Abstract
Many common genetic variants have been associated with non-Hodgkin lymphoma (NHL), but individual study results are often conflicting. To confirm the role of putative risk alleles in B-cell NHL etiology, we performed a validation genotyping study of 67 candidate single nucleotide polymorphisms within InterLymph, a large international consortium of NHL case-control studies. A meta-analysis was performed on data from 5633 B-cell NHL cases and 7034 controls from 8 InterLymph studies. rs3789068 in the proapoptotic BCL2L11 gene was associated with an increased risk for B-cell NHL (odds ratio = 1.21, P random = 2.21 × 10−11), with similar risk estimates for common B-cell subtypes. PRRC2A rs3132453 in the HLA complex class III region conferred a reduced risk of B-cell NHL (odds ratio = 0.68, P random = 1.07 × 10−9) and was likewise evident for common B-cell subtypes. These results are consistent with the known biology of NHL and provide insights into shared pathogenic components, including apoptosis and immune regulation, for the major B-cell lymphoma subtypes.
Introduction
Non-Hodgkin lymphoma (NHL) comprises a heterogeneous group of malignancies of lymphoid tissues. Familial aggregations1 and NHL candidate gene and genome-wide association studies support the influence of common gene variants on NHL risk.2-7 We conducted a validation study within the large international consortium of lymphoma, InterLymph, to confirm associations of 67 putative B-cell NHL risk alleles previously identified in smaller studies through a meta-analysis of 5633 B-cell NHL cases and 7034 controls.
Methods
Participating InterLymph studies
Data were made available from the British Columbia NHL study8 ; the Connecticut Women's NHL Study from Yale University9 ; the European EpiLymph multicenter study10 ; the Mayo Clinic NHL study11 ; the National Cancer Institute/Surveillance, Epidemiology, and End Results Multi-Center Case-Control Study (NCI-SEER)12 ; the New South Wales (NSW) lymphoma study13 ; and the University of California, San Francisco (UCSF1)/University of California, Berkeley (UCSF2) studies of NHL14,15 (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Demographic data, including age at diagnosis for cases and age at interview for controls, sex, self-reported race and Hispanic/Latino ethnicity, HIV status, histologic subtype classification and/or International Classification of Diseases for Oncology code for lymphoma diagnoses, and genotype data were obtained for each study.
Cases were diagnosed with incident lymphoma between 1989 and 2008. Analyses were restricted to participants ≥ 18 years of age who were HIV-negative, non-Hispanic whites diagnosed with B-cell NHL. Histologic subtypes were grouped for analyses using the pathology coding scheme developed by InterLymph collaborators and pathologists based on the current World Health Organization classification.16 Outcomes for which results are presented include B-NHL and specific subtypes, including diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), chronic lymphocytic leukemia (CLL)/small lymphocytic lymphoma (SLL), mantle cell lymphoma, and marginal zone lymphoma. The institutional review boards of participating centers approved each study, which were conducted in accordance with the Declaration of Helsinki.
Genotyping
Ninety-four single nucleotide polymorphisms (SNPs) were originally nominated based on a previously identified association with NHL in at least a single study of moderate size. The list of candidate variants was generated by consensus among the members of the InterLymph Genetics Working Group. In total, 67 unique SNPs were genotyped in 4 or more studies, which was our inclusion criterion (supplemental Table 2). Genotyping was performed using Illumina Golden Gate technology in the British Columbia, EpiLymph, Mayo Clinic, NCI-SEER, NSW, and Yale studies and the Illumina CNV370 array was used in UCSF2. TaqMan (Applied Biosystems) was used in the UCSF1 study and for partial or all genotyping in the NCI-SEER, NSW, Yale, and UCSF2 studies.
Statistical analysis
For each study, odds ratios (ORs) were estimated from multivariate logistic regression analysis under an additive model using PLINK and adjusting for age and sex, except for the Yale study, for which only summary statistics and no individual data were available. Deviations from Hardy-Weinberg equilibrium in controls were tested in each study; SNPs with a Hardy-Weinberg P < .001 were removed from that particular study before meta-analysis (supplemental Table 2). ORs from individual studies were combined in a meta-analysis under fixed- and random-effects inverse variance models using the metagen function from the “meta” package in R. Heterogeneity across studies was tested with the Cochran Q test and the I2 heterogeneity index using the same R package. Associations were considered significant at a meta-analysis P threshold of 1.32 × 10−3, corresponding to a false discovery rate of 5%.
Results and discussion
In the present study, the largest validation study of NHL, our meta-analysis validated the association of 9 of the 67 candidate SNPs, which showed statistically significant P values for the random-effects model for at least one B-cell NHL subtype (supplemental Table 3). The strongest associations were seen for rs3789068 in BCL2L11 (BCL-2 interacting mediator of cell death or BIM) and rs3132453 in PRRC2A (proline-rich coiled-coil 2A or BAT2). Interestingly, the effects of both SNPs were seen consistently across the 3 major B-cell lymphoma subtypes (Q > 0.10, supplemental Figure 1), contrary to several other previously identified risk loci that showed subtype specificity.
