Initial) | RAEBT | 46,XY | P95H | N-RAS, RUNX1, |
| 33 mo | AML | 45,XY,-4, der(17),t(4;17)(q12;p11.2) | P95H | N-RAS, RUNX1, ASXL1 | |
| 2 | (Initial) | CMML | 46,XY | P95H | N-RAS, RUNX1, |
| 6 mo | CMML | 46,XY | P95H | N-RAS, RUNX1, | |
| 3 | (Initial) | RAEBT | 46,XY | P95H | ASXL1 |
| 4 mo | AML | 46,XY | P95H | K-RAS, ASXL1, | |
| 4 | (Initial) | RARS | 46,XX | P95H | IDH2 |
| 17 mo | RARS | 46,XX | P95H | ASXL1, IDH2, | |
| 5 | (Initial) | RAEB | 46,XY | P95L | RUNX1, ASXL1 |
| 26 mo | AML | Not done | P95L | RUNX1, ASXL1 | |
| 31 mo | AML | 46,XY | P95L | RUNX1, ASXL1 | |
| 6 | (Initial) | RA | 46,XY | P95_R102del | RUNX1 |
| 3 mo | RAEB | 50,XY,del(11)(q13q23),+13,+15,+18 | P95_R102del | RUNX1 | |
| 7 | (Initial) | RA | 46,XX,i(17)(q10) | P95_R102del | |
| 12 mo | AML | 46,XY,-7,i(17)(q10) | P95_R102del | ASXL1, | |
| 8 | (Initial) | RA | 47,XY,+8 | P95_D97del | ASXL1, RUNX1, IDH2 |
| 6 mo | AML | 46,XY | P95_D97del | RUNX1, IDH2 |
| Patient no. . | Time after the initial BM sampling . | Diagnosis . | Karyotype . | SRSF2 mutation . | Other mutations . |
|---|---|---|---|---|---|
| 1 | (Initial) | RAEBT | 46,XY | P95H | N-RAS, RUNX1, |
| 33 mo | AML | 45,XY,-4, der(17),t(4;17)(q12;p11.2) | P95H | N-RAS, RUNX1, ASXL1 | |
| 2 | (Initial) | CMML | 46,XY | P95H | N-RAS, RUNX1, |
| 6 mo | CMML | 46,XY | P95H | N-RAS, RUNX1, | |
| 3 | (Initial) | RAEBT | 46,XY | P95H | ASXL1 |
| 4 mo | AML | 46,XY | P95H | K-RAS, ASXL1, | |
| 4 | (Initial) | RARS | 46,XX | P95H | IDH2 |
| 17 mo | RARS | 46,XX | P95H | ASXL1, IDH2, | |
| 5 | (Initial) | RAEB | 46,XY | P95L | RUNX1, ASXL1 |
| 26 mo | AML | Not done | P95L | RUNX1, ASXL1 | |
| 31 mo | AML | 46,XY | P95L | RUNX1, ASXL1 | |
| 6 | (Initial) | RA | 46,XY | P95_R102del | RUNX1 |
| 3 mo | RAEB | 50,XY,del(11)(q13q23),+13,+15,+18 | P95_R102del | RUNX1 | |
| 7 | (Initial) | RA | 46,XX,i(17)(q10) | P95_R102del | |
| 12 mo | AML | 46,XY,-7,i(17)(q10) | P95_R102del | ASXL1, | |
| 8 | (Initial) | RA | 47,XY,+8 | P95_D97del | ASXL1, RUNX1, IDH2 |
| 6 mo | AML | 46,XY | P95_D97del | RUNX1, IDH2 |
The result of sequential analysis of the 58 patients without SRSF2 mutation at diagnosis is not shown; none of these patients acquired SRSF2 mutation. Thirty-one of them had disease progression, including 23 patients with transformation to AML during the clinical course.
Discussion
After the discovery of recurrent somatic mutations in spliceosome complex genes in a significant proportion of adult MDS patients by whole exome sequencing studies,4-6 it is of interest to know whether there are distinct clinical features in the patients harboring these mutations. The mutations affecting the spliceosome complex were found to be apparently mutually exclusive in prior studies of Yoshida et al4 and Makishima et al.10 The clinical correlations of the other 2 prevalent mutations involving the splicing machinery, U2AF354,6,10 and SF3B1,4,5,7,11-14,35 have been explored, whereas there have been few similar reports about SRSF2 mutation. Here, we confirmed that SRSF2 mutation was more prevalent in CMML than in other MDS subtypes, as reported previously 4,10 ; however, it occurred with similar frequency in patients with lower risk and higher risk MDS. In addition, to the best of our knowledge, we demonstrated for the first time the distinct association of SRSF2 mutation with male sex; older age; and mutations of RUNX1, IDH2, and ASXL1 genes.
In the current cohort, SRSF2 mutation was detected in 14.6% of patients. Similar to prior reports, P95H was the most frequent mutation.4,10 Besides, a novel mutation, P95_D97del (c.283_291del), was identified. The universal involvement of Pro95 in SRSF2 mutation indicates a critical role of this residue on the protein function, which needs to be explored by further studies. Recently, Allain and colleagues determined the solution structure of SRSF2 in the free-state and in complex with a pyrimidine- and a more purine-rich RNA sequence.36 They demonstrated that Pro95 of SRSF2 forms extensive contact with the RNA through stacking. Following this clue, we speculate that Pro95 is essential for SRSF2 binding to its target RNAs. Mutation of Pro95 may reduce the RNA binding affinity of SRSF2 protein and thus affects the function of SRSF2.
The serial study of SRSF2 mutation demonstrated the stability of this mutation during disease progression. All patients without SRSF2 mutation remained germ line of the gene during clinical follow-ups, whereas all SRSF2-mutated patients retained their original mutations. These findings suggested that though SRSF2 mutation may play a role in the development of MDS, it is not associated with the disease evolution. To further test this hypothesis, the time-to-AML-transformation was analyzed in low and intermediate-1 IPSS risk patients of the current cohort. We found no significant impact for SRSF2 mutation on the time-to-AML-transformation (P = .061; supplemental Figure 2). The multivariate Cox regression analysis also showed the same result (supplemental Table 3). These findings are in agreement with the proposed major role of SRSF2 mutation in MDS development rather than in MDS progression.
In this study, we found that SRSF2 mutation were strongly associated with male sex and older age. The same was also true if the analysis was performed only in the 171 MDS patients based on WHO classification. Intriguingly, a recent study surveying the spliceosome gene mutations in pediatric patients with MDS also showed that these mutations were extremely uncommon in the pediatric cohort,12 a finding compatible with our result.
The analysis of 12 other gene mutations in the current cohort demonstrated that SRSF2 mutation was closely associated with RUNX1, IDH2, and ASXL1 mutations. Intriguingly, SRSF2 mutation rarely occurred alone; all but 5 patients with SRSF2 mutation had at least 1 concurrent mutation of other genes, a finding compatible with the report of Nagata et al who found that there was frequent accompaniment of additional mutations in patients with SRSF2 mutation.37 This finding is in agreement with the concept that the development of MDS may require concerted interactions among different genetic alterations in the hematopoietic progenitor compartment.8,38,39 In addition, among the 66 patients enrolled for serial analysis in this study, 4 (50%) of the 8 SRSF2-mutated patients acquired additional mutations of other genes subsequently (Table 4), whereas 11 (19%) of the 58 SRSF2-wild patients did so during follow-ups (data not shown); although the sample size was small, there was a trend of statistical difference in the incidence of genetic evolution between these 2 groups (P = .071 by Fisher exact test). Xiao et al demonstrated that SRSF2 is essential for genomic stability in their SRSF2-knockout mouse model.40 Because mutation of SRSF2 is supposed to result in loss of its protein function, the frequent occurrence of additional gene mutations at diagnosis and acquisition of new mutations during follow-ups in patients with SRSF2 mutation in the current study support the claim that depletion of SRSF2 may lead to DNA hypermutability.40 Conversely, Schnittger et al reported that SRSF2 mutation were mutually exclusive with EZH2 mutation in CMML patients.41 In the current cohort, the same finding also was observed in patients with CMML, but not in those with other subtypes: all 10 CMML patients with SRSF2 mutation had wild-type EZH2 (data not shown). This interesting finding is worthy of further confirmation by studies on more patients.
The prognostic impact of mutations of spliceosome genes in MDS patients is still controversial. SF3B1 gene mutation has been reported to have significant impact on OS in 2 studies,5,35 but not in others.11,13 The U2AF1 mutation did not show influence on OS in one study,6 whereas it was associated with shorter survival in another study.10 In the current study, SRSF2 mutation possessed negative impact on OS in MDS patients, especially for those with lower risk disease, similar to the finding of Makishima et al.10 However, we found that the SRSF2 mutation was not an independent poor prognostic factor, and its impact on OS might be a reflection of the close association of this mutation with old age. In addition, we also observed no survival impact of SRSF2 mutations in patients with CMML in which this mutation is more prevalent. It is true that the sample size of CMML patients in the current cohort is too small to make a solid conclusion. However, similar finding was also reported by other groups. For example, Schnittger et al reported that no survival impact was observed in their larger CMML cohort41 ; Makishima et al also reported that SRSF2 mutations did not affect outcomes in CMML patients.10 Further analyses on larger patient cohorts are needed to confirm these findings.
In summary, this study demonstrated that SRSF2 mutation could be detected in a substantial number of patients with MDS. The mutation was closely associated with male sex; older age; CMML; and mutations of RUNX1, ASXL1, and IDH2, and it was stable during disease evolution. Patients with SRSF2 mutation had an inferior OS, an impact that might be related to the predominant occurrence of the mutation in older patients.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was partially sponsored by grants from the National Science Council (NSC 99-2314-B-002-026-MY2, NSC 100-2325-B-002-032, and NSC 100-2314-B-002-112-MY3); Department of Health (DOH100-TD-C-111-001); and Department of Medical Research, National Taiwan University Hospital (NTUH 100-001692 and 101-002014), Taiwan, Republic of China.
Authorship
Contribution: S.-J.W. was responsible for literature collection, data management and interpretation, statistical analysis, and manuscript writing. L.-Y.L., M.-H.T., C.-F.H., F.-Y.L., M.-C.L., and C.-W.L. performed the gene mutation and chromosome studies; C.-T.L., H.-A.H., C.-Y.C., W.-C.C., M.Y., S.-Y.H., B.-S.K., J.-L.T., and W.T. contributed patient samples and clinical data; and H.-F.T. and Y.-Y.K. planned, designed, and coordinated the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hwei-Fang Tien, Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan; e-mail: hftien@ntu.edu.tw; or Yuan-Yeh Kuo, Graduate Institute of Oncology, College of Medicine, National Taiwan University, Taipei, Taiwan; e-mail: d90442001@ntu.edu.tw.