In this issue of Blood, Gawad et al provide an unprecedented look at the clonal diversity of pediatric B-precursor acute lymphoblastic leukemia (ALL).1
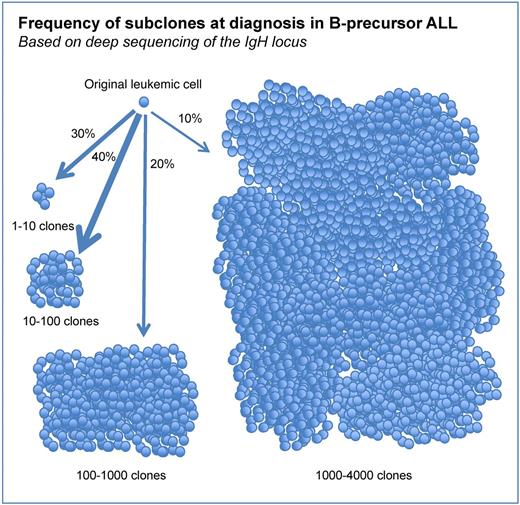
Although the presence of coexisting subclones in B-precursor ALL at diagnosis has been previously well reported, using deep sequencing of the IgH locus Gawad and colleagues demonstrate that individual patient samples had up to 4000 unique IgH sequences, suggesting an unexpected degree of clonal heterogeneity (see figure). Interestingly, clonal evolution often showed evidence of allelic exclusion, retained stable JH sequences with divergent VH, and adjacent NDN regions, consistent with prolific aberrant rearrangement during early leukemogenesis.
Frequency of subclones at diagnosis in B-precursor ALL. Each cell shown represents a distinct subclone based on IgH sequencing. Arrow thickness and percentage reflect the relative fraction of patient samples with the indicated number of distinct IgH clones.
Frequency of subclones at diagnosis in B-precursor ALL. Each cell shown represents a distinct subclone based on IgH sequencing. Arrow thickness and percentage reflect the relative fraction of patient samples with the indicated number of distinct IgH clones.
The critical contribution of this study is the revelation that B-precursor ALL is commonly composed of low frequency subclones, in some cases numbering in the thousands. Relevant to this finding, a report by Mullighan et al revealed that 52% of relapsed ALL clones were derived from minor “ancestral” subclones present at diagnosis, using genomic copy number abnormalities (CNAs) and lesion-specific (PCR) at diagnosis and relapse.2 The findings of Gawad et al provide strong evidence that clonal heterogeneity at diagnosis is the rule, rather than the exception, and when combined with the findings of Mullighan et al suggests that for most ALL patients treatment success should not be measured by loss of the predominant clone at diagnosis, but rather by effects on numerous under-appreciated subclones. At this time we cannot predict whether these subclones will result in increased relapse, and whether the dramatic heterogeneity in IgH sequences represents biologically distinct subclones that have additional heterogeneity beyond the IgH sequences. Indeed, we do not yet identify these rare but frequent subclones prospectively in patients.
If relapse clones are commonly derived from minor subclones at diagnosis, are current MRD techniques adequate to measure future relapse clones? Gawad et al report that 99% of the subclones were present at a low frequency of less than 0.1%. As a point for discussion, it is unclear what implications these findings have on our current techniques for detection of minimal residual disease (MRD). We currently use flow cytometry or PCR-based MRD monitoring, which depend on identification of distinct phenotypic and IgH rearrangements, respectively. These techniques attempt to identify a single major clone representing the leukemia at diagnosis and at MRD time points. Gawad and colleagues found that IgH subclones were associated with distinct flow cytometry–based phenotypic subsets and IgH PCR-based MRD techniques select limited unique primer sets, suggesting current MRD techniques are not capable of assessing even a fraction of the heterogeneity of ALL. Interestingly, a recent paper by Denys et al revealed discordance of MRD-based risk group assignment comparing 6-color flow cytometry and PCR-based assessments in 32% of ALL patient samples.3 Indeed, flow cytometry–based MRD measurements generally reported lower MRD levels than PCR-based techniques. IgH PCR measurements have greater sensitivity than flow cytometry in most cases, and IgH PCR techniques may be modified to detect multiple subclones, suggesting that IgH PCR-based techniques may have an advantage over flow cytometry–based techniques. On the other hand, new multiparameter-based flow cytometry techniques may allow much higher resolution and detection of rare ALL subclones at diagnosis.
If indeed 70% of patients carry tens to thousands of subclones of B-precursor ALL at diagnosis, as suggested by Gawad et al, and these subclones have additional biologic heterogeneity, it would be plausible that a significant percentage of treatment failures are due to a selection of the resistant subclones. Then we are not treating one leukemia, but rather an often-massive collection of subtly divergent subclones. In this case, identification of these subclones and an understanding of their biologic heterogeneity will be necessary to develop therapeutic approaches that will be able to eradicate all B-precursor ALL subclones.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■