A total of 5330 B-cell NHL cases and 6260 controls from 7 InterLymph studies were available for the pooled analysis of rs3789068 in BCL2L11. In the combined analysis, this variant was associated with a 1.21-fold (P random = 2.21 × 10−11) increased risk for B-cell NHL and similar risk estimates for FL, CLL/SLL, and DLBCL. No significant heterogeneity was found among studies (Figure 1 and supplemental Table 4). Combined risk estimates remained robust after exclusion of the NCI-SEER, NSW, Yale, and Mayo Clinic studies, which provided the first evidence of an association of rs3789068 specifically with FL risk17,18 (supplemental Table 4 rows 8 and 9).
ORs associated with rs3789068. Shown are ORs for the risk of B-cell NHL (A), DLBCL (B), FL (C), and CLL/SLL (D) associated with rs3789068 (G > A; BCL2L11) genotypes in individual studies (squares) and a pooled analysis (diamonds). The sizes of the rectangles for individual studies are proportional to the weight (sample size) of each individual study on the pooled estimate.
ORs associated with rs3789068. Shown are ORs for the risk of B-cell NHL (A), DLBCL (B), FL (C), and CLL/SLL (D) associated with rs3789068 (G > A; BCL2L11) genotypes in individual studies (squares) and a pooled analysis (diamonds). The sizes of the rectangles for individual studies are proportional to the weight (sample size) of each individual study on the pooled estimate.
BCL2L11, located at chromosome 2q13, is a member of the BCL2 family and is a key player in regulation of apoptosis in T- and B-cell homeostasis. BCL2L11 binds with high affinity to prosurvival Bcl-2-like proteins and plays a prominent role in the initiation of apoptosis. Evidence of a crucial role for BCL2L11 in lymphomagenesis is accumulating in mice and in humans.19,20
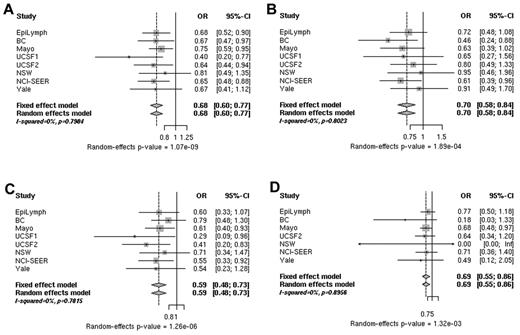
Analysis of data from 5633 B-cell NHL cases and 7034 controls for rs3132453 in PRRC2A revealed a highly statistically significant association. In the combined analysis, rs3132453 was associated with a decreased risk for B-cell NHL (OR = 0.68, P random = 1.07 × 10−9) consistently across the main B-cell NHL subtypes, FL, DLBCL, and CLL/SLL (supplemental Figure 1). Significant heterogeneity among studies was not found (Figure 2 and supplemental Table 5). After excluding the Mayo Clinic study, which originally reported a reduced risk of NHL associated with rs3132453,11 risk estimates remained essentially unchanged (supplemental Table 5 rows 8 and 9).
ORs associated with rs3132453. Shown are ORs for the risk of B-cell NHL (A), DLBCL (B), FL (C), and CLL/SLL (D) associated with rs3132453 A > C (PRRC2A) genotypes in individual studies (squares) and a pooled analysis (diamonds). The sizes of the rectangles for individual studies are proportional to the weight (sample size) of each individual study on the pooled estimate.
ORs associated with rs3132453. Shown are ORs for the risk of B-cell NHL (A), DLBCL (B), FL (C), and CLL/SLL (D) associated with rs3132453 A > C (PRRC2A) genotypes in individual studies (squares) and a pooled analysis (diamonds). The sizes of the rectangles for individual studies are proportional to the weight (sample size) of each individual study on the pooled estimate.
rs3132453 lies within PRRC2A and 2.7 kb downstream of BAG6 (or BAT3) in a cluster of HLA-B associated transcripts (BAT1-5) in the HLA complex class III region.21 PRRC2A and BAG6 are, respectively, 42 and 61 kb downstream of TNF. However, there is no linkage disequilibrium (r2 = 0.027 in HapMap-CEU rel#27) between rs3132453 and a previously reported DLBCL risk allele, TNF-308G > A (rs1800629)2,22 rs3132453 is a nonsynonymous variant in PRRC2A that results in an amino acid substitution (V1895L), but in silico analyses using SIFT and PolyPhen2 did not reveal any putative structurally relevant effect. However, it is intriguing that rs3132453 and additional PRRC2A SNPs are associated with other immune-related diseases such as rheumatoid arthritis23 and atopic asthma.24 Despite several disease associations with SNPs in or near PRRC2A, the gene's biologic function remains largely unknown. More is known of the involvement of a nearby apoptotic regulator gene, BAG6 (BAT3), in immune function and tumorigenesis. Bat3 is important for DNA-damage–induced apoptosis25 and for successful killing of DLBCL cells via BCL6 inhibitors.26
We have also verified in the present study that 3 variants in the TNF/LTA locus (rs2239704, rs909253, and rs2844482) were specifically associated with DLBCL (supplemental Table 3) and significant heterogeneity among common B-cell NHL subtypes was observed for rs2239704 and rs909253 (Q < 0.10, supplemental Figure 1). These 3 SNPs are in moderate to low linkage disequilibrium to the established DLBCL risk locus, TNF-308G > A (r2 = 0.18, 0.53, and 0.07, respectively). These data add to previously published data on the relevance of the TNF/LTA locus on DLBCL etiology and call for fine mapping efforts to further clarify the role of this locus in DLBCL susceptibility. Further validated SNPs associated with B-NHL and FL risk were located in CASP8 (rs3769821) and in TLR1 (rs4833103 and rs10008492) and in NAT2 (rs1799930), respectively (supplemental Table 3).
In summary, pooled analyses in the framework of the InterLymph consortium including studies from North American, Europe, and Australia have provided robust evidence of associations between SNPs in PRRC2A and BCL2L11 and the risk of B-cell lymphoma across the main subtypes. These findings highlight the relevance of genetic variation in the HLA region and in apoptotic regulators in the pathogenesis of B-cell lymphoma.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the José Carreras Leukemia Foundation (grant DJCLS-R04/08) and the German Federal Ministry of Education and Research (BMBF-01-EO-0803; A.N., principal investigator; EpiLymph Germany genetic study); the National Institutes of Health (NIH; grants CA122663, CA154643-01A1, and CA104682; C.F.S., principal investigator; UCSF1/UCSF2 studies and grants CA45614 and CA89745; P.M.B., principal investigator; UCSF1/UCSF2 studies); the National Cancer Institute of Canada; the Canadian Institutes of Health Research, and the Michael Smith Foundation for Health Research (British Columbia study); the Intramural Research Program of the NIH (P.H., principal investigator; NCI-SEER study); the European Commission (grant QLK4-CT-2000-00422; EpiLymph study); NIH contract NO1-CO-12400 (EpiLymph Italy study); Compagnia di San Paolo-Programma Oncologia (P.C., principal investigator; EpiLymph Italy study); the Federal Office for Radiation Protection (grants StSch4261 and StSch4420; N.B., principal investigator; EpiLymph Germany study); the Spanish Ministry of Health (grant references CIBERESP, FIS principal investigator11/01810, RCESP C03/09, RTICESP C03/10, and RTIC RD06/0020/0095) and the Agència de Gestió d'Ajuts Universitaris i de Recerca-Generalitat de Catalunya (2009SGR1465; S.d.S., principal investigator; EpiLymph Spain study); the National Health and Medical Research Council of Australia, the Cancer Council New South Wales, and the University of Sydney Medical Foundation Program (B.A., principal investigator; NSW study); the Intramural Research Program of the NCI (NSW study); NCI grant CA62006 and the Intramural Research Program of the NCI (T.Z., principal investigator; Yale study); NIH grant CA92153 (J.C., principal investigator; Mayo Clinic study); the Health Research Board, Ireland (EpiLymph Ireland study); and Cancer Research, Ireland (InterLymph; A.S., principal investigator; EpiLymph Ireland study).
National Institutes of Health
Authorship
Contribution: The project was conceived and led by cochairs of the InterLymph Genetic Working Group: A.N., S.L.S, A.B., S.S.W., N.R., and C.F.S. Investigators who carried out and provided data from each study are as follows: for the British Columbia study, J.J.S. and A.B.; for EpiLymph-Germany, N.B., K. B., and A.N.; for EpiLymph-France, M.M.; for EpiLymph-Spain, S.D.S. and Y.B.; for EpiLymph-Italy, P.L.C.; for EpiLymph-Ireland, A.S.; and for EpiLymph-Czech Republic, L.F. (EpiLymph is coordinated by P. Brennan and P. Boffetta); for the Mayo Clinic, J.R.C., S.L.S., T.D.S., A.J.N., S.M.A., T.D.S, and A.H.W.; for NCI-SEER, P.H., S.S.W., J.R.C., W.C., R.K.S., A.J.D.R., and L.M.; for NSW, B.A., A.K., C.M.V., and A.G; for UCSF1/UCSF2, E.H., P.M.B., M.T.S., D.R.S., A.J.N., J.R., S.A., L.A., L.C., and C.F.S.; and for Yale, Q.L., M.P., T.Z., and Y.Z. The statistical analysis was carried out by L.C. Data were analyzed and interpreted by A.N., L.C., S.L.S., A.B., L.M., S.S.W., N.R., and C.F.S. The manuscript was written and revised by A.N., L.C., and C.F.S. and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alexandra Nieters, Centre of Chronic Immunodeficiency, University Medical Center Freiburg, Engesser Strasse 4, D-79108 Freiburg, Germany; e-mail: alexandra.nieters@uniklinik-freiburg.de.
References
Author notes
A.N., L.C., S.L.S., A.B.-W., and L.M. contributed equally to this work.
S.S.W., N.R., and C.F.S. share senior authorship.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal