Abstract
To identify genomic alterations in chronic lymphocytic leukemia (CLL), we performed single-nucleotide polymorphism–array analysis using Affymetrix Version 6.0 on 353 samples from untreated patients entered in the CLL8 treatment trial. Based on paired-sample analysis (n = 144), a mean of 1.8 copy number alterations per patient were identified; approximately 60% of patients carried no copy number alterations other than those detected by fluorescence in situ hybridization analysis. Copy-neutral loss-of-heterozygosity was detected in 6% of CLL patients and was found most frequently on 13q, 17p, and 11q. Minimally deleted regions were refined on 13q14 (deleted in 61% of patients) to the DLEU1 and DLEU2 genes, on 11q22.3 (27% of patients) to ATM, on 2p16.1-2p15 (gained in 7% of patients) to a 1.9-Mb fragment containing 9 genes, and on 8q24.21 (5% of patients) to a segment 486 kb proximal to the MYC locus. 13q deletions exhibited proximal and distal breakpoint cluster regions. Among the most common novel lesions were deletions at 15q15.1 (4% of patients), with the smallest deletion (70.48 kb) found in the MGA locus. Sequence analysis of MGA in 59 samples revealed a truncating mutation in one CLL patient lacking a 15q deletion. MNT at 17p13.3, which in addition to MGA and MYC encodes for the network of MAX-interacting proteins, was also deleted recurrently.
Introduction
Chronic lymphocytic leukemia (CLL) has a highly variable clinical course, with some patients surviving for decades without treatment and others developing rapid disease progression and early requirement for treatment. Biologic disease characteristics have been identified that predict the outcome of patients. In addition to the mutational status of the IGHV gene, genomic abnormalities are among the strongest prognostic markers.1,2 The current gold standard for detecting genomic aberrations is fluorescence in-situ hybridization (FISH). A hierarchical prognostic model was established identifying del(17)(p13) and del(11)(q22.3) as independent predictors of faster disease progression and inferior survival, whereas 13q deletion occurring as the sole abnormality is associated with a favorable outcome.1 The prognostic value of this model was confirmed in prospective trials.3,4 However, these genomic abnormalities fail to explain the heterogeneity of the disease to the full extent because patients with a favorable genetic profile can also present with rapid disease progression, treatment failure, and poor outcome.
So far, the target genes of only few of these recurrent lesions have been identified. The unfavorable prognosis of patients with del(17)(p13) is supposed to be due to alteration of the tumor protein 53 gene (TP53).5,6 Loss of function of the remaining TP53 allele by somatic mutations can be found in approximately 80% of 17p-deleted cases.6,7 In del(11)(q22.3), the ataxia telangiectasia mutated gene (ATM) is the most likely target gene.8,9 Somatic ATM mutations could be found in approximately one-third of 11q-deleted CLL cases.10 Concerning del(13)(q14), the 2 miRNAs miR-15a and miR-16-1 have been implicated in pathogenesis.11 Mice carrying a deletion of the Dleu2/miR-15a/miR-16-1 cluster develop lymphoproliferative disorders partly resembling human CLL.12,13 Genes