Abstract
E-proteins are critical transcription factors in B-cell lymphopoiesis. E2A, 1 of 3 E-protein–encoding genes, is implicated in the induction of acute lymphoblastic leukemia through its involvement in the chromosomal translocation 1;19 and consequent expression of the E2A-PBX1 oncoprotein. An interaction involving a region within the N-terminal transcriptional activation domain of E2A-PBX1, termed the PCET motif, which has previously been implicated in E-protein silencing, and the KIX domain of the transcriptional coactivator CBP/p300, critical for leukemogenesis. However, the structural details of this interaction remain unknown. Here we report the structure of a 1:1 complex between PCET motif peptide and the KIX domain. Residues throughout the helical PCET motif that contact the KIX domain are important for both binding KIX and bone marrow immortalization by E2A-PBX1. These results provide molecular insights into E-protein–driven differentiation of B-cells and the mechanism of E-protein silencing, and reveal the PCET/KIX interaction as a therapeutic target for E2A-PBX1–induced leukemia.
Introduction
The E-proteins are a family of class I basic helix-loop-helix (bHLH) mammalian transcription factors that function in cell lineage-specific developmental processes, including indispensable and particularly well characterized roles in lymphopoiesis.1,2 Family members include E12 and E47, which are alternatively spliced products of the E2A gene (hereafter referred to collectively as E2A), HEB, and E2-2. Each member contains a C-terminal bHLH domain responsible for protein dimerization and recognition of the E-box DNA sequence CANNTG as well as 2 activation domains, AD1 and AD2, which function either independently or cooperatively depending on cell type to regulate target gene transcription through the recruitment of transcriptional coactivators and corepressors.1,3-5
A highly conserved 17-residue region within AD1 at the N-terminus of E-proteins is the target of the eTAFH domain from the transcriptional corepressor ETO and the oncogenic fusion protein AML-ETO.6-9 This interaction leads to E-protein silencing by preventing the binding of the transcriptional coactivators p300/CBP to the same site on AD1, which has been termed the “p300/CBP and ETO target in E-proteins” (PCET) motif.9 The PCET motif is composed of 2 overlapping protein recognition sites: a Leu-x-x-Leu-Leu (LXXLL) sequence and a Leu-Asp-Phe-Ser (LDFS) sequence. The LXXLL sequence and the more generic φ-x-x-φ-φ sequence, where φ is a bulky hydrophobic residue and x is any amino acid residue, have been identified in several transcription factors, including CREB, c-Myb, MLL, p53, and E2A,10,11 and shown to bind the KIX domain of CBP/p300 at 1 of 2 sites.10,12-15 The LDFS sequence of yeast bHLH transcription factors was shown to be important for recruiting the SAGA histone acetyltransferase complex,16 and more recently its importance in E2A recruitment of human histone acetyltransferase complexes, such as CBP/p300, has been proposed.1,17,18
Among the E-proteins, E2A is critical for the differentiation of B-lymphoid progenitors through direct activation of multiple target genes, primarily EBF,19,20 with E2A−/− mice displaying a blockage at the earliest stages of B-lymphoid specification.21 Given the importance of E2A in lymphoid differentiation, it is perhaps not surprising that in 4%-12% of pediatric acute lymphoblastic leukemia cases, regions of the E2A and PBX1 genes are fused by a chromosomal translocation between chromosomes 1 and 19 [t(1;19)(q23;p13.3)].22,23 These leukemias predominantly show a pre-B cell immunophenotype24 and are associated with expression of the proto-oncogene product E2A-PBX1.25 This chimeric protein includes a large N-terminal region of E2A containing both AD1 and AD2 fused with most of PBX1, including its C-terminal DNA-binding homeodomain. Although the exact molecular mechanisms by which E2A-PBX1 contribute to the leukemic transformation of B-lymphoid progenitor cells remain uncertain, 2 potential mechanisms are consistent with the literature: recruitment of transcriptional coregulators to target gene loci bound directly by the E2A-PBX1 homeodomain and/or sequestration of coregulators away from wild-type E-proteins and other factors by E2A-PBX1.5
Both of these general mechanisms invoke a critical role for AD1 of E2A-PBX1 in recruiting transcriptional coregulators. Consistent with either of these scenarios, AD1 is indispensable for E2A-PBX1 mediated oncogenesis.12,26 Furthermore, we previously demonstrated that AD1 interacts directly with the KIX domain of CBP/p300 and that this interaction is required in the immortalization of primary hematopoietic cells in vitro.12 Mutation of the common leucine residue of the overlapping LXXLL and LDFS sequences in the PCET motif of AD1 markedly impaired both KIX binding and leukemogenesis by E2A-PBX1 in a murine bone marrow transplantation model.17 A detailed structural characterization of the PCET/KIX interaction will provide molecular insights into both the critical role of E-proteins to lymphopoiesis and that of E2A-PBX1 in leukemia induction.
Here we report the 3-dimensional structure of the PCET/KIX complex. The structure revealed that residues throughout the entire PCET motif of E-proteins contacting KIX. We confirm the functional importance of these contacts in biophysical and cell-based functional analyses. In particular, mutation of these KIX-interacting residues in the context of the E2A-PBX1 oncoprotein indicated their requirement in the immortalization of primary hematopoietic cells in vitro.
Methods
NMR spectroscopy
All nuclear magnetic resonance (NMR) spectra were acquired on either a Bruker 800 MHz or Varian 600 MHz NMR spectrometer equipped with triple resonance cryoprobe at 25°C on samples prepared in 20mM MES, pH 6.0, 90% H2O/10% D2O. The NMR samples for structure determination of the HEB-PCET/KIX complex was composed of 600μM 15N/13C-labeled KIX with 1.2mM unlabeled HEB-PCET or 300μM 15N/13C-labeled HEB-PCET with 1mM unlabeled KIX. All the NMR spectra were processed and analyzed using NMRPipe27 and NMRView,28 respectively. Production and preparation of recombinant protein and peptide constructs are described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Backbone and side chain resonance assignments for 15N/13C-labeled KIX in the presence of unlabeled HEB-PCET and for 15N/13C-labeled HEB-PCET saturated with unlabeled KIX were made using standard triple resonance experiments. Distance restraints for the KIX domain were identified in 3D 15N-NOESY-HSQC (120 ms mixing time) and aliphatic (100 ms mixing time) and aromatic (120 ms mixing time) 3D 13C-NOESY-HSQC datasets and for HEB-PCET in a 3D 15N-NOESY-HSQC dataset (100 ms mixing time). Identification of intermolecular distance restraints was provided by 14N-filtered/15N-edited NOESY-HSQC (120 ms mixing time) and aliphatic (100 ms mixing time) and aromatic (120 ms mixing time) 12C-filtered/13C-edited NOESY-HSQC datasets. Steady-state {1H}-15N NOE data were collected on 15N-labeled HEB-PCET in complex with KIX and 15N-labeled KIX in complex with HEB-PCET with and without 2.5 seconds of proton saturation and with a total recycle delay of 5 seconds. {1H}-15N NOE values were calculated from the ratio of peak intensities with and without saturation for the HEB-PCET and KIX resonances.
Structure calculation
From the NOESY experiments, 1018 interproton distance restraints were manually assigned and classified based on peak intensity into upper bounds of 3.0, 5.0, or 6.0 Å. Distance restraints were calibrated from peak intensities using known distances in α-helical [dαN (i,i+3), dαN(i,i+4)] and loop regions [dNN(i,i+1)]. Backbone dihedral angle restraints were found by analysis of 13Cα, 13Cβ, 13C', 1Hα, and 15N chemical shifts using TALOS29 and applied to helical regions of the structure with the restraints restricted to ± 15° or the error from the TALOS output, whichever was greater. The program CNS 1.2 was used to generate 200 trial structures using a simulated annealing protocol.30 Structures with no interproton distance restraint violations > 0.3 Å or dihedral angle restraint violations > 5° were analyzed with MOLMOL31 to yield a final ensemble of 20 low-energy structures. The final ensemble was evaluated using PROCHECK.32 The structure coordinates have been deposited in the Protein Data Bank under accession no. 2KWF.
Mammalian 2-hybrid assay
Plasmids conferring mammalian expression of GAL4-KIX or VP16-E2A(1-483) fusion proteins were assembled as previously described,12 with E2A mutations generated using the QuikChange site directed mutagenesis kit (Stratagene). The assay was performed in 293T cells as previously described,17 except for the quantities of plasmid DNA used (0.01 μg/well pCMV-GAL4 construct, 0.01 μg/well pCMV-2xVP16 construct, 0.7 μg/well p5xGAL luciferase reporter, and 0.1 μg/well pCMV-Renilla internal control). Statistical significance was measured using a one-way ANOVA with Tukey posthoc test.
Coimmunoprecipitation
Log-phase 293T cells were transfected in 100-mm tissue culture dishes with 10 μg each of plasmids that confer expression of GAL4-E2A fusion proteins and FLAG-tagged, full-length CBP using the calcium phosphate method and expression vectors described previously.12,33 Two days later, the cells were detached from the dishes by trypsinization, washed twice in PBS solution, resuspended in 1 mL of lysis buffer (0.5% NP-40, 150mM NaCl, 50mM Tris, pH 7.4, and protease inhibitors) and incubated on ice for 10 minutes. Cell suspensions were sonicated with 5 one-second pulses and then centrifuged to remove particulate matter. A total of 200-400 μL of cell lysate was used for each immunoprecipitation reaction depending on the relative abundance of transfected protein. The volume was made up to 500 μL with lysis buffer, 2 μg of anti-FLAG antibody (M2, Sigma-Aldrich) was added, and the tubes were rocked overnight at 4°C. A total of 20 μL of A/G agarose beads (A/G agarose PLUS; Santa Cruz Biotechnology) was added, and the suspension was incubated at 4°C for 2 hours. The beads were then washed 5 times in lysis buffer and resuspended in 50 μL of 2× protein sample buffer containing SDS and DTT. The suspension was then boiled and centrifuged, and 25 μL was loaded onto an 8% polyacrylamide gel for immunoblotting.
Retroviral transduction and bone marrow immortalization
The cDNA encoding E2A-PBX1b was engineered into a GFP-expressing pMIEV retroviral backbone plasmid using NotI and SalI restriction sites. Generation of Glu15Ala, Leu16Ala, Asp21Ala, and Asp21Ala/Phe22Ala E2A-PBCX1b mutants was performed using a previously described PCR-based method34 with Pfu Ultra DNA polymerase from Stratagene. To generate virus, pMIEV and MCV-ecopac (ectopic packaging plasmid) were cotransfected into 293T cells via calcium phosphate precipitation. Virus was secreted into IMDM/15% FBS at 32°C for 1 day. Viral supernatant was filter sterilized, frozen in liquid nitrogen, and stored at −80°C. The titer of the virus was measured by infecting NIH 3T3 fibroblasts with a serial dilution series of viral supernatant and assessing GFP and E2A-PBX1b expression by flow cytometry of live cells and immunoblotting of cell lysates,12 where the secondary antibodies used were a 1/2000 dilution of anti-E2A yae mouse monoclonal antibody (Santa Cruz Biotechnology) and a 1/2000 dilution of mouse monoclonal anti-GFP (Roche Diagnostics).
To assay immortalization, bone marrow was harvested from the femurs of 6- to 8-week-old female BALB/c mice that had been given 2 mg of 5-fluorouracil 4 days earlier. Whole bone marrow was resuspended in ACK lysis buffer to lyse the erythrocytes, and the remaining cells were resuspended in prestimulation media (IMDM, 20% FBS, 2mM l-glutamine, 140μM β-mercaptoethanol, 10 ng/mL IL-3, 10 ng/mL IL-6, 100 ng/mL murine stem cell factor, 100 U/mL penicillin G, 100 μg/mL streptomycin, 250 ng/mL amphotericin B, and 50 μg/mL gentamicin). Cells were cultured in prestimulation media in a humidified 5% CO2 atmosphere at 37°C for 2 days and split into groups of 3 × 105 cells in 1 mL each of an appropriate dilution of retroviral supernatant based on the titer of the virus. Samples were incubated on ice with 5 μg/mL polybrene for 10 minutes, transferred into a 24-well tissue culture plate, and spin infected at 1140g for 2 hours at 28°C. Cells were resuspended in myeloid stimulation media (same as prestimulation media, except FBS is at 10% and the cytokines are replaced with 10 ng/mL GM-CSF), grown at 37°C, and counted and refed every 3 or 4 days for 35 days after transduction. On day 6, a sample of each transduction group was analyzed by flow cytometry to confirm the emergence of a small subpopulation of GFP+ cells.
Results
Structure of the PCET/KIX complex
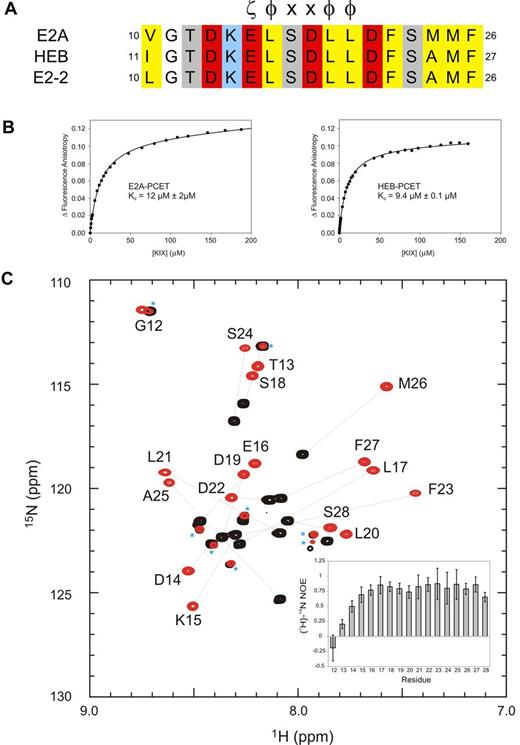
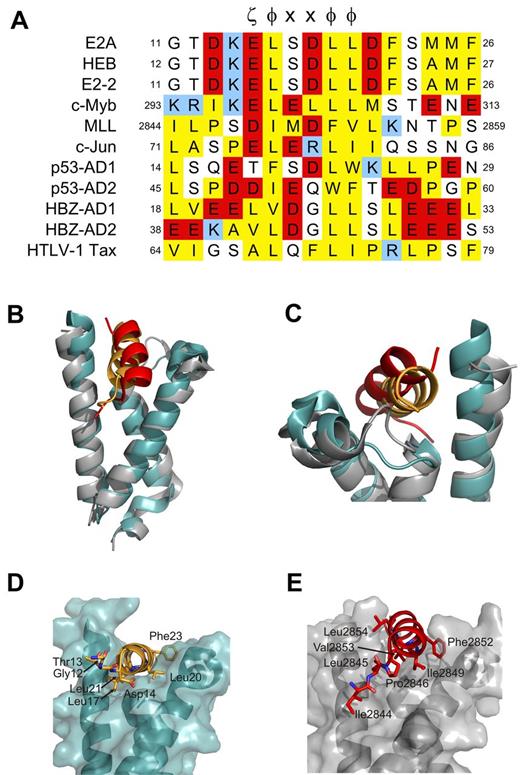
The highly conserved PCET motif within activation domain 1 of E-proteins (residues 10-26 of E2A and E2-2, residues 11-27 of HEB; Figure 1A) plays fundamental roles in E-protein–mediated lymphopoiesis and leukemogenesis. In part, this occurs through recruitment of the coactivator CBP/p300 via the CBP/p300 KIX domain.9,12,35 Fluorescence anisotropy experiments indicated that the PCET motifs from E2A and HEB bind KIX with very similar affinities (dissociation constants [Kd] of 12μM and 9μM, respectively; Figure 1B; supplemental Methods). Given this observation, and our ability to produce milligram quantities of isotopically labeled HEB-PCET (residues 11-28) in a bacterial system, we chose HEB as the representative E-protein PCET motif for use in NMR structural studies. The 1H-15N HSQC spectrum of HEB-PCET displayed poor dispersion of HN resonances, indicative of an intrinsically disordered peptide (Figure 1C). Addition of CBP KIX to saturating concentrations increased the dispersion of resonances, with analysis of backbone chemical shifts36 predicting a helical conformation from Lys15 to Phe27.
The conserved PCET motifs of E2A and HEB bind the KIX domain of CBP/p300. (A) Sequence alignment of the PCET motifs from E2A, HEB, and E2-2. The ζ-φ-x-x-φ-φ consensus sequence is indicated, and numbering is in accordance to the native protein sequence. ζ indicates an acidic amino acid; φ, a bulky hydrophobic amino acid; and x, any amino acid. Conserved hydrophobic amino acid residues are colored yellow, acid residues red, and basic residues blue. (B) Representative binding curves showing the change in fluorescence anisotropy of 5-carboxyfluorescein tagged E2A-PCET (12-25) and HEB-PCET (11-24) peptides with increasing total concentration of KIX. (C) Overlay of 1H-15N HSQC spectra of 0.4mM 15N-labeled HEB-PCET in the absence (black) and presence (red) of 0.75mM KIX in 20mM MES, pH 6.0. Resonances are labeled by position in full-length HEB. Lines connect the resonances of free PCET (black) and the corresponding resonance when bound to KIX (red). Asterisks identify resonances that arise from impurities in the sample. Inset: Plot of {1H}-15N NOE values as a function of HEB-PCET residue number at 18.8 T.
The conserved PCET motifs of E2A and HEB bind the KIX domain of CBP/p300. (A) Sequence alignment of the PCET motifs from E2A, HEB, and E2-2. The ζ-φ-x-x-φ-φ consensus sequence is indicated, and numbering is in accordance to the native protein sequence. ζ indicates an acidic amino acid; φ, a bulky hydrophobic amino acid; and x, any amino acid. Conserved hydrophobic amino acid residues are colored yellow, acid residues red, and basic residues blue. (B) Representative binding curves showing the change in fluorescence anisotropy of 5-carboxyfluorescein tagged E2A-PCET (12-25) and HEB-PCET (11-24) peptides with increasing total concentration of KIX. (C) Overlay of 1H-15N HSQC spectra of 0.4mM 15N-labeled HEB-PCET in the absence (black) and presence (red) of 0.75mM KIX in 20mM MES, pH 6.0. Resonances are labeled by position in full-length HEB. Lines connect the resonances of free PCET (black) and the corresponding resonance when bound to KIX (red). Asterisks identify resonances that arise from impurities in the sample. Inset: Plot of {1H}-15N NOE values as a function of HEB-PCET residue number at 18.8 T.
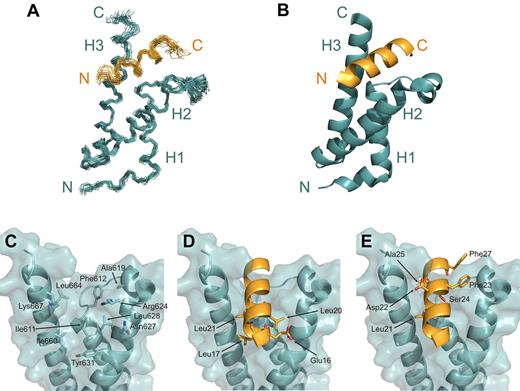
The HEB-PCET/KIX complex structure was determined using 1018 NOE-derived distance restraints and 150 TALOS-derived dihedral angle restraints from heteronuclear NMR data collected on samples containing 1 uniformly 15N/13C-labeled constituent in complex with the other unlabeled constituent. Of 200 total generated structures, 102 structures displayed no NOE violations > 0.3 Å and no backbone dihedral angle violations > 5°. The final ensemble of 20 low-energy structures displayed a well-defined 1:1 PCET/KIX complex (Figure 2A-B) with good overall structural statistics and low pairwise root mean squared deviation values for the isolated PCET peptide, the KIX domain, and for the complex (Table 1).
Structure of the PCET/KIX complex. (A) Overlay of the 20 low-energy structures of the complex, with the backbone (N, Cα, C′ atoms) of residues Lys15-Ser28 of HEB-PCET (orange) and residues 589-685 of KIX (teal) displayed. (B) Ribbon diagram of the energy-minimized average structure of the HEB-PCET/KIX complex. The helices of KIX are labeled H1, H2, and H3, and the N- and C-termini are indicated. (C) Transparent surface of KIX displaying the backbone ribbon and the residues involved in intermolecular van der Waals contacts. (D) Ribbon representation of HEB-PCET (in orange) showing the position of residues Glu16, Leu17, Leu20, and Leu21 on the surface of KIX. (E) Ribbon representation of HEB-PCET (in orange) showing the position of the LDFS sequence (Leu21, Asp22, Phe23, and Ser24) as well as Ala25 and Phe27 on the surface of KIX. Residues are numbered by position in full-length CBP or HEB.
Structure of the PCET/KIX complex. (A) Overlay of the 20 low-energy structures of the complex, with the backbone (N, Cα, C′ atoms) of residues Lys15-Ser28 of HEB-PCET (orange) and residues 589-685 of KIX (teal) displayed. (B) Ribbon diagram of the energy-minimized average structure of the HEB-PCET/KIX complex. The helices of KIX are labeled H1, H2, and H3, and the N- and C-termini are indicated. (C) Transparent surface of KIX displaying the backbone ribbon and the residues involved in intermolecular van der Waals contacts. (D) Ribbon representation of HEB-PCET (in orange) showing the position of residues Glu16, Leu17, Leu20, and Leu21 on the surface of KIX. (E) Ribbon representation of HEB-PCET (in orange) showing the position of the LDFS sequence (Leu21, Asp22, Phe23, and Ser24) as well as Ala25 and Phe27 on the surface of KIX. Residues are numbered by position in full-length CBP or HEB.
NMR restraints and structural statistics for HEB-PCET/KIX complex
| Restraints used for structure calculation . | Values . |
|---|---|
| Distance restraints | |
| Sequential | KIX: 375; HEB-PCET: 43 |
| Medium range (1 < i-j ≤ 5) | KIX: 352; HEB-PCET: 8 |
| Long range ( i-j > 5) | KIX: 184; HEB-PCET: 0 |
| Intermolecular | 56 |
| Total | 1018 |
| Dihedral angle restraints* | |
| Φ | KIX: 64; HEB-PCET: 11 |
| Ψ | KIX: 64; HEB-PCET: 11 |
| Total | 150 |
| RMS deviations for constraints | |
| Distance constraints, Å | 0.0163 ± 0.0007 |
| Dihedral angles, degrees | 0.30 ± 0.06 |
| RMS deviations from covalent geometry | |
| Bond lengths, Å | 0.0018 ± 0.000 05 |
| Bond angles, degrees | 0.35 ± 0.01 |
| Ramachandran statistics,†% | |
| Residues in most favorable regions | 87.8 |
| Residues in additional allowed regions | 10.1 |
| Residues in generously allowed regions | 1.0 |
| Residues in disallowed regions | 1.0 |
| RMS deviations of ordered regions from mean structure, Å | KIX (589-669), HEB-PCET (16-26) |
| Backbone atoms | 0.62 ± 0.09 |
| All heavy atoms | 1.07 ± 0.08 |
| Restraints used for structure calculation . | Values . |
|---|---|
| Distance restraints | |
| Sequential | KIX: 375; HEB-PCET: 43 |
| Medium range (1 < i-j ≤ 5) | KIX: 352; HEB-PCET: 8 |
| Long range ( i-j > 5) | KIX: 184; HEB-PCET: 0 |
| Intermolecular | 56 |
| Total | 1018 |
| Dihedral angle restraints* | |
| Φ | KIX: 64; HEB-PCET: 11 |
| Ψ | KIX: 64; HEB-PCET: 11 |
| Total | 150 |
| RMS deviations for constraints | |
| Distance constraints, Å | 0.0163 ± 0.0007 |
| Dihedral angles, degrees | 0.30 ± 0.06 |
| RMS deviations from covalent geometry | |
| Bond lengths, Å | 0.0018 ± 0.000 05 |
| Bond angles, degrees | 0.35 ± 0.01 |
| Ramachandran statistics,†% | |
| Residues in most favorable regions | 87.8 |
| Residues in additional allowed regions | 10.1 |
| Residues in generously allowed regions | 1.0 |
| Residues in disallowed regions | 1.0 |
| RMS deviations of ordered regions from mean structure, Å | KIX (589-669), HEB-PCET (16-26) |
| Backbone atoms | 0.62 ± 0.09 |
| All heavy atoms | 1.07 ± 0.08 |
Backbone dihedral angle restraints generated with TALOS for residues in helical regions when 9 of 10 predictions fell in the same region of the Ramachandran plot. The resulting Φ and Ψ angles were restricted to ± 15 degrees or the error from the TALOS output, whichever was greater.
Ramachandran statistics generated with PROCHECK-NMR
The HEB-PCET peptide in the complex adopted an amphipathic α-helical conformation from residues Lys15 to Phe27, with the solvent exposed side chains of Lys15 and Asp19 oriented so as to allow formation of a salt bridge. The N-terminal residues Gly12 and Thr13 displayed disordered and flexible properties that do not appreciably contact KIX, as indicated by significantly reduced {1H}-15N NOE values (< 0.25), strong intraresidue NOEs and a lack of identifiable medium-range, long-range, or intermolecular NOEs.
The KIX domain was composed of 3 α-helices (H1: Gln597-Ile611; H2: Arg623-Glu641; H3: Arg646-Arg669), a short N-terminal 310-helix (G1: Trp591-His594), and an intervening loop (L12: Phe612-Pro617) and 310-helix (G2: Ala618-Asp622) between helices H1 and H2. Helix H3 of KIX extended to Arg669, which is consistent with the chemical shift index data36 (data not shown) and observed sequential and medium NOE patterns [dαN (i,i+3), dαN(i,i+4)], and is the same length as that observed in the c-Myb/KIX/MLL complex.13 Further congruent with the in the complex structure, {1H}-15N NOE analysis of KIX revealed reduced {1H}-15N NOE values relative to the mean for the core (0.82 ± 0.16) for the 5 N-terminal residues (Gly586-Gly590), Thr614 and Asp616 of the L12 loop, and the 3 C-terminal residues (Ser670-Leu672; supplemental Figure 1). Overall, the KIX structures in complex presented here and in the c-Myb/KIX/MLL complex13 are similar, displaying a backbone root mean squared deviation value of 1.55 for the core residues (Lys589-Arg669).
PCET/KIX interface
The helical region of HEB-PCET occupies 930 Å2 of solvent accessible surface area on KIX in a deep hydrophobic cleft between helices H1 and H3, composing Ile611 (H1), Phe612 (L12), Ala619 (G2), the aliphatic regions of Arg624, Leu628, and Tyr631 (H2), Ile660, Leu664, and the aliphatic region of Lys667 (H3; Figure 2C). Extensive contacts to KIX were observed along the entire length of the PCET helix, including the hydrophobic residues Leu17, Leu20, and Leu21 from the LXXLL sequence, Phe23, from the overlapping LDFS sequence, and Ala25 and Phe27 (Figure 2D-E). The side chain of Leu17 from HEB-PCETmade van der Waals contacts to Ile611, Tyr631, Ile660, and the aliphatic region of Glu663 of KIX, while Leu20 of HEB-PCET inserted into a shallow cavity formed by the aliphatic component of Arg624, the methyl groups of Ile611 and Leu628, and the aromatic ring of Tyr631. Leu21 from HEB-PCET, for which the oncogenic importance of the analogous residue in E2A (Leu20) has been previously demonstrated,16 associated with a hydrophobic pocket on the surface of KIX formed by the side chains of Ile611, Ile660, Leu664, and the aliphatic region of Lys667. Beyond the LXXLL sequence of HEB-PCET, the aromatic side chain of Phe23 in the LDFS sequence interacted with Phe612, Ala619, the nonpolar component of Asp622, and the guanidinium group of Arg624, which was positioned to make a possible cation-π interaction. The side chain methyl group of Ala25 of HEB-PCET made hydrophobic contacts with Leu664, Lys667, and Arg668 whereas Phe27 makes contacts with Phe612 of loop L12.
These nonpolar interactions appeared to be complemented by hydrogen bonding and electrostatic interactions. The glutamate immediately preceding the LXXLL sequence of HEB-PCET (Glu16) was oriented to allow for a possible hydrogen bond with Asn627 from helix H2 of KIX (dOϵ1-Nδ2 = 4.0 Å). Asp22 was in sufficient proximity to Lys667 of helix H3 of KIX to form a salt bridge (dOδ2-Nζ = 3.1 Å), and Ser24 was positioned to form hydrogen bonds with Ile611 (dOγ-HN = 3.5 Å) and Thr614 (dOγ-Oγ1 = 4 Å) in the minimized average structure of KIX.
Mutations throughout the E-protein PCET motif attenuate KIX binding
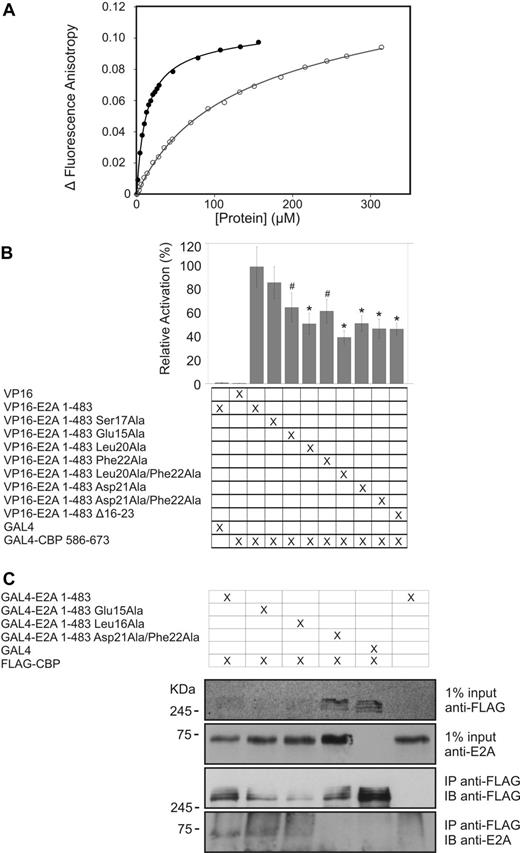
The HEB-PCET/KIX structure informed subsequent experiments in which mutagenesis, fluorescence anisotropy, and isothermal titration calorimetry were used to assess the importance of various PCET residues in KIX recognition (Figure 3A; Table 2; supplemental Methods). An alanine mutation of the glutamate residue preceding the LXXLL sequence (Glu16 in HEB and Glu15 in E2A) resulted in a subtle decrease in affinity (Kd of 25μM), whereas an alanine substitution of the leucine residue occupying the point of overlap between the LXXLL and LDFS sequences of PCET (Leu21 in HEB and Leu20 in E2A), which abrogated immortalization of myeloid cells in vitro and impaired leukemogenesis in a whole animal model,17 showed no detectable binding to KIX. Mutation of the neighboring aspartate residue within the LDFS sequence of PCET, which is in the vicinity of Lys667 from helix H3 of KIX, decreased the affinity of the interaction by ∼ 3-fold (Kd of 25μM). The reciprocal Lys667Ala KIX mutant showed a similar effect, whereas a Lys667Glu KIX charge-repulsion mutant resulted in an ∼ 10-fold decrease in affinity (Kd of 95μM). An alanine mutation of the phenylalanine within the LDFS sequence of the PCET motif impaired KIX binding by ∼ 4-fold. A quadruple KIX mutant (Phe612Ala/Asp622Ala/Arg624Ala/Lys667Glu) targeting multiple points throughout the binding cleft displayed a 22-fold weaker interaction with the PCET motif (Table 2).
PCET mutations impair KIX binding. (A) Representative binding curves showing the change in fluorescence anisotropy of 5-carboxyfluorescein tagged HEB-PCET (12-25) peptide with increasing total concentration of either wild-type (●) or Lys667Glu mutant (○) KIX. (B) In a mammalian 2-hybrid assay, the KIX domain (residues 586-673 of CBP) fused to a GAL4 DNA-binding domain can recruit E2A to the GAL4 binding sites on a reporter plasmid, driving expression of firefly luciferase. The luciferase expression was normalized against expression of Renilla driven by a constitutively active internal control plasmid used to control for variations in transfection efficiency. Renilla-normalized luciferase expression of E2A (1-483) PCET mutants were measured relative to that seen with wild-type E2A (1-483), with the result presented being the mean from at least 3 independent transfections ± the SD (represented by vertical error bars). #P < .015 vs wild-type 2 × VP16-E2A (1-483). *P < .001 vs wild-type 2 × VP16-E2A (1-483). The results shown in the figure are the mean value from at least 3 independent transfections; error bars represent SD. Statistical significance was measured using a 1-way ANOVA with Tukey post hoc test. (C) SV293T cells were transfected with FLAG-CBP and GAL4-E2A (1-483) or GAL4-E2A (1-483) PCET mutants. Lysates were subjected to immunoprecipitation with anti-FLAG and immunoblotted with anti-E2A yae or anti-FLAG.
PCET mutations impair KIX binding. (A) Representative binding curves showing the change in fluorescence anisotropy of 5-carboxyfluorescein tagged HEB-PCET (12-25) peptide with increasing total concentration of either wild-type (●) or Lys667Glu mutant (○) KIX. (B) In a mammalian 2-hybrid assay, the KIX domain (residues 586-673 of CBP) fused to a GAL4 DNA-binding domain can recruit E2A to the GAL4 binding sites on a reporter plasmid, driving expression of firefly luciferase. The luciferase expression was normalized against expression of Renilla driven by a constitutively active internal control plasmid used to control for variations in transfection efficiency. Renilla-normalized luciferase expression of E2A (1-483) PCET mutants were measured relative to that seen with wild-type E2A (1-483), with the result presented being the mean from at least 3 independent transfections ± the SD (represented by vertical error bars). #P < .015 vs wild-type 2 × VP16-E2A (1-483). *P < .001 vs wild-type 2 × VP16-E2A (1-483). The results shown in the figure are the mean value from at least 3 independent transfections; error bars represent SD. Statistical significance was measured using a 1-way ANOVA with Tukey post hoc test. (C) SV293T cells were transfected with FLAG-CBP and GAL4-E2A (1-483) or GAL4-E2A (1-483) PCET mutants. Lysates were subjected to immunoprecipitation with anti-FLAG and immunoblotted with anti-E2A yae or anti-FLAG.
Affinity of the PCET/KIX interaction
| Ligand . | Kd, μM . |
|---|---|
| PCET peptides titrated with KIX> | |
| E2A-PCET | 12 ± 2* |
| HEB-PCET | 9.4 ± 0.1* |
| E2A-PCET Leu20Ala | ND† |
| E2A-PCET Phe22Ala | 71 ± 2† |
| HEB-PCET Glu16Ala | 25 ± 1* |
| HEB-PCET Asp22Ala | 25 ± 1* |
| KIX constructs titrated into HEB-PCET | |
| KIX | 9.4 ± 0.1* |
| KIX Lys667Ala | 28 ± 2* |
| KIX Lys667Glu | 95 ± 1* |
| KIX Phe612Ala/Asp622Ala/Arg624Ala/Lys667Ala | 204 ± 7* |
| Ligand . | Kd, μM . |
|---|---|
| PCET peptides titrated with KIX> | |
| E2A-PCET | 12 ± 2* |
| HEB-PCET | 9.4 ± 0.1* |
| E2A-PCET Leu20Ala | ND† |
| E2A-PCET Phe22Ala | 71 ± 2† |
| HEB-PCET Glu16Ala | 25 ± 1* |
| HEB-PCET Asp22Ala | 25 ± 1* |
| KIX constructs titrated into HEB-PCET | |
| KIX | 9.4 ± 0.1* |
| KIX Lys667Ala | 28 ± 2* |
| KIX Lys667Glu | 95 ± 1* |
| KIX Phe612Ala/Asp622Ala/Arg624Ala/Lys667Ala | 204 ± 7* |
ND indicates no detectable binding.
Kd values determined by fluorescence anisotropy titration.
Kd values determined by isothermal titration calorimetry.
In vivo effects of PCET mutations on the E2A/KIX interaction
To test the contributions of PCET residues in KIX recognition in the context of full-length E2A and an intracellular environment, a mammalian 2-hybrid assay involving the region of E2A found in E2A-PBX1 (residues 1-483) was performed (Figure 3B). The Glu15Ala substitution in E2A 1-483 (analogous to Glu16Ala in HEB) produced 65% of wild-type luciferase activation (P = .014), whereas a Ser17Ala mutant (analogous to Ser18Ala in HEB) had no significant effect on intracellular KIX binding relative to wild-type E2A 1-483, an observation consistent with the solvent exposed position of this serine residue in our PCET/KIX complex structure and with a Ser17Ala E2A-PBX1 mutant displaying the same cell immortalization potential and ability to interact with CBP as wild-type E2A-PBX1.17
Complementing the biophysical binding data, substitutions of individual residues throughout the PCET motif also impaired the ability of E2A 1-483 to induce the expression of a luciferase reporter gene, consistent with impaired recruitment of endogenous CBP/p300 (Figure 3B). The Leu20Ala and Asp21Ala mutants displayed 51% and 52%, wild-type activity, respectively, whereas Phe22Ala E2A show 62% wild-type activity. The Leu20Ala/Phe22Ala E2A mutant showed the largest decrease with only 40% wild-type activity. None of these constructs gave activity that was significantly greater than that associated with deleting the overlapping LXXLL and LDFS sequences (Leu16-Ser23). To investigate the effect of PCET mutations on binding to full-length CBP, 293T cells were transiently transfected with E2A 1-483 or engineered PCET motif variants, expressed as GAL4-fusion proteins, and FLAG-tagged CBP. Immunoprecipitation was carried out with anti-FLAG and the immobilized proteins evaluated by immunoblotting (Figure 3C). Leu16Ala appears to reduce binding slightly and the Asp21Ala/Phe22Ala double mutation abrogates detectable binding. Although no impairment of binding is evident consequent to the Glu15Ala substitution, this may simply reflect the inability of this relatively imprecise assay to detect a modest reduction in binding affinity. These data illustrate that amino acids within the PCET motif that were identified as important for KIX binding in our structural and biophysical studies also contribute to KIX binding by the E2A portion of E2A-PBX1, namely residues 1-483, in the context of intact mammalian cells and full-length CBP.
Residues spanning the PCET motif contribute to E2A-PBX1 induced cell immortalization
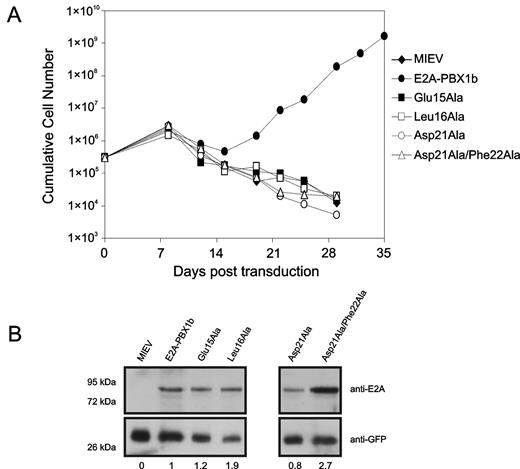
Previously, we had shown that enforced expression of E2A-PBX1 in primary murine hematopoietic progenitors resulted in the outgrowth of a rapidly proliferating population of immortalized myeloid cells and that a Leu20Ala PCET motif mutant abrogated this effect.17 Our interest in the molecular mechanisms of E2A-PBX1 in acute lymphoblastic leukemia prompted us to use this immortalization assay to investigate the implications of our structural and mutational analysis of the PCET/KIX interaction for neoplastic transformation by E2A-PBX1 (Figure 4).
Residues throughout the E2A-PCET motif are critical for oncogenesis. (A) Proliferation of myeloid progenitors associated with retroviral-mediated expression of wild-type E2A-PBX1b and engineered E2A-PBV1b variants. For each construct, 3 × 105 bone marrow cells were cultured in GM-CSF immediately after retroviral infection and counted for 35 days. (B) Western blot of lysates from NIH 3T3 fibroblasts infected with retroviruses forcing expression of the indicated E2A-PBX1b constructs, confirming expression of all the different mutants of E2A-PBX1b. The values presented below the blot represent expression levels of the various E2A-PBX1 mutant constructs, relative to wild-type E2A-PBX1, after normalization according to GFP expression.
Residues throughout the E2A-PCET motif are critical for oncogenesis. (A) Proliferation of myeloid progenitors associated with retroviral-mediated expression of wild-type E2A-PBX1b and engineered E2A-PBV1b variants. For each construct, 3 × 105 bone marrow cells were cultured in GM-CSF immediately after retroviral infection and counted for 35 days. (B) Western blot of lysates from NIH 3T3 fibroblasts infected with retroviruses forcing expression of the indicated E2A-PBX1b constructs, confirming expression of all the different mutants of E2A-PBX1b. The values presented below the blot represent expression levels of the various E2A-PBX1 mutant constructs, relative to wild-type E2A-PBX1, after normalization according to GFP expression.
Infection of primary bone marrow cells with a retrovirus conferring expression of wild-type E2A-PBX1 gave rise to a population of myeloid progenitors with sustained exponential growth (Figure 4). In contrast, transduction with engineered E2A-PBX1 bearing the Glu15Ala, Leu16Ala, Asp21Ala, or Asp21Ala/Phe22Ala substitutions conferred no apparent growth advantage relative to cells infected with the empty retroviral vector (Figure 4). Therefore, the capacity of E2A-PBX1 to immortalize primary myeloid progenitors is exquisitely susceptible to focal perturbations within the PCET motif.
Discussion
Structure-function studies have indicated that an interaction between the PCET motif within activation domain 1 of E-proteins and the KIX domain of CBP/p300 is involved in B-lymphopoiesis,3,12 is targeted in a mechanism of E-protein silencing,9 and is essential for E2A-PBX1 oncogenesis.16 KIX also binds activation domains of several other mammalian transcription factors10,13,37,38 and viral proteins39-41 involved in the regulation of lymphopoiesis through their LXXLL sequence or more generic φ-x-x-φ-φ sequence described by Lee et al,10 which is typically preceding by an acidic residue (ζ; Figure 5A).6,11 Our PCET/KIX complex and associated mutagenesis studies described here provide structural insights into these functions.
Residues adjacent to ζ-φ-x-x-φ-φ motif dictate mode of KIX recognition. (A) Sequence alignment of ζ-φ-x-x-φ-φ containing KIX-binding proteins. ζ indicates an acidic amino acid; φ, a bulky hydrophobic amino acid; and x, any amino acid. Conserved hydrophobic amino acid residues are colored yellow, acid residues red, and basic residues blue. Positions within the respective protein sequences are indicated as numbers. (B-G) Comparison of the orientation of PCET (orange ribbon) and MLL (red ribbon) on the KIX domain (gray ribbon/transparent surface for KIX bound to MLL and teal for KIX bound to HEB-PCET). Noteworthy residues from each activation domain are represented as sticks. (E-G) Views of panels B-D, respectively, that have been rotated clockwise 70 degrees about their x-axes. The KIX/MLL complex is from PDB accession 2AGH.
Residues adjacent to ζ-φ-x-x-φ-φ motif dictate mode of KIX recognition. (A) Sequence alignment of ζ-φ-x-x-φ-φ containing KIX-binding proteins. ζ indicates an acidic amino acid; φ, a bulky hydrophobic amino acid; and x, any amino acid. Conserved hydrophobic amino acid residues are colored yellow, acid residues red, and basic residues blue. Positions within the respective protein sequences are indicated as numbers. (B-G) Comparison of the orientation of PCET (orange ribbon) and MLL (red ribbon) on the KIX domain (gray ribbon/transparent surface for KIX bound to MLL and teal for KIX bound to HEB-PCET). Noteworthy residues from each activation domain are represented as sticks. (E-G) Views of panels B-D, respectively, that have been rotated clockwise 70 degrees about their x-axes. The KIX/MLL complex is from PDB accession 2AGH.
The ability of KIX to recognize multiple activation domains of regulators of lymphopoiesis that display low sequence identity beyond the general ζ-φ-x-x-φ-φ consensus sequence suggests that it possesses considerable structural plasticity, as proposed by De Guzman et al.13 Comparison of our PCET/KIX to the c-Myb/KIX/MLL ternary complex13 illustrates this plasticity. PCET and MLL bind the same cleft between helices H2 and H3 of KIX (Figure 5B-C); however, subtle differences exist in the orientation of the 2 activation domains, including in the L12 loop region and the G2 helix of KIX (Figure 5C) and in the positioning of the KIX-contacting hydrophobic residues in the ζ-φ-x-x-φ-φ consensus sequence (Figure 5D-E). The L12 loop in the PCET/KIX complex adopts a conformation more similar to that observed in the c-Myb/KIX complex42 to accommodate the aromatic side chain of Phe23 in PCET. Structural differences also exist in the sequences N-terminal to the ζ-φ-x-x-φ-φ of PCET and MLL. The N-terminus of PCET (Gly12-Asp14) is disordered and makes no detectable contacts with KIX, whereas the corresponding region of MLL (Ile2844, Leu2845, and Pro2846) is ordered and binds to a hydrophobic groove on KIX (Figure 5D-E).13 These differences appear to account, at least in part, for the slight difference in orientation of MLL versus PCET relative to KIX and the higher affinity reported for the MLL:KIX interaction38 (Kd of 2.8μM vs 9.0μM for PCET/KIX). We therefore propose that the residues adjacent to the ζ-φ-x-x-φ-φ consensus sequence of activation domains significantly influence their binding modes and corresponding affinities for KIX.
A sequence alignment of PCET, MLL, and other KIX-binding activation domains illustrates this concept and allows for a prediction of binding modes. PCET and c-Myb display a high degree of identity within the ζ-φ-x-x-φ-φ consensus sequence (Figure 5A), yet bind to unique sites on opposite faces of KIX. It is the differences in the physicochemical properties of the N- and C-terminal adjacent residues in the activation domains (ie, Ile295 and Met303 of c-Myb13,42 ; Asp14 and Asp22 of HEB) and their complementarity to their respective binding surfaces that dictate KIX binding specificity. The c-Jun activation domain, the AD2 activation domain of p53 (p53-AD2), and the viral protein HTLV-1 Tax, each of which has been previously shown by NMR spectroscopy to bind the general PCET/MLL site on KIX,10,37,41 contain multiple hydrophobic residues N-terminal to the ζ-φ-x-x-φ-φ sequence similar to MLL and lack sequence similarity to KIX-contacting residues C-terminal to the ζ-φ-x-x-φ-φ sequence in PCET (Figure 5A), suggesting that these proteins bind KIX in a manner more similar to that of MLL than PCET. In contrast, the N-terminus of the first activation domain of p53 (p53-AD1) is less hydrophobic while at the same time displaying conservation of a hydrophobic residue (Leu25) at the analogous position as Phe23 of PCET C-terminal to the ζ-φ-x-x-φ-φ sequence, which our mutagenesis data suggest plays an important role in KIX recognition. We therefore predict that p53-AD1 binds KIX in a manner more similar to PCET. Indeed, NMR-based chemical shift perturbation studies have shown that p53-AD1 preferentially binds to the PCET/MLL site over the c-Myb site and that phosphorylation enhances this selectivity.10 The KIX-interactive activation domain of the oncoprotein HBZ, which is involved in HTLV1-induced adult T-cell leukemia/lymphoma,43 has hydrophobic residues both N-terminal and C-terminal to the ζ-φ-x-x-φ-φ sequence that align with KIX-interactive residues of MLL (Ile2842, Leu2843, and Pro2844) and PCET (Phe23 and Phe27). HBZ activation domain 1 appears to combine the binding modes of both MLL and PCET, a proposal supported by reported higher affinity (Kd of 130nM) interaction with KIX.40
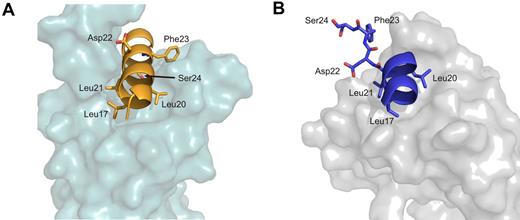
In addition to binding coactivators, such as the KIX domain of CBP/p300, PCET binds the transcriptional corepressor ETO via its eTAFH domain.7-9 This represents a proposed mechanism by which normal E-protein function is silenced by the oncoprotein AML1-ETO in cases of acute myeloid leukemia.9 Furthermore, because wild-type E proteins appear capable of transcriptional suppression in certain cellular contexts, recruitment of wild-type ETO or other corepressors by PCET could also contribute to physiologic transcriptional regulation by E-proteins.3,20,44 A comparison of our PCET/KIX structure with the previously determined PCET/eTAFH complex structure6 illustrates that, although both eTAFH and KIX bind the PCET motif through hydrophobic interactions with the ζ-φ-x-x-φ-φ sequence, there are notable differences in the PCET conformation and interactions with residues outside the consensus sequence (Figure 6). When bound to the eTAFH domain, the helical conformation of HEB-PCET (Lys15 to Leu21) is disrupted by a kink at Asp22,6 while in complex with KIX the PCET helix extends to Phe27. Phe23 within the LDFS sequence of HEB-PCET plays a critical role in KIX recognition as evidenced by our structural, biophysical, biochemical, and mammalian 2-hybrid data. However, Park et al observed no contacts between this residue and the eTAFH domain and demonstrated that a truncation of HEB-PCET lacking Phe23 had minimal effect on eTAFH binding.6 In normal myelopoeisis, the greater affinity observed of the PCET/eTAFH interaction would allow ETO to outcompete CBP/p300 for E-proteins, leading to the repression of lymphoid E-protein target genes. In cases of acute myelogenous leukemia associated with t(8;21) and AML1-ETO fusion protein expression, constitutive E-protein silencing may be driven not only by relative cofactor abundance as proposed by Zhang et al,9 but also by the greater affinity of the eTAFH domain of AML1-ETO relative to the KIX domain for E-proteins.
Comparison of PCET/KIX with PCET/eTAFH. (A) The HEB-PCET structure bound to KIX. (B) eTAFH. Leucine residues from the LXXLL sequence (Leu17, Leu20, and Leu21) as well as Asp22, Phe23, and Ser24 from the LDFS sequence are represented as sticks. HEB-PCET is colored orange in the complex with KIX and blue in the complex with eTAFH. The HEB-PCET/eTAFH complex is from PDB accession number 2KNH.
Comparison of PCET/KIX with PCET/eTAFH. (A) The HEB-PCET structure bound to KIX. (B) eTAFH. Leucine residues from the LXXLL sequence (Leu17, Leu20, and Leu21) as well as Asp22, Phe23, and Ser24 from the LDFS sequence are represented as sticks. HEB-PCET is colored orange in the complex with KIX and blue in the complex with eTAFH. The HEB-PCET/eTAFH complex is from PDB accession number 2KNH.
The PCET motif has been linked to B-cell development by E2A and leukemogenesis by E2A-PBX1.3,21 It contains 2 previously proposed protein recognition sequences: an LXXLL sequence and an overlapping LDFS sequence.7,16-18 We had previously identified an overlapping leucine residue (Leu20) as critical for coactivator recruitment by E2A-PBX1, but this residue was found in both sequences so the issue over which sequence was involved remained unresolved.17 We have now shown that both sequences are involved, forming a contiguous helical binding motif. This structure is consistent with the importance of E2A Leu20 as the analogous residue in HEB-PCET (Leu21) is in the center of the PCET helix and anchored into the binding cleft on KIX by numerous hydrophobic contacts.
The second activation domain of E2A, AD2, can interact with CBP/p300 independently of the PCET motif of AD1 in pull-down experiments.12 Furthermore, we have shown that the 2 activation domains can function cooperatively both in CBP/p300 binding and transactivating a reporter gene in transient cotransfection experiments. Bhalla et al subsequently confirmed cooperative trans-activation by AD1 and AD2 and showed that deletion of either domain abrogates the ability of E12 to induce B-lymphoid differentiation in 70Z/3m cells.3 As with the PCET motif, recruitment of CBP/p300 by AD2 requires the KIX domain.12,33 Therefore, available evidence supports a model in which PCET motif of AD1 and AD2 function cooperatively to bind to the KIX domain, recruit CBP/p300, and promote gene transcription and B-lymphoid commitment. Although the focus of our current work on the PCET motif is justified by its obvious role in oncogenesis, it nonetheless remains true that a full understanding of CBP/p300 recruitment by E-proteins will require detailed characterization of the interactions involving AD2.
Ectopic expression of E2A-PBX1 in murine bone marrow cells by retroviral transduction has been shown to give rise to immortalized GM-CSF–dependent progenitor cells in vitro or a myeloproliferative disease if transduced bone marrow is transplanted into syngeneic recipient mice.45,46 Both of these oncogenic capabilities were abolished by the E2A Leu20Ala mutation17 ; and in the present study, we have correlated this to decreased in vitro affinity of PCET for the KIX domain of the transcriptional coactivator CBP/p300. Consistent with the structure presented here, alanine substitutions of residues throughout the PCET helix were correlated with decreased affinity by titration and mammalian 2-hybrid assays and abrogation of bone marrow immortalization. The one mutation that did not impair immortalization was E2A Ser17Ala,17 consistent with our observations that it did not significantly impair KIX binding in mammalian 2-hybrid assays. Therefore, the newly identified PCET binding site on KIX is a candidate drug target. It is noteworthy that small molecules based on an isoxazolidine scaffold have been shown to bind KIX selectively at this site.47,48 The importance of this site for E2A-PBX1–driven oncogenesis, and the accompanying structure of the PCET/KIX complex, opens the exciting possibility of designing novel therapeutics for E2A-PBX1+ acute lymphoblastic leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Peter Wright (Scripps Institute, La Jolla, CA) for the KIX expression plasmid and Mr Kim Munro (Protein Function Discovery Facility, Queen's University) for technical assistance in the collection of the fluorescence anisotropy and ITC data.
This study was supported by research funding from the Canadian Institutes of Health Research (S.P.S., M.I., and D.P.L.). C.M.D. was supported by the Natural Sciences and Engineering Research Council of Canada (CGS-D Scholarship). M.I. holds a Canada Research Chair in Cancer Structural Biology.
Authorship
Contribution: C.M.D. designed and performed the research, analyzed data, and wrote the manuscript; S.C. performed research and wrote the manuscript; M.J.P. and F.W. designed and performed research; P.T., S.L., and H.L.S. performed research; M.I. contributed vital new reagents and wrote the manuscript; and D.P.L. and S.P.S. designed research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven P. Smith, Department of Biomedical and Molecular Sciences, Queen's University, Kingston, ON, Canada K7L 3N6; e-mail: steven.smith@queensu.ca; and David P. LeBrun, Department of Pathology and Molecular Medicine, Queen's University, Kingston, ON, Canada K7L 3N6; e-mail: lebrun@cliff.path.queensu.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal