Abstract
The expression of Pax5 commits common lymphoid progenitor cells to B-lymphoid lineage differentiation. Little is known of possible variations in the levels of Pax5 expression and their influences on hematopoietic development. We have developed a retroviral transduction system that allows for the study of possible intermediate stages of this commitment by controlling the levels of Pax5 expressed in Pax5-deficient progenitors in vitro and in vivo. Retroviral transduction of Pax5-deficient pro-/pre-B cell lines with a doxycycline-inducible (TetON) form of the human Pax5 (huPax5) gene yielded cell clones that could be induced to different levels of huPax5 expression. Clones inducible to high levels developed B220+/CD19+/IgM+ B cells, while clones with low levels differentiated to B220+/CD19−/CD11b+/Gr-1− B-lymphoid/myeloid biphenotypic cells in vitro and in vivo. Microarray analyses of genes expressed at these lower levels of huPax5 identified C/ebpα, C/ebpδ, Pu.1, Csf1r, Csf2r, and Gata-3 as myeloid-related genes selectively expressed in the pro-/pre-B cells that can develop under myeloid/lymphoid conditions to biphenotypic cells. Therefore, reduced expression of huPax5 during the induction of early lymphoid progenitors to B-lineage–committed cells can fix this cellular development at a stage that has previously been seen during embryonic development and in acute lymphoblastic lymphoma–like biphenotypic acute leukemias.
Introduction
During hematopoiesis from pluripotent hematopoietic stem cells, the transcription factor Pax5 is selectively expressed in B-lymphoid lineage cell development.1 Pax5 directly activates the signaling component of the B-cell receptor (BcR; Ig), Igα, the costimulatory receptor CD19, and the signaling adaptor Blnk.2,3
At the same time, Pax5 represses the multilineage and homing potential of the multilineage-primed4 Pax5−/− pro-/pre-B cells by shutting down the expression of T lineage (eg, Notch-1),5 NK lineage (eg, 2B4, CD244),6 and myeloid cell lineage directed (eg, Csf1r, c-fms),7 B-lymphoid–inappropriate signaling genes.6,8 Furthermore, Pax5 represses expression of Flt3, a tyrosine kinase receptor that is required to develop common lymphoid progenitors (CLPs) from pluripotent hematopoietic stem cells.9 When Pax5 expression is turned off at the transition from B cells to plasma cells, some of the inappropriate gene expression programs are reactivated,8 as it happens when Pax5 is inactivated by cre-recombinase–mediated deletion in mature B cells.10
Pax5-deficient mice are blocked in early B-cell development11 at a B220+/CD19−/Flt3+/c-kit+/IgM− pro-/pre-B-cell–like state of differentiation4 while the development of all other hematopoietic lineages is unaltered. Cloned Pax5−/− cells show remarkable flexibility of hematopoietic developmental choices. As CLP-similar, lymphoid-primed cells, they can be induced in vitro to precursor and immature T cells, to natural killer (NK) cells, and to different types of myeloid cells.4 They can also be transplanted into Rag-1– or Rag-2–deficient, severe combined immunodeficient recipient mice and will populate all thymocyte, mature T-cell, and NK-cell compartments within 3 weeks, while all myeloid cell compartments are filled within 2 months.12 Unlike wild-type pre-B-I cells, Pax5−/− pro-/pre-B cells home to the bone marrow from where they can be reisolated and repropagated in vitro.13 Hence, Pax5−/− pro-/pre-B cells maintain self-renewal, multipotency, and homing potential for many successive transplantations.
Pax5 can play a divergent role in lymphomagenesis. Different groups have identified the PAX5 gene in some B-cell non-Hodgkin lymphomas. The coding regions of the PAX5 gene are translocated to, and its expression controlled by, regulatory elements of the IgH locus.14,15 Frequent somatic mutations in the PAX5 locus leading to haploinsufficiency and hypomorphic alleles or complete loss of function can be observed in paediatric acute lymphoblastic lymphomas (ALL).16-18 In addition, conditional deletion of PAX5 in CD19-CRE;Pax5fl/− mice led to the development of B-progenitor lymphomas,19 whereas Pax5+/− heterozygous mice never develop malignancies.20
Restoration of Pax5 expression in Pax5-deficient mice and pro-/pre-B cells has been shown to repress B-lymphoid–inappropriate gene expression, and to promote development of B lymphocytes.4 To rescue Pax5 expression and B-cell development, we insert here the huPax5 gene into a retroviral transactivator (rtTA)–dependent TetON-expression vector21 that allows doxycycline-inducible huPax5 expression in Pax5−/− pro-/pre-B cells. Retrovirally transduced cells are then cloned to generate lines that express huPax5 at different levels because different sites of insertion into the genome restrict retroviral gene expression to different extents.22 This allows us to investigate the influence of graded concentrations of huPax5 on molecular and cellular changes induced by huPax5 expression from a primed lymphoid-CLP–like to a B lymphoid–committed state. At low expression levels of huPax5, a B-lymphoid/myeloid biphenotypic state can be stabilized in appropriately doxycycline-dosed cells in vitro and in vivo.
Methods
Mice
Pax5−/−/Ly5.2+ mice, a kind gift of M. Busslinger (Research Institute of Molecular Pathology, Vienna, Austria), and Rag-2−/−/Ly5.1+ mice were bred in the laboratory animal facility of the Max Planck Institute for Infection Biology (Berlin, Germany) under specific pathogen-free (SPF) conditions.
All experiments complied with national regulations for the care and use of laboratory animals (protocol G0099/08, approved by the Landesamt für Gesundheit und Soziales, Berlin, Germany).
Cell culture
Two-week-old, bone marrow–derived Pax5−/− pro-/pre-B cells4,12 were grown in IMDM/2% FCS/0.03% primatone-RL (Quest) on γ-irradiated OP9 stromal cells and mouse IL-7.23,24 OP9 and OP9-Δl-1 cells (a kind gift of J.-C. Zúñiga-Pflücker, University of Toronto, Toronto, ON) were cultured in αMEM/2% FCS/0.03% primatone-RL. The Phoenix-eco virus producer cells (Orbigen Inc) were cultured in RPMI/10% FCS.
Cytokine supernatants were produced by using the appropriate hybridoma cell lines: IL-3 (X63/IL-3), IL-6 (X63/IL-6), IL-7(J558L/IL-7), SCF (CHO-SCF, a kind gift of Thorsten Feyerabend, University of Ulm, Ulm, Germany), Flt3l (Sp2.0-Flt3l, a kind gift of Paulo Vieira, Institute Pasteur, Paris, France), GM-CSF (X63-GM-CSF, kindly provided by Antonius Rolink, University of Basel, Basel, Switzerland). All cytokine supernatants were used at optimal concentrations determined against the respective purified recombinant cytokines.
Retroviral TetON constructs
A 1313-bp NotI-huPax5-NotI-PCR fragment was amplified from the MSCV-huPax5-huCD2t plasmid (kindly provided by M. Busslinger, Research Institute of Molecular Pathology, Vienna, Austria) with the primers NotI-huPax5: forward 5′-gatgcggccgcTTTTCCCTGTCCATTCCATC-3′, reverse 5′-agtgcggccgcTGGGCTCTCTGGCTATCTTC-3′. The NotI-huPax5-NotI-PCR fragment was inserted upstream of the Tet-responsive CMVmin promoter of the retroviral vector pSR-TRE.21
Generation of pre-B-cell lines
Viruses were produced by transient transfection of Phoenix-eco cells using Lipofectamine (Invitrogen) according to manufacturer's protocol. Pax5−/− pro-/pre-B cells were spin-infected at 30°C with 1150g for 3 hours 30 minutes, thereafter cultured on IL7/OP9. Two independent rtTA-expressing pro-B-cell lines, independently infected with the retroviral huPax5-expression vector, were generated. Double-transgenic (pSR-rtTA/pSR-TRE-huPax5) polyclonal cell lines were cloned by limiting dilutions in IL-7/OP9, yielding clones with different expression levels of huPax5 on doxycycline-stimulation in vitro. Selection of clone 4 (bulk culture 1) and clones 16 and 20 (both bulk culture 2) was performed according to Pax5 and CD19 expression levels, respectively.
In vitro differentiation of Pax5−/− pro-/pre-B cells into CD19−/B220+/CD11b+ biphenotypic cells
huPax5-TetON–transduced Pax5−/− pro-/pre-B cells were induced to express huPax5 by addition of 1000 ng/mL, 300 ng/mL, and 10 ng/mL (or without) doxycycline for 3 days on IL7/OP9. Differentiation of 2 × 106 Pax5−/− pro-/pre-B cells into biphenotypic CD19−/B220+/CD11b+ cells was induced on γ-irradiated OP9 cells in IMDM/2% FCS/2% Flt3l/3% SCF at different doxycycline levels.
In vivo differentiation of Pax5−/− pro-/pre-B cells into CD19+ pre-B- and CD19−/B220+/CD11b+ biphenotypic cells
Six- to 12-week-old Rag-2−/− (Ly5.1+) mice were γ-irradiated (400 cGy) 24 hours before transplantation. Pax5−/− pro-/pre-B cells (5 × 106 cells/mouse) were injected intravenously. In vivo differentiation of huPax5-TetON–transduced Pax5−/− pro-/pre-B cells into CD19+ pre-B- or CD19−/B220+/CD11b+ biphenotypic cells was induced in vivo 1 week after transplantation by 0.2 g/L doxycycline dissolved with 5% sucrose in acidified (pH 3.0) drinking water.
Antibodies and flow cytometry
If not stated otherwise, all anti–mouse antibodies were obtained from eBioscience Inc. Antibodies used for cell staining were Flt3-PE (A2F10), CD19-PE-Cy7 (1D3), biotinylated or allophycocyanin (APC)–conjugated B220 (RA3-6B2), CD11b-APC (M1/70), c-kit-APC (ACK4), Gr-1–Pacific Blue (RB6-8C5). Streptavidin-Qdot605 (Molecular Probes) was used to visualize biotin-conjugated primary antibodies. Cells were incubated with anti–mouse CD16/CD32 followed by staining with the conjugated antibodies in PBS/2% FCS. Dead cell discrimination was performed by DAPI (Carl Roth). Analyses were done on a LSR-II flow cytometer (BD Biosciences) equipped with a 488-, 405-, 633-, and 355-nm laser.
Semiquantitative RT-PCR and quantitative real-time RT-PCR
Control- and Pax5−/− pro-/pre-B cells, induced in vitro by doxycycline to huPax5-dependent differentiation, were harvested and incubated for 45 minutes on a tissue-culture flask in IL-7/doxycycline to remove contaminating OP9 cells. RNA was prepared by using TRIzol reagent (Invitrogen). For semiquantitative RT-PCR, cDNA was synthesized with random hexamer primers and Superscript-III-RT (Invitrogen). PCRs were performed with a 4-fold dilution series (1:5) prepared of each cDNA sample.
Expression of mRNAs was quantitatively assessed by real-time RT-PCR using the QuantiTect SYBR Green PCR Kit (QIAGEN) in a 7900HT fast real-Time PCR system (Applied Biosystems). Each sample was assayed in triplicate for every run, and results were normalized against GAPDH mRNA expression.
Immunoblotting
Control and Pax5−/− pro-/pre-B cells, induced in vitro by doxycycline to huPax5-dependent differentiation and OP9 separated, were lysed with modified RIPA buffer (25mM Tris-HCL,150mM NaCl, 1% NP40, 2% Triton X100, 2% CHAPS, 0.1% SDS, 1× protease inhibitor cocktail; Roche). Cell lysates (10 μg of protein/lane) were loaded on 4%-20% Mini-PROTEAN TGX gels (Bio-Rad), separated by electrophoresis, and blotted onto a nitrocellulose membrane. The membrane was blocked in 1× PBS with 0.1% Tween 20 (PBS-T) with 5% skim milk for 1 hour and incubated with a Pax5- or β-actin–specific antibody (Abcam, clone 12000, or GenScript) in 1× PBS-T with 5% skim milk overnight at 4°C. After wash in 1× PBS-T, membranes were incubated with a HRP-labeled polyclonal secondary anti–rabbit-specific Ab (Jackson ImmunoResearch Laboratories). Chemiluminescence was detected with the ECL Prime kit (PerkinElmer) using an LAS4000 imaging system (Fujifilm).
Results
Doxycycline-inducible Pax5-deficient pro-/pre-B-cell lines
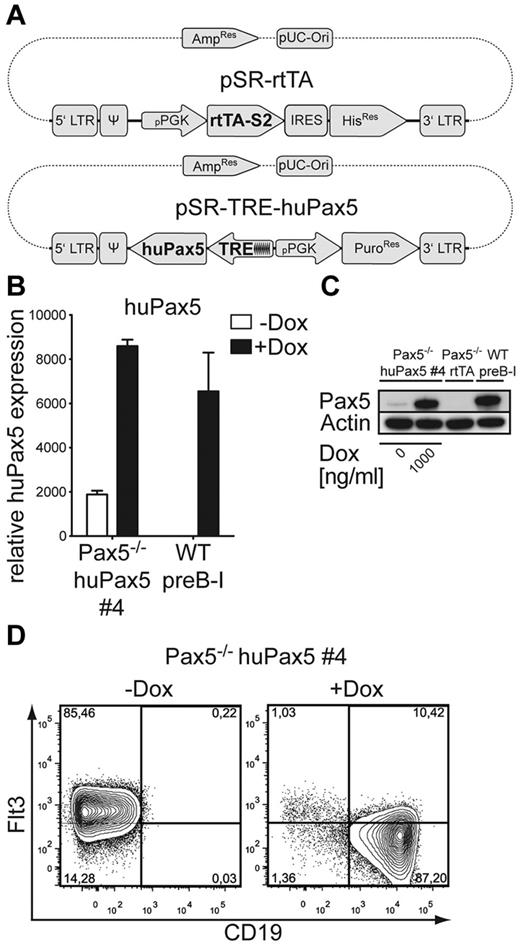
Bone marrow–derived pro-/pre-B cells were first transfected with the retroviral vector pSR-rtTA21 expressing the gene for the TetON doxycycline-sensitive rtTA (Figure 1A). The rtTA-transduced Pax5−/− cells were thereafter transfected with a self-inactivating (SIN) retroviral vector containing the human form of the Pax5 (huPax5) gene under the control of a minimal CMV promoter containing an upstream Tet-responsive element (TRE) that binds rtTA (Figure 1A). When bound by doxycycline, rtTA can bind to the CMVmin promoter and activate huPax5 expression. Several double-transgenic, that is, rtTA/huPax5-transfected, polyclonal cell lines (pools) were established, induced for 3 days with 1000 ng/mL doxycycline in IL-7/OP9 and analyzed for CD19 expression. The majority of cells of 2 individual polyclonal cell lines up-regulate CD19 expression in a doxycycline-concentration-dependent manner (not shown). Interestingly, these pools contained cells with individually different, that is, negative, low, intermediate, and high CD19 expression (not shown), suggesting that individually different huPax5 expression might be stabilized by different integration sites in the genome. To investigate the consequences of different levels of huPax5 on Pax5−/− pro-/pre-B cells, several clonal cell lines (clones 4, 16, and 20) were established from 2 pools of such rtTA-huPax5–transfected cells. Thereafter, huPax5 was induced by addition of maximal concentrations of doxycycline (1000 ng/mL) to the cultures, and were tested for doxycycline-inducible expression of huPax5 by RT-PCR (Figures 1B, 2A) and Western blot analysis (Figures 1C, 2B).
Pax5−/− huPax5 pro-/pre-B-cell clone 4 expresses CD19 and down-regulates Flt3 surface expression on doxycycline-induced huPax5 expression in vitro. (A) The retroviral huPax5-TetON-expression system. huPax5 will be transcribed after binding of the doxycycline-dependent reverse transactivator rtTA-S2 to the transactivator-dependent TRE-element containing a pCMVmin. (B) Quantitative RT-PCR for huPax5 mRNA levels in doxycycline-induced Pax5−/− huPax5 pro-/pre-B-cell clone 4 before and 3 days after doxycycline administration (1000 ng/mL) in vitro. Each bar represents the mean ± SEM (error bars) of 3 individual experiments. (C) Western blot analysis with a Pax5-specific antibody of whole cellular lysates of Pax5−/− huPax5 pro-/pre-B-cell clone 4 before and 3 days after doxycycline administration (1000 ng/mL) in vitro in comparison to wild-type pre-B-I cells and Pax5−/− rtTA pro-/pre-B cells. β-actin–specific antibody was used as a loading control. (D) Monitoring of huPax5-dependent CD19 and Flt3 expression by FACS analysis of doxycycline-induced Pax5−/− huPax5 pro-/pre-B-cell clone 4. The numbers represent percentages of Flt3+ or CD19+ pro-/pre-B cells before and 3 days after doxycycline administration (1000 ng/mL) in vitro. Data are representative for 3 individual experiments.
Pax5−/− huPax5 pro-/pre-B-cell clone 4 expresses CD19 and down-regulates Flt3 surface expression on doxycycline-induced huPax5 expression in vitro. (A) The retroviral huPax5-TetON-expression system. huPax5 will be transcribed after binding of the doxycycline-dependent reverse transactivator rtTA-S2 to the transactivator-dependent TRE-element containing a pCMVmin. (B) Quantitative RT-PCR for huPax5 mRNA levels in doxycycline-induced Pax5−/− huPax5 pro-/pre-B-cell clone 4 before and 3 days after doxycycline administration (1000 ng/mL) in vitro. Each bar represents the mean ± SEM (error bars) of 3 individual experiments. (C) Western blot analysis with a Pax5-specific antibody of whole cellular lysates of Pax5−/− huPax5 pro-/pre-B-cell clone 4 before and 3 days after doxycycline administration (1000 ng/mL) in vitro in comparison to wild-type pre-B-I cells and Pax5−/− rtTA pro-/pre-B cells. β-actin–specific antibody was used as a loading control. (D) Monitoring of huPax5-dependent CD19 and Flt3 expression by FACS analysis of doxycycline-induced Pax5−/− huPax5 pro-/pre-B-cell clone 4. The numbers represent percentages of Flt3+ or CD19+ pro-/pre-B cells before and 3 days after doxycycline administration (1000 ng/mL) in vitro. Data are representative for 3 individual experiments.
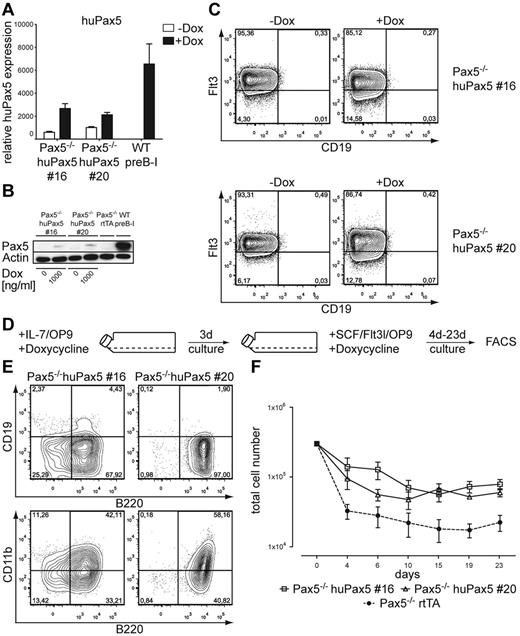
Selected Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 can be induced to develop into biphenotypic cells if they express low levels of huPax5 after high-level doxycycline induction in vitro. (A) Quantitative RT-PCR for huPax5 mRNA levels in doxycycline-induced Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 before and 3 days after doxycycline administration (1000 ng/mL) in vitro. Each bar represents the mean ± SEM (error bars) of 3 individual experiments. (B) Western blot analysis with a Pax5-specific antibody of whole cellular lysates of Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 before and 3 days after doxycycline administration (1000 ng/mL) in vitro in comparison to wild-type pre-B-I cells and Pax5−/− rtTA pro-/pre-B cells. β-actin–specific antibody was used as a loading control. (C) Monitoring of huPax5-dependent CD19 and Flt3 expression by FACS analysis of doxycycline-induced Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 (n = 3). The numbers represent percentages of Flt3+ or CD19+ pro-/pre-B cells before and 3 days after doxycycline administration (1000 ng/mL) in vitro. (D) Experimental overview: Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 were induced to express low levels of huPax5 by high levels of doxycycline (1000 ng/mL) in IL-7/OP9 for 3 days in vitro and subsequently cultivated in SCF/Flt3l/OP9 with constant high levels of doxycycline for 23 days. (E) Representative FACS analysis (n = 3) of B220, CD19, and CD11b surface expression of Pax5−/− pro-/pre-B-cell clones 16 and 20 cultivated for 4 days in SCF/Flt3l/OP9 at high levels of doxycycline (1000 ng/mL) in vitro. The numbers represent percentages of cells. B220+/CD19−/CD11b+ biphenotypic cells develop at high doxycycline concentrations (1000 ng/mL). (F) Growth curves of Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 induced to express huPax5 by high levels of doxycycline (1000 ng/mL) in IL-7/OP9 that were subsequently cultivated in SCF/Flt3l/OP9 (d0) at the same doxycycline concentration for 23 days.
Selected Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 can be induced to develop into biphenotypic cells if they express low levels of huPax5 after high-level doxycycline induction in vitro. (A) Quantitative RT-PCR for huPax5 mRNA levels in doxycycline-induced Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 before and 3 days after doxycycline administration (1000 ng/mL) in vitro. Each bar represents the mean ± SEM (error bars) of 3 individual experiments. (B) Western blot analysis with a Pax5-specific antibody of whole cellular lysates of Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 before and 3 days after doxycycline administration (1000 ng/mL) in vitro in comparison to wild-type pre-B-I cells and Pax5−/− rtTA pro-/pre-B cells. β-actin–specific antibody was used as a loading control. (C) Monitoring of huPax5-dependent CD19 and Flt3 expression by FACS analysis of doxycycline-induced Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 (n = 3). The numbers represent percentages of Flt3+ or CD19+ pro-/pre-B cells before and 3 days after doxycycline administration (1000 ng/mL) in vitro. (D) Experimental overview: Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 were induced to express low levels of huPax5 by high levels of doxycycline (1000 ng/mL) in IL-7/OP9 for 3 days in vitro and subsequently cultivated in SCF/Flt3l/OP9 with constant high levels of doxycycline for 23 days. (E) Representative FACS analysis (n = 3) of B220, CD19, and CD11b surface expression of Pax5−/− pro-/pre-B-cell clones 16 and 20 cultivated for 4 days in SCF/Flt3l/OP9 at high levels of doxycycline (1000 ng/mL) in vitro. The numbers represent percentages of cells. B220+/CD19−/CD11b+ biphenotypic cells develop at high doxycycline concentrations (1000 ng/mL). (F) Growth curves of Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 induced to express huPax5 by high levels of doxycycline (1000 ng/mL) in IL-7/OP9 that were subsequently cultivated in SCF/Flt3l/OP9 (d0) at the same doxycycline concentration for 23 days.
All clonal cell lines showed low, but significant, levels of huPax5 mRNA expression even in the absence of doxycycline, compared with control wild-type pre-B-I or Pax5−/−rtTA cells (Figures 1B, 2A). However, analysis of the doxycycline (1000 ng/mL)–induced huPax5 protein level of clone 4 displays a high expression, comparable with wild-type pre-B-I cells. Even in the absence of doxycycline, a faint, Pax5-specific protein band could be detected (Figure 1C). In contrast, treatment of clones 16 and 20 with the same high doxycycline concentration in vitro resulted in low huPax5 protein expression, comparable in levels to clone 4 cultivated without doxycycline in vitro (Figure 2B).
Phenotypes of doxycycline-induced clones
To study the effect of different levels of huPax5 on the phenotype of clones 4, 16, and 20, surface expression of Pax5-dependent genes, Flt3,9 CD19,9 c-kit, and CD11b was assayed by FACS before and 3 days after doxycycline induction. B220 and c-kit were expressed on all cells of all cell clones before and after doxycycline induction (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). CD19 was not expressed in the absence of doxycycline on any of the cell clones. In the presence of high levels of doxycycline, a large majority of the induced cells of clone 4 expressed CD19 (Figure 1D) but none of the induced cells of clones 16 and 20 did (Figure 2C). All clones (ie, nos. 4, 16, and 20) expressed Flt3 before induction (Figure 1C). After induction, clone 4 lost Flt3 expression (Figure 1D) whereas clones 16 and 20 continued to express Flt3 (Figure 2C). All cell clones did not express CD11b before or after doxycycline induction (supplemental Figure 1).
Development of biphenotypic cells by graded huPax5 expression induced by different levels of doxycycline in vitro
Because normal levels of Pax5 induce the development to CD19+/Flt3l− pre-B-I cells,2,5 we reasoned that submaximal levels of Pax5 might develop myeloid and lymphoid cells at intermediate stages of their development. We used clone 4, which is fully induced to CD19+ cells by high concentrations (1000 ng/mL) of doxycycline, and applied varying doxycycline concentrations between 0 ng/mL and 1000 ng/mL in vitro on OP9/IL7 (Figure 3A).
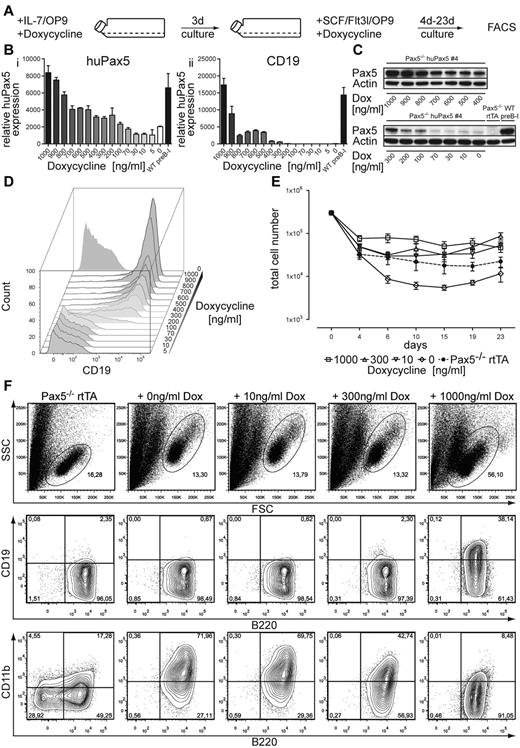
Pax5−/− huPax5 pro-/pre-B-cell clone 4 can be induced to develop into biphenotypic cells by graded huPax5 expression induced by different levels of doxycycline in vitro. (A) Experimental overview: Pax5−/− huPax5 pro-/pre-B-cell clone 4 was induced to express huPax5 by different levels of doxycycline (0-1000 ng/mL) in IL-7/OP9 for 3 days in vitro and was subsequently cultivated in SCF/Flt3l/OP9 with the same levels of doxycycline for 23 days. (B) Quantitative RT-PCR for huPax5 (i) and CD19 (ii) mRNA levels of Pax5−/− huPax5 pro-/pre-B-cell clone 4 induced by different levels of doxycycline (0-1000 ng/mL) in IL-7/OP9 in vitro. Results were normalized against GAPDH expression and plotted relative to control Pax5−/− rtTA pro-/pre-B cells. Each bar represents the mean ± SEM (error bars) of 3 individual experiments. (C) Western blot analysis with a Pax5-specific antibody of whole cellular lysates of Pax5−/− huPax5 pro-/pre-B-cell clone 4 before and 3 days after graded doxycycline administration (0-1000ng/mL) in vitro in comparison to wild-type pre-B-I cells and Pax5−/− rtTA pro-/pre-B cells. β-actin–specific antibody was used as a loading control. (D) Representative FACS analysis (n = 3) of CD19 surface expression of Pax5−/−huPax5 pro-/pre-B-cell clone 4 induced by different levels of doxycycline (0-1000 ng/mL) in vitro. CD19 surface expression was detected at doxycycline levels more than 100 ng/mL. (E) Growth curves of Pax5−/−huPax5 pro-/pre-B-cell clone 4 induced to express huPax5 by different levels of doxycycline (0, 10, 300, and 1000 ng/mL) in IL-7/OP9 that were subsequently cultivated in SCF/Flt3l/OP9 (d0) at the same doxycycline concentrations for 23 days. (F) Representative FACS analysis (n = 3) of B220, CD19, and CD11b surface expression of Pax5−/−huPax5 pro-/pre-B-cell clone 4 cultivated for 4 days in SCF/Flt3l/OP9 at different levels of doxycycline (0, 10, 300, and 1000 ng/mL) in vitro. The numbers represent percentages of cells. B220+/CD19−/CD11b+ biphenotypic cells develop at low doxycycline concentrations (0-300 ng/mL).
Pax5−/− huPax5 pro-/pre-B-cell clone 4 can be induced to develop into biphenotypic cells by graded huPax5 expression induced by different levels of doxycycline in vitro. (A) Experimental overview: Pax5−/− huPax5 pro-/pre-B-cell clone 4 was induced to express huPax5 by different levels of doxycycline (0-1000 ng/mL) in IL-7/OP9 for 3 days in vitro and was subsequently cultivated in SCF/Flt3l/OP9 with the same levels of doxycycline for 23 days. (B) Quantitative RT-PCR for huPax5 (i) and CD19 (ii) mRNA levels of Pax5−/− huPax5 pro-/pre-B-cell clone 4 induced by different levels of doxycycline (0-1000 ng/mL) in IL-7/OP9 in vitro. Results were normalized against GAPDH expression and plotted relative to control Pax5−/− rtTA pro-/pre-B cells. Each bar represents the mean ± SEM (error bars) of 3 individual experiments. (C) Western blot analysis with a Pax5-specific antibody of whole cellular lysates of Pax5−/− huPax5 pro-/pre-B-cell clone 4 before and 3 days after graded doxycycline administration (0-1000ng/mL) in vitro in comparison to wild-type pre-B-I cells and Pax5−/− rtTA pro-/pre-B cells. β-actin–specific antibody was used as a loading control. (D) Representative FACS analysis (n = 3) of CD19 surface expression of Pax5−/−huPax5 pro-/pre-B-cell clone 4 induced by different levels of doxycycline (0-1000 ng/mL) in vitro. CD19 surface expression was detected at doxycycline levels more than 100 ng/mL. (E) Growth curves of Pax5−/−huPax5 pro-/pre-B-cell clone 4 induced to express huPax5 by different levels of doxycycline (0, 10, 300, and 1000 ng/mL) in IL-7/OP9 that were subsequently cultivated in SCF/Flt3l/OP9 (d0) at the same doxycycline concentrations for 23 days. (F) Representative FACS analysis (n = 3) of B220, CD19, and CD11b surface expression of Pax5−/−huPax5 pro-/pre-B-cell clone 4 cultivated for 4 days in SCF/Flt3l/OP9 at different levels of doxycycline (0, 10, 300, and 1000 ng/mL) in vitro. The numbers represent percentages of cells. B220+/CD19−/CD11b+ biphenotypic cells develop at low doxycycline concentrations (0-300 ng/mL).
Increasing concentrations of doxycycline led to increasing levels of huPax5 mRNA and protein levels (Figure 3B-C). Parallel to this doxycycline-dose–dependent up-regulation of huPax5 levels, CD19 mRNA and protein expression increased as well (Figure 3B,D). Above 200 ng/mL doxycycline, CD19 protein expression became detectable (Figure 3D). Maximal levels on the highest numbers of induced cells were reached at 1000 ng/mL doxycycline, which is comparable with the expression level of wild-type pre-B-I cells (Figure 3B-D). Doxycycline concentrations between 0 and 200 ng/mL induced huPax5 mRNA levels of not more than one-third of the wild-type control (Figure 3B), whereas huPax5 protein levels were approximately 100 times lower than the wild-type control when stimulated with 0-70 ng/mL doxycycline. The huPax5 protein levels increased slightly at 100-200 ng/mL doxycycline, but these levels of induced huPax5 did not lead to significant CD19 expression within 3 days of culture (Figure 3B,D). At that time, the microarray analyses described in the following paragraphs were done.
We have previously studied in vitro culture conditions for progenitors with myelopoietic and lymphopoietic potencies.25 From these experiments, we reasoned that SCF and Flt3l might favor survival and/or growth of differentiating B-lymphoid progenitor cells expressing different levels of huPax5.
After replating the cells at the same low doxycycline concentrations between 0 and 300 ng/mL on SCF/Flt3l/OP9 (Figure 3A), B220+/CD19−/CD11b+ biphenotypic cells developed within the next 4 days (Figure 3E-F). For the next 19 days, these cells survived in SCF/Flt3l-containing cultures, remained biphenotypic but did not expand by proliferation (Figure 3E). Negative control Pax5−/− rtTA cells did not express detectable levels of huPax5 or CD19 mRNA or protein (Figure 3B-D) and were not able to develop a B220+/CD19−/CD11b+ phenotype (Figure 3E-F). By contrast, at high concentrations of doxycycline, clone 4 cells fully differentiated to B220+/CD19+/CD11b− pre-B-I cells, and did not develop biphenotypic cells (Figure 3E).
Note that clone 4 used in these studies expressed low background levels of huPax5 in comparison to the Pax5−/− rtTA control cells even in the absence of doxycycline (Figure 3B-C), apparently at sufficient levels to allow the huPax5-transduced cells to survive in the differentiation conditions (Figure 3E), and to exhibit the biphenotypic markers even in the absence of doxycycline induction (Figure 3F). These results suggest that very low levels of huPax5, well below those needed for the expression of CD19, appear to be sufficient to generate biphenotypic cells that can be stabilized in vitro on SCF/Flt3l/OP9.
Development of biphenotypic cells from Pax5-deficient progenitors expressing low levels of huPax5 after induction with high levels of doxycycline in vitro
Because we had found that the levels of uninduced or doxycycline-induced huPax5 expression appeared to determine the development of biphenotypic cells, we reasoned that clones 16 and 20 that expressed only low levels of Pax5 should be able to develop biphenotypic cells even at high concentrations of doxycycline in culture. The levels of huPax5 mRNA expression of clones 16 and 20 induced by doxycycline at high concentrations (1000 ng/mL, Figure 2A) were comparable with those in clone 4 cultured with 0 and 300 ng/mL (Figure 3B). Furthermore, Western blot analysis of huPax5 levels detected in clones 16 and 20 cultured under high doxycycline concentrations (Figure 2B) were comparable with those of clone 4 stimulated with low levels of doxycycline (Figure 3C).
These results suggest that submaximal huPax5 expression can be stabilized by retroviral integration at selected sites in the genome. We reasoned that such clones should be able to develop biphenotypic cells, even at high doxycycline concentrations
To test this hypothesis, we performed an in vitro differentiation experiment of clones 16 and 20 cultured with high doxycycline concentrations on SCF/Flt3l/OP9 (Figure 2D). B220+/CD19−/CD11b+ biphenotypic cells developed from these clones within 4 days of differentiation (Figure 2E). Again they did not expand by proliferation, but survived for the next 19 days on SCF/Flt3l/OP9 and remained biphenotypic (Figure 2F).
Lack of in vitro proliferation of biphenotypic cells in cytokine environments favoring either myeloid, T-lymphoid, or B-lymphoid cells
The development of biphenotypic cells from Pax5-deficient pro-/pre-B cells expressing low levels of huPax5 in vitro led us to the hypothesis that these biphenotypic cells may have the capacity to proliferate, and possibly differentiate in cytokine environments favoring either myeloid (ie, OP9/M-CSF, OP9/GM-CSF, or OP9/IL-3), T-lymphoid (ie, OP9-Δl-1/Flt3l/IL7), or B-lymphoid (ie, OP9/Flt3l/IL-7) cell development.
To test this, we induced the development of biphenotypic cells from clone 4 (see conditions described in “Development of biphenotypic cells by graded huPax5 expression induced by different levels of doxycycline in vitro”). Biphenotypic cells were then replated into cytokine environments favoring either T-, B-lymphoid, or myeloid cell development, in the presence of 0, 10, and 300 ng/mL doxycycline. We monitored the proliferation and surface marker expression of the biphenotypic cells for the next 24 days of culture in these different conditions. As shown in supplemental Figure 2, we could only observe a proliferative expansion of cells in the presence of IL-7, which favors B-lymphoid (OP9) or T-lymphoid (OP9-Δl-1) development. Under lymphoid culture conditions, biphenotypic cells proliferated as B220+ cells and lost their B220+/CD11b+ biphenotypic expression (not shown). In contrast, all conditions favoring myeloid cell development did not lead to a proliferative expansion of biphenotypic cells, or any other myeloid cell type (supplemental Figure 2).
In conclusion, we did not find tissue-culture conditions that would have allowed not only an increased survival, but a proliferative expansion of these biphenotypic cells, or of cells differentiating from them along the myeloid pathway of differentiation.
In vivo induction of huPax5 in transplanted huPax5-transduced cell clones
Because doxycycline can be fed with the drinking water, the induction of huPax5-transduced Pax5−/− pro-/pre-B-cell lines and clones (all Ly5.2+) along the B-lineage pathway of differentiation can also be monitored in vivo with cells transplanted into Rag-2−/−/Ly5.1+ recipient mice. One parameter of this differentiation in vivo is a loss of the homing properties of the transplanted Pax5−/− cells to the bone marrow. The feeding of mice with doxycycline can only be controlled with the highest concentrations, that is, at 0.2 g/L, which is equivalent in its inductive effect of full CD19 expression to 1000 ng/mL doxycycline in vitro.
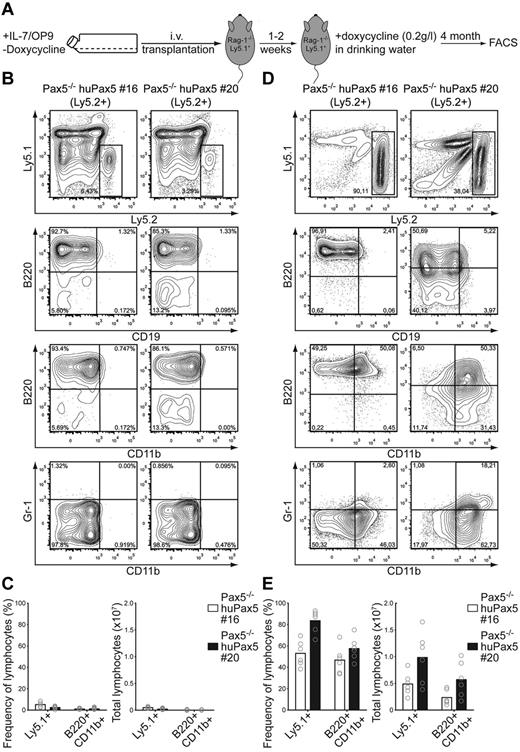
Transplantation of clones 4, 16, and 20 (all Ly5.2+) into Rag-2−/−/Ly5.1+ recipient mice in the absence of doxycycline allowed homing of the Pax5−/− cells into bone marrow, reisolatable 2 weeks after transplantation as puromycin-resistant cells proliferating on OP9/IL7 (not shown). These bone marrow reconstitution experiments showed that approximately 5% of total bone marrow lymphocytes were reconstituted with Ly5.2+ cells, that is, to approximately 1 × 105 total cells (Figure 4A-C). Furthermore, almost all Ly5.2+ cells showed a B220+/CD19−/CD11b−/Gr-1− phenotype (Figure 4B).
Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 can be induced to develop into biphenotypic cells if they express low levels of huPax5 after high-level doxycycline induction in vivo. (A) Experimental overview: Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 (all Ly5.2+) were transplanted into sublethal γ-irradiated Rag2−/−/Ly5.1+ hosts that were fed with doxycycline (0.2 g/L) in the drinking water 1 week after transplantation for the next 15 weeks. (B) Representative FACS analysis of 2 individual experiments (n = 3) of Ly5.2+ bone marrow cells for B220, CD19, and CD11b surface expression 2 weeks after transplantation. The numbers represent percentages of cells. Almost no Ly5.1+ and B220+/CD19−/CD11b+ biphenotypic cells were found 2 weeks after transplantation in the bone marrow of Rag2−/−/Ly5.1+ hosts. (C) Summary of the frequencies and total cell numbers of Ly5.1+ and B220+/CD19−/CD11b+ biphenotypic cells 2 weeks after transplantation detected in the bone marrow of Rag2−/−/Ly5.1+ hosts. Circles represent individual mice. (D) Representative FACS analysis of 2 individual experiments (n = 3) of Ly5.2+ bone marrow cells for B220, CD19, and CD11b surface expression 2 weeks after transplantation. The numbers represent percentages of cells. High numbers of Ly5.1+ and B220+/CD19−/CD11b+ biphenotypic cells were found 16 weeks after transplantation in the bone marrow of Rag2−/−/Ly5.1+ hosts. (E) Summary of the frequencies and total cell numbers of Ly5.1+ and B220+/CD19−/CD11b+ biphenotypic cells 16 weeks after transplantation detected in the bone marrow of Rag2−/−/Ly5.1+ hosts. Circles represent individual mice.
Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 can be induced to develop into biphenotypic cells if they express low levels of huPax5 after high-level doxycycline induction in vivo. (A) Experimental overview: Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 (all Ly5.2+) were transplanted into sublethal γ-irradiated Rag2−/−/Ly5.1+ hosts that were fed with doxycycline (0.2 g/L) in the drinking water 1 week after transplantation for the next 15 weeks. (B) Representative FACS analysis of 2 individual experiments (n = 3) of Ly5.2+ bone marrow cells for B220, CD19, and CD11b surface expression 2 weeks after transplantation. The numbers represent percentages of cells. Almost no Ly5.1+ and B220+/CD19−/CD11b+ biphenotypic cells were found 2 weeks after transplantation in the bone marrow of Rag2−/−/Ly5.1+ hosts. (C) Summary of the frequencies and total cell numbers of Ly5.1+ and B220+/CD19−/CD11b+ biphenotypic cells 2 weeks after transplantation detected in the bone marrow of Rag2−/−/Ly5.1+ hosts. Circles represent individual mice. (D) Representative FACS analysis of 2 individual experiments (n = 3) of Ly5.2+ bone marrow cells for B220, CD19, and CD11b surface expression 2 weeks after transplantation. The numbers represent percentages of cells. High numbers of Ly5.1+ and B220+/CD19−/CD11b+ biphenotypic cells were found 16 weeks after transplantation in the bone marrow of Rag2−/−/Ly5.1+ hosts. (E) Summary of the frequencies and total cell numbers of Ly5.1+ and B220+/CD19−/CD11b+ biphenotypic cells 16 weeks after transplantation detected in the bone marrow of Rag2−/−/Ly5.1+ hosts. Circles represent individual mice.
Transplantation of clone 4 (Ly5.2+) into doxycycline-fed Rag-2−/−/Ly5.1+ mice and subsequent in vitro culture of bone marrow cells of the Rag-2−/−/Ly5.1+ recipients 16 weeks after transplantation did not recover any donor-derived cells as puromycin-resistant cells capable of in vitro proliferation on OP9/IL7 (not shown).
Transplantation of clones 16 or 20 (both Ly5.2+) into doxycycline-fed Rag-2−/−/Ly5.1+ mice (Figure 4A) allowed the isolation of puromycin-resistant cells from bone marrow 4 months after transplantation (not shown). The bone marrow of the reconstituted mice exhibited a dramatic expansion of the Ly5.2+ cell population of approximately 50% to 90% of all bone marrow lymphocytes, which is equivalent to approximately 5 × 106 to 1 × 107 total cells (Figure 4D-E). Interestingly, approximately 50% of the Ly5.2+ cell population was B220+/CD19−/CD11b+/Gr-1− (Figure 4D), hence, showed the biphenotype also seen in an in vitro differentiation experiment on OP9/SCF/Flt3l with high-dose doxycycline induction (Figure 2E).
From these experiments, we conclude that doxycycline-induced huPax5 expression from retroviral integration sites allowing only submaximal expression levels of huPax5 stabilizes a highly proliferative biphenotypic cell stage in vivo.
Our observation that the abundance of Pax5 is critical for the stabilization of biphenotypic cells in vivo offers the possibility that we can find in the bone marrow of Pax5+/− mice accumulated B220+/CD19−/CD11b+ cells. However, bone marrow of adult wild-type (Pax5+/+) and heterozygous (Pax5+/−) mice have almost no B220+/CD19−/CD11b+ biphenotypic cells (supplemental Figure 3). It should be noted that Pax5+/− and Pax5+/+ mice develop comparable numbers of B220+/CD19+/CD11b− B lymphocytes in the bone marrow (supplemental Figure 3) and secondary lymphoid organs.11,20,26 Hence, Pax5 expression levels in progenitor and mature B lymphocytes of Pax5+/− mice are sufficient to induce CD19 expression. Because our results show that B220+/CD19−/CD11b+ biphenotypic cells develop from cells that express huPax5 at low levels that are insufficient to induce CD19 surface expression, it is reasonable to conclude that Pax5 levels in Pax5+/− mice are too high to allow the development and stabilization of biphenotypic cells.
Concomitant transcription of myeloid- and lymphoid-related genes in Pax5−/−-rtTA-huPax5–transduced cells induced by low levels of doxycycline
To study the repression and activation of genes induced by different levels of huPax5 through different concentrations of doxycycline, gene expression profiling was done either with Pax5−/− rtTA cells not transduced with huPax5, or with clone 4, transduced with huPax5 and induced to different levels of huPax5 by 0, 10, 300, or 1000 ng/mL doxycycline for 3 days in IL-7/OP9. Table 1 summarizes a collection of B-lymphocyte lineage-related genes previously detected in analyses performed on similar stages of B-cell development from Pax5−/− pro-/pre-B cells to more mature forms by Schebesta et al.6 The comparison of this previous analysis with the expression profiles obtained with our huPax5-transduced clone 4 at high (1000 ng/mL) levels of doxycycline documents a widespread agreement in the detected genes (supplemental Table 1). Table 2 depicts characteristic examples, showing that CD19, CD79α, and Blnk as B lineage–related genes are up-regulated, while Notch-1 as a T cell–related gene, Id2 as a NK lineage–related gene, and Csf1r as a myeloid lineage–related gene are down-regulated.
Summary of a collection of B-lymphocyte lineage-related genes
| Function and gene(s) . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Secreted proteins . | Cell-surface receptors and adhesion proteins . | Signal transducers . | Transporters and channels . | Transcription factors and nuclear proteins . | Cell cycle and proliferation . | Protein turnover and proliferation . | Protein trafficking and secretion . | Cytoskeleton-associated proteins . | Cellular metabolism . | Other functions . |
| Dkk3 | Enpep | Blnk | Atp1b1 | Ifi209 | Caprin1 | Uchl1 | Cplx2 | Mylip | Gldc | Bcl2l1 |
| CD19 | Bcar3 | Bach2 | Btg1 | Ifi30 | Dlgh3 | Fhod3 | Mgst1 | |||
| Bst1 | Prkd2 | Klf2 | Ccnd3 | Capn2 | Gsn | Akp2 | ||||
| VpreB3 | T2bp | E2f2 | Otub2 | Myh10 | Tpstl | |||||
| Cd79a | Dusp | Arntl | ||||||||
| Siglecg | Nedd9 | Id3 | ||||||||
| Slamf6 | Pea15 | Irf8 | ||||||||
| Igk | Eps8 | Lef1 | ||||||||
| Ms4a6c | Pard3 | Tcf712 | ||||||||
| Lphn2 | Rapgef5 | Rb1 | ||||||||
| Cd97 | Lcp2 | Ebf1 | ||||||||
| Tnfrsf19 | Prkcb1 | Irf4 | ||||||||
| Cd44 | Sit1 | Ikzf3 | ||||||||
| Edg1 | Plekha2 | Spib | ||||||||
| Slamf7 | ||||||||||
| Sdc4 | ||||||||||
| H2-Ob | ||||||||||
| Igh-VJ558 | ||||||||||
| Tnfrrsf13c | ||||||||||
| CD55 | ||||||||||
| Function and gene(s) . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Secreted proteins . | Cell-surface receptors and adhesion proteins . | Signal transducers . | Transporters and channels . | Transcription factors and nuclear proteins . | Cell cycle and proliferation . | Protein turnover and proliferation . | Protein trafficking and secretion . | Cytoskeleton-associated proteins . | Cellular metabolism . | Other functions . |
| Dkk3 | Enpep | Blnk | Atp1b1 | Ifi209 | Caprin1 | Uchl1 | Cplx2 | Mylip | Gldc | Bcl2l1 |
| CD19 | Bcar3 | Bach2 | Btg1 | Ifi30 | Dlgh3 | Fhod3 | Mgst1 | |||
| Bst1 | Prkd2 | Klf2 | Ccnd3 | Capn2 | Gsn | Akp2 | ||||
| VpreB3 | T2bp | E2f2 | Otub2 | Myh10 | Tpstl | |||||
| Cd79a | Dusp | Arntl | ||||||||
| Siglecg | Nedd9 | Id3 | ||||||||
| Slamf6 | Pea15 | Irf8 | ||||||||
| Igk | Eps8 | Lef1 | ||||||||
| Ms4a6c | Pard3 | Tcf712 | ||||||||
| Lphn2 | Rapgef5 | Rb1 | ||||||||
| Cd97 | Lcp2 | Ebf1 | ||||||||
| Tnfrsf19 | Prkcb1 | Irf4 | ||||||||
| Cd44 | Sit1 | Ikzf3 | ||||||||
| Edg1 | Plekha2 | Spib | ||||||||
| Slamf7 | ||||||||||
| Sdc4 | ||||||||||
| H2-Ob | ||||||||||
| Igh-VJ558 | ||||||||||
| Tnfrrsf13c | ||||||||||
| CD55 | ||||||||||
Summary of a collection of B-lymphocyte lineage-related genes previously detected in analyses performed on similar stages of B-cell development from Pax5−/− pro-/pre-B cells to more mature forms by Schebesta et al.6
Prime candidate genes for the determination and stabilization of the B-lymphoid-myeloid biphenotypic state of cellular development
| Accession no. . | UniGene no. . | Description . | FC of 0 ng/mL Dox vs Pax5−/− rtTA . | FC of 0 ng/mL Dox vs 10 ng/mL Dox . | FC of 0 ng/mL Dox vs 300 ng/mL Dox . | FC of 0 ng/mL Dox vs 1000 ng/mL Dox . |
|---|---|---|---|---|---|---|
| NM_009844 | Mm.4360 | CD19 antigen | 1.20 | −1.14 | 3.98 | 8.68 |
| NM_007655 | Mm.1355 | CD79A antigen (immunoglobulin-associated alpha) | −2.26 | −1.85 | 1.15 | 3.59 |
| NM_008528 | Mm.9746 | BLNK, B-cell linker | −11.56 | −1.77 | 6.07 | 18.78 |
| NM_008714 | Mm.290610 | Notch1, Notch gene homolog 1 (Drosophila) | −2.90 | 1.01 | −2.51 | −6.96 |
| NM_010496 | Mm.34871 | Id2, inhibitor of DNA-binding 2 | −1.05 | 1.03 | −2.85 | −3.85 |
| NM_009970 | Mm.287228 | Csf2ra, Mus musculus colony-stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) | 2.78 | 2.28 | 2.20 | −1.16 |
| NM_008091 | Mm.313866 | Gata3, GATA-binding protein 3 | 2.83 | 2.02 | 1.37 | −1.01 |
| BC058161 | Mm.349667 | Cebpa, CCAAT/enhancer-binding protein (C/EBP), alpha | 1.40 | 1.70 | 1.50 | −1.44 |
| NM_007679 | Mm.347407 | Cebpd, CCAAT/enhancer-binding protein (C/EBP), delta | 1.65 | 1.87 | 1.59 | −1.39 |
| NM_008037 | Mm.24684 | Fosl2, Fos-like antigen 2 | 1.63 | 2.30 | 2.80 | −1.15 |
| NM_011355 | Mm.1302 | PU.1, Sfpi1, SFFV proviral integration 1 | −2.00 | −1.19 | −2.49 | −1.65 |
| NM_008091 | Mm.313866 | GATA3, GATA-binding protein 3 | −2.83 | −2.02 | −1.37 | −1.01 |
| NM_07779 | Mm.22574 | Csf1r, Mcsf1r (macrophage), colony-stimulating factor 1 receptor | −1.45 | −1.85 | −3.10 | −1.67 |
| Accession no. . | UniGene no. . | Description . | FC of 0 ng/mL Dox vs Pax5−/− rtTA . | FC of 0 ng/mL Dox vs 10 ng/mL Dox . | FC of 0 ng/mL Dox vs 300 ng/mL Dox . | FC of 0 ng/mL Dox vs 1000 ng/mL Dox . |
|---|---|---|---|---|---|---|
| NM_009844 | Mm.4360 | CD19 antigen | 1.20 | −1.14 | 3.98 | 8.68 |
| NM_007655 | Mm.1355 | CD79A antigen (immunoglobulin-associated alpha) | −2.26 | −1.85 | 1.15 | 3.59 |
| NM_008528 | Mm.9746 | BLNK, B-cell linker | −11.56 | −1.77 | 6.07 | 18.78 |
| NM_008714 | Mm.290610 | Notch1, Notch gene homolog 1 (Drosophila) | −2.90 | 1.01 | −2.51 | −6.96 |
| NM_010496 | Mm.34871 | Id2, inhibitor of DNA-binding 2 | −1.05 | 1.03 | −2.85 | −3.85 |
| NM_009970 | Mm.287228 | Csf2ra, Mus musculus colony-stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) | 2.78 | 2.28 | 2.20 | −1.16 |
| NM_008091 | Mm.313866 | Gata3, GATA-binding protein 3 | 2.83 | 2.02 | 1.37 | −1.01 |
| BC058161 | Mm.349667 | Cebpa, CCAAT/enhancer-binding protein (C/EBP), alpha | 1.40 | 1.70 | 1.50 | −1.44 |
| NM_007679 | Mm.347407 | Cebpd, CCAAT/enhancer-binding protein (C/EBP), delta | 1.65 | 1.87 | 1.59 | −1.39 |
| NM_008037 | Mm.24684 | Fosl2, Fos-like antigen 2 | 1.63 | 2.30 | 2.80 | −1.15 |
| NM_011355 | Mm.1302 | PU.1, Sfpi1, SFFV proviral integration 1 | −2.00 | −1.19 | −2.49 | −1.65 |
| NM_008091 | Mm.313866 | GATA3, GATA-binding protein 3 | −2.83 | −2.02 | −1.37 | −1.01 |
| NM_07779 | Mm.22574 | Csf1r, Mcsf1r (macrophage), colony-stimulating factor 1 receptor | −1.45 | −1.85 | −3.10 | −1.67 |
Microarray analysis was performed on total RNA extracted from Pax5−/− huPax5 pro-/pre-B-cell clone 4, induced to express huPax5 at different levels of doxycycline (0, 10, 300, and 1000 ng/mL) in IL-7/OP9 for 3 days in vitro. Presented are characteristic examples for the differential expression of B-, T-, NK-, and myeloid-lineage–related genes at different levels of doxycycline.
FC indicates fold change; Dox, doxycycline; and rtTA, retroviral transactivator.
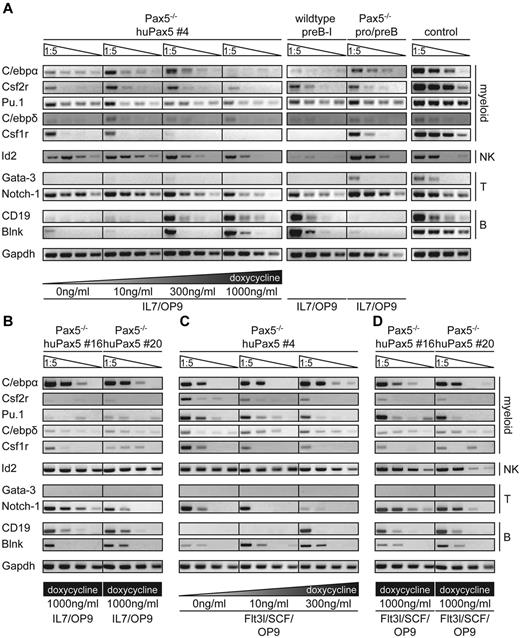
Importantly, however, the myeloid-related genes C/ebpα, C/ebpδ, Gata-3, FosL-2, and Csf2r remained up-regulated between 0 and 300 ng/mL doxycycline, only to be down-regulated at the highest (1000 ng/mL) doxycycline concentration. These microarray analyses were confirmed by semiquantitative RT-PCR for C/ebpα, C/ebpδ, Pu.1, Csf2r, Csf1r, Gata-3, Notch-1, CD19, CD79α, and Blnk (Figure 5A).
Coexpression of genes characteristic for both lymphoid and myeloid cells in Pax5−/− huPax5 pro-/pre-B-cell clones 4, 16, and 20 before and on induction with low levels of doxycycline in vitro. (A) Pax5−/− huPax5 pro-/pre-B-cell clone 4 was induced to express huPax5 by different levels of doxycycline (0, 10, 300, and 1000 ng/mL) in IL-7/OP9 for 3 days in vitro. Semiquantitative RT-PCR analysis for the expression of lymphoid (Id2, Gata-3, Notch-1, CD19, and Blnk) and myeloid (C/ebpα, Csf2r, Pu.1, C/ebpδ, and Csf1r) related genes was performed on total RNA extracted after 3 days of conditional huPax5 expression at low and high levels of doxycycline. At low levels of doxycycline stimulation (0-300 ng/mL), the expression of lymphoid (Notch-I, Id-2) as well as myeloid (C/ebpα, Csf2r, Pu.1, and Csf1r) related genes can be observed. However, at high levels of doxycycline stimulation (1000 ng/mL) especially, B-lineage–specific genes are expressed and myeloid-related genes are suppressed. (B) Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 were induced to express huPax5 by 1000 ng/mL doxycycline in IL-7/OP9 for 3 days in vitro. Gene transcription levels are comparable with clone 4 stimulated with low doxycycline levels. (C-D) Analysis for FACS-sorted B220+/CD19−/CD11b+ biphenotypic cells on day 4 of differentiation in Flt3l/SCF/OP9 conditions stimulated with 0, 10, or 300 ng/mL doxycycline (clone 4) or 1000 ng/mL doxycycline (clone 16 and 20). Data are representative of 3 individual experiments.
Coexpression of genes characteristic for both lymphoid and myeloid cells in Pax5−/− huPax5 pro-/pre-B-cell clones 4, 16, and 20 before and on induction with low levels of doxycycline in vitro. (A) Pax5−/− huPax5 pro-/pre-B-cell clone 4 was induced to express huPax5 by different levels of doxycycline (0, 10, 300, and 1000 ng/mL) in IL-7/OP9 for 3 days in vitro. Semiquantitative RT-PCR analysis for the expression of lymphoid (Id2, Gata-3, Notch-1, CD19, and Blnk) and myeloid (C/ebpα, Csf2r, Pu.1, C/ebpδ, and Csf1r) related genes was performed on total RNA extracted after 3 days of conditional huPax5 expression at low and high levels of doxycycline. At low levels of doxycycline stimulation (0-300 ng/mL), the expression of lymphoid (Notch-I, Id-2) as well as myeloid (C/ebpα, Csf2r, Pu.1, and Csf1r) related genes can be observed. However, at high levels of doxycycline stimulation (1000 ng/mL) especially, B-lineage–specific genes are expressed and myeloid-related genes are suppressed. (B) Pax5−/− huPax5 pro-/pre-B-cell clones 16 and 20 were induced to express huPax5 by 1000 ng/mL doxycycline in IL-7/OP9 for 3 days in vitro. Gene transcription levels are comparable with clone 4 stimulated with low doxycycline levels. (C-D) Analysis for FACS-sorted B220+/CD19−/CD11b+ biphenotypic cells on day 4 of differentiation in Flt3l/SCF/OP9 conditions stimulated with 0, 10, or 300 ng/mL doxycycline (clone 4) or 1000 ng/mL doxycycline (clone 16 and 20). Data are representative of 3 individual experiments.
Semiquantitative RT-PCR for the same set of lineage marker genes of clones 16 and 20 (3 days in IL-7/OP9 + 1000 ng/mL doxycycline) showed an almost comparable gene expression profile to clone 4 induced to huPax5 expression with low levels of doxycycline (Figure 5B).
Therefore, these genes are prime candidates for the determination and stabilization of the B-lymphoid-myeloid biphenotypic state of cellular development. To further support this hypothesis, we have analyzed FACS-purified biphenotypic cells generated within 4 days from clone 4 (+0, 10, and 300 ng/mL doxycycline) as well as from clones 16 and 20 (+1000 ng/mL doxycycline). Semiquantitative RT-PCRs were performed for the set of lineage marker genes that we had previously tested (Figure 5C-D). Interestingly, almost the same RT-PCR expression patterns were observed, as the ones obtained with the unpurified mixture of clone 4 cells induced to huPax5 expression with low levels of doxycycline on IL-7/OP9 (Figure 5C-D).
In conclusion, these findings suggest that concomitant transcription of myeloid- and lymphoid-related genes in Pax5−/− rtTA-huPax5 biphenotypic cells is induced by low levels of huPax5. This expression pattern can, in fact, be stabilized in a cytokine milieu favoring lymphopoiesis as well as myelopoiesis.
Discussion
During development of B lymphocytes from hematopoietic stem cells, the transcription factor Pax5 is essential for committing common lymphoid progenitors, the precursors of B-lymphoid, T-lymphoid, NK, and dendritic cells to enter the B-lymphocyte pathway.27 Pax5 exerts a dual role by activating the expression of B cell–specific genes6 and by repressing B-lymphocyte–inappropriate genes, such as those specific for T lymphocyte–, NK cell–, or myeloid cell–specific functions.8 Our differential microarray expression and semiquantitative RT-PCR analyses of Pax5−/− pro-/pre-B-cell clones and with the same clones induced by doxycycline-dependent, high-level expression of huPax5 and of CD19 confirm these analyses. We add here the differential microarray and semiquantitative RT-PCR analyses of the same clones induced by low doxycycline concentrations to low levels of huPax5 expression levels that do not allow expression of CD19.
While many of the genes that are up-regulated, respectively, down-regulated by high huPax5 expression are already on their expected way of change in expression, a selected group of genes remain expressed, or are even up-regulated. These genes (Figure 5) are not specific for B-lymphocyte functions, but appear in majority specific for myeloid cell development and function. We take this as indication that they are involved in the development of the B-lymphoid/myeloid B220+/CD11b+/CD19− biphenotypic cells that we observe at this level of huPax5 expression both in vitro as well as in vivo. The capacity of these biphenotypic cells to develop myeloid cells needs to be investigated in greater detail in future experiments. However, it is already evident that they might not be potent as precursors for all myeloid cell types because the microarray and semiquantitative RT-PCR analyses show that granulocyte-macrophage Csf2 receptor keeps being up-regulated, but the macrophage Csf1 receptor is down-regulated.
The gene encoding the transcription factor C/ebpα, one of the genes selectively up-regulated in our biphenotypic cells has been found to be essential for granulocyte development and is expressed in immature myeloid cells.28,29 It appears to inhibit monocyte formation, an observation that could be in line with the possibility that not all myeloid lineage cells might develop from our biphenotypic cells.28 Transduction of C/ebpα, together with Pu.1, has been found to reprogram fibroblasts to macrophage-like cells,30 again suggesting that the expression of C/ebpα is essential for the development of some of the myeloid cells.
B-lymphoid/myeloid biphenotypic cells with the capacity to develop to B cells or myeloid cells have been repeatedly detected. Common lymphoid progenitors31 as well as CD4+/B220+/CD19−/CD93+ progenitors from bone marrow32 can develop dendritic cells, indicating that this myeloid cell type can be generated from a progenitor cell capable of pan-lymphoid B-lymphoid development. B220+/CD11b+ cells expressing also CD117, CD24, CD43, the Csf1 receptor, Rag-2, TdT and, notably, Pax5 were grown in vitro from bone marrow in the presence of Flt3 ligand. They could be developed to either macrophages or to B cells, depending on the cytokine conditions of the cultures.33 Not by phenotype, but by function, our biphenotypic cells are reminiscent of those progenitors that are seen early in fetal development, namely bipotential CD93+/B220−/CD19− cells that can differentiate either to B cells or to macrophages.34 In this report, a CD93+/B220+/CD11b+ cell was also identified where the capacity to develop to B-lymphoid–like or to macrophage-like cells was 1:4. Similar cells could be contained in bone marrow cell populations that have the capacity to develop to B-lymphoid and to myeloid cells that are CD93+/B220+/CD117+/CD19−.35,36
Our findings that low levels of huPax5 expression induce and, in the presence of SCF/Flt3l/OP9, stabilize such a lymphoid/myeloid progenitor poses the question of whether biphenotypes observed by others, for example, by ectopic expression of Pax5,37 might have resulted from low-level Pax5 expression.
CLPs, and the Pax5−/− CLP-like progenitors, as well as the Pax5-induced differentiated cells, such as CD19+ pre-B-I cells and the B220+/CD19−/CD11b+ biphenotypic cells express Rag-1 and Rag-2, hence, are genetically unstable, prone to abnormal chromosomal translocations. Therefore, if abnormal levels of Pax5 arrest progenitors at abnormal stages of development, Pax5 may have the effect of an oncogene, as seen in ALLs or acute myeloid leukemias (AMLs).38,39 Interestingly, a genome-wide screening of genetic alterations in pediatric ALLs identified copy number changes in the Pax5 gene in approximately 30% of B-progenitor ALL cases by monoallelic and biallelic loss, chromosomal translocations, and point mutations.16-18 Furthermore, several investigations reported an aberrant expression of Pax5 in nonhematopoietic tumors.40-42
Pax5 expression levels in progenitor and mature B ymphocytes of heterozygous Pax5+/− mice are sufficient to induce CD19 expression, and apparently too high to allow the stable development of biphenotypic cells. In our experiments, B220+/CD19−/CD11b+ biphenotypic cells develop from cells that express huPax5 at levels that are insufficient for CD19 expression. However, leukemias arise in CD19-CRE;Pax5fl/− mice19 as well as Stat5b-CA;Pax5+/− mice.17 Interestingly, Stat5b-CA;Pax5+/− leukemias show reduced expression of both Ebf1 and Pax5. Consequently, Ebf-1– and Pax5-regulated genes were found significantly altered in their expression, and among them were also potential tumor suppressors and oncogenes. However, we would not expect biphenotypic cells to be the target cells, in which these oncogenic transformations would take place.
Interestingly, 5% of all ALLs are defined as an uncommon subtype, called biphenotypic acute lymphoblastic leukemia (BAL). In 60%-70% of these biphenotypic ALLs, the leukemic blast cells coexpress B220 and CD11b.43-45 Translocations resulting in an in-frame fusion of the CALM and AF10 genes, detected in some of the ALLs, can cause BAL with a B220+/CD11b+/CD19− phenotype in a transgenic mouse model.46 Therefore it is conceivable, but has yet to be tested, that low-level Pax5 expression generates a B220+/CD11b+/CD19− biphenotypic progenitor during B-cell development. If low-level Pax5 expression is stabilized by an oncogenic deregulation that does not allow high-level Pax5 expression, the stabilized biphenotypic progenitor cells could become the cellular sites in which RAG-1 and RAG-2 gene activity at the stage of DH to JH rearrangements in the IgH chain locus contributes to genetic instability that increases the risk of translocations as those between the CALM and the AF10 gene.46 Our observations that abnormally proliferating biphenotypic cells develop late, but not early after transplantation into doxycycline-fed mice, allowing long periods of low level Pax5 expression in the genetically unstable cells might point to secondary transforming events. Unfortunately, our limited experimental capacities have not allowed us to search for such genomic alterations by genome-wide screens.
The results of our experiments emphasize that gene expression programs determining cell fates depend not only on the qualitative up- or down-regulation of transcription factor expression, here of huPax5, but on the level of their expression. It suggests that, like in the earliest stages of embryonic development,47 different affinities to promoter and enhancer elements of different, critically important genes regulate hematopoietic cell development. Our results with huPax5 are reminiscent of findings by Nutt et al,48 Dakic et al,49 and DeKoter and Singh50 that show differential commitment to myeloid or lymphoid cell differentiation, dependent on the concentration of the transcription factor Pu.1 expressed in hematopoietic progenitor cells.
With our in vitro and in vivo doxycycline-controllable huPax5 expression system in Pax5-deficient pro-/pre-B cells, we have developed a cell-culture system that can, in principle, be adopted to study the in vitro and in vivo effects of any protein or miRNA on central and peripheral B-cell development.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Meinrad Busslinger (Institute for Molecular Pathology, Vienna, Austria) for the Pax5-deficient mouse strain and for the cDNA of the human form of the Pax5 gene; Hermann Bujard (Center for Research and Higher Education in Molecular Biology and Biomedicine, University of Heidelberg, Germany) for reagents and advice to use the TetON-gene expression system; and Jana Winckler, Nicole Dittberner, Patricia Vegh, and Ina Wagner (all at Max Planck Institute for Infection Biology, Berlin, Germany) for skillful technical help.
Authorship
Contribution: S.S. performed most of the experiments, analyzed the data, and wrote the manuscript; M.K., C.D., and I.W. participated in some of the experiments and discussed their outcomes; H.-J.M. performed the microarray analyses; C.B. provided the retroviral expression vectors; and F.M. designed the research, supervised the experiments, and wrote and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.S. is Immunology Frontier Research Center, Laboratory of Cellular Dynamics, Osaka University, Osaka, Japan.
Correspondence: Fritz Melchers, Max Planck Institute for Infection Biology, Lymphocyte Development Group, Charitéplatz 1, 10117 Berlin, Germany, e-mail: melchers@mpiib-berlin.mpg.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal