Abstract
Abstract 4193
T cell repertoire diversity is generated by recombination of variable (V), diversity (D) and joining (J) segments in the T cell receptor (TCR) locus. Further variability and antigen recognition capacity is introduced by nucleotide insertion (NI) in the recombined sequences resulting in a complex repertoire, the organization of which is poorly understood. We postulate that TCR b D, J and V gene segment usage in an individual would result in a TCR repertoire with a fractal, self-similar frequency distribution of T cell clones with respect to gene segment usage. To determine this, the TCR repertoire of donors and recipients of HLA matched-related and unrelated allogeneic stem cell transplantation (SCT) was evaluated by high-throughput (HT) sequencing of the CDR3 region of TCR b.
Ten SCT donor-recipient pairs were selected for HT-TCR b sequencing. cDNA was isolated from T cells obtained from the donors at baseline and recipients at day 100, 1 year post SCT or at the time of graft-versus-host disease (GVHD) diagnosis. HT-TCR β sequencing was performed and analyzed using the ImmunoSEQ analyzer tool (Adaptive Biotechnologies, Seattle, WA). TCR b clone frequencies were used to determine the TCR self-similarity score (SSS = log clone frequency × log scale), evaluating clonal frequency at each gene segment-scale (J, VJ and VJ+NI). This revealed a TCR SSS with a relatively narrow distribution of 1.64 ± 0.1 (mean ± SD) for J, 1.69 ± 0.2 for VJ, and 1.41 ± 0.01 for VJ+NI, which was consistent between all normal stem cell donors analyzed.
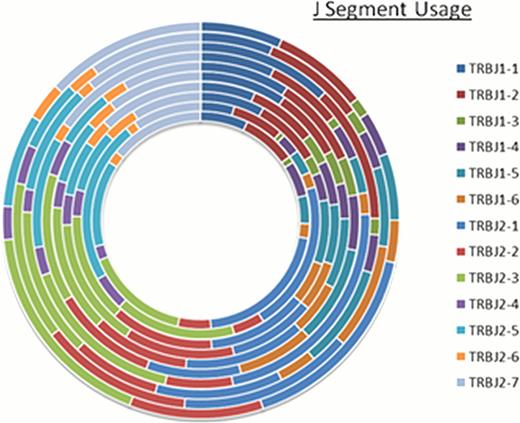
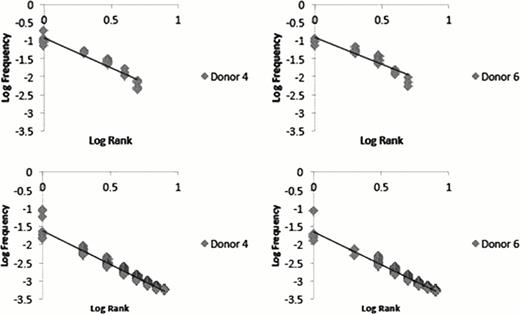
Relative proportional distribution (RPD) graphs were then generated to allow for the depiction of the TCR b D, J, V distribution for simultaneous comparison at an individual level. Representative data in Figure 1 shows the relatedness across donors at the TCR b J segment level. Relative clonal frequency was determined for each unique TCR clone at the specified segment level and plotted according to frequency-rank. Slope of the resulting linear regression lines from log-log plots of rank-frequency was used to determine self-similarity and fractal dimension. These plots were comparable among donors with resulting slopes of 1.6 ± 0.01 and 1.8 ± 0.1, respectively for TCR J and VJ containing clones (Figure 2). The plots also revealed a hierarchy of T cell clones, with few dominant clones occupying the high ranks and a multitude of clones in the later ranks.
For donor-recipient comparisons, we examined the dominant ranking clones which would be most likely to be involved in GVHD and response to infection. The ordered TCR clonal frequency distribution seen in donors was perturbed in recipients following SCT, with recipients demonstrating a lower level of complexity in their TCR repertoire, and a large shift in the frequency distribution of the dominant T cell clones compared to the donor. When dominant clones were compared between donors and recipients, recipients shared only a small proportion of TCR clonotypes with their donors despite full donor T cell chimerism. This difference did not change over time, suggesting an alternate T cell clonal hierarchy and repertoire develops in transplant recipients when compared with their donors.
Using simple mathematical analysis, we demonstrate that the TCR b repertoire has a fractal, self-similar pattern with a hierarchy of dominant and minor clones. We note that the complexity of the TCR repertoire is diminished and TCR b hierarchy is altered following SCT. Restoration of this order may serve as a marker for post-SCT immune reconstitution. Further, by demonstrating shifts in TCR clonal dominance, fractal analysis comparing donor and recipient T cell repertoire may allow for more accurate monitoring of immunotherapy of malignancies in general, beyond allogeneic SCT.
Graphical representation of self-similarity in TCR b J gene segment usage across segments and individuals. Relative-proportional-distribution (RPD) graph depicting TCR b J segment usage in ten hematopoietic stem cell donors. Each ring in the graph represents J segment usage frequency in a single donor, showing similarity in J segment frequency across ten different donors.
Graphical representation of self-similarity in TCR b J gene segment usage across segments and individuals. Relative-proportional-distribution (RPD) graph depicting TCR b J segment usage in ten hematopoietic stem cell donors. Each ring in the graph represents J segment usage frequency in a single donor, showing similarity in J segment frequency across ten different donors.
Log-Log plot depicting linear distribution of frequency ranked TCR b DJ (top panels) and VDJ (bottom panels) clones in two stem cell donors.
Log-Log plot depicting linear distribution of frequency ranked TCR b DJ (top panels) and VDJ (bottom panels) clones in two stem cell donors.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal