Abstract
Abstract 1537
With current rituximab-based treatment about 60 % of patients with diffuse large B-cell lymphoma (DLBCL) can be cured. However, for patients with early relapse or being refractory to such therapy outcome is very poor. The International Prognostic Index (IPI) is still the only prognostic tool used in daily clinical practice to risk stratify DLBCL patients. Yet, IPI has a limited ability to identify patients with risk of early relapse or refractory disease. Thus, there is a great need for reliable biomarkers capable of identifying high-risk patients for whom alternative therapies could be considered. Even though gene expression profiling can differentiate between DLBCL tumor subtypes, several factors influence RNA translation to a final protein. Consequently, there could be a considerable discrepancy between RNA expression levels and the corresponding protein level. Therefore, we hypothesized that global protein expression profiling, using proteomic analysis, could be a tool to provide additional prognostic information.
To compare differences in global protein expression from two groups of DLBCL patients: i) early relapse/refractory patients, and ii) cured patients, in order to find candidate prognostic biomarkers and also potential drug targets.
All de novo DLBCL patients diagnosed in the Gothenburg region between 2004 and 2008 treated with curative intent (R-CHOP) were retrospectively analyzed. Two subgroups were determined; A) patients with progressive disease or relapse within 1 year after completion of treatment and B) patients considered cured, i.e. progression-free with a follow-up of at least 5 years. Five age- and gendermatched patients from each subgroup were selected and protein solutions were extracted from fresh frozen tumor tissue. In parallel, five established DLBCL cell lines were metabolically labeled with 13C6-arginine and 13C6-lysine. Lysates from these labeled cell lines were combined and used as an internal protein standard for relative protein quantification in the so-called Super-SILAC approach. Proteins were separated by SDS-PAGE and in-gel trypsin digested fractions were analyzed by LC-MS using an LTQ-FT/MS instrument. The MaxQuant software was used for data-analysis.
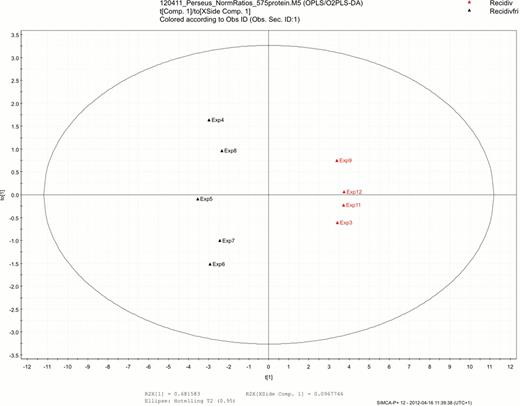
In the tumor samples 2523 different proteins were identified, and 1953 of these could be quantified. When comparing the groups, 41 separate proteins with statistically different expression were identified. Furthermore, 18 proteins were solely expressed in the early relapse/refractory group; most of these proteins were associated with cell proliferation (P54, snoRNP GAR1, SCOT), anti-apoptosis (TRAP1, AHA1), cell migration (Caveolin-1, ELMO1) and cell adhesion (VAP-1). We also performed a multivariate analysis (Principal Components Analysis), which produced a panel of 17 proteins that completely separated the groups (Figure 1).
Among the specific proteins separating the groups are e.g. proteins involved in cell proliferation/tumor growth (Erp-72, glutaredoxin, ACTG1, superoxid dismutase 1, DEAD box protein p72), tumor invasion (Moesin), adhesion (hnRNP M receptor) and anti-apoptosis (PTB1, ANT2).
We have identified a substantial amount of proteins with potential prognostic value to identify high-risk DLBCL patients. Some of these proteins could expectedly be found in patients with refractory disease, however for many of them their role in therapy resistance remains to be determined. Further studies of both the individual and clustered proteins may also result in the identification of potential targets for the development of novel therapies for DLBCL.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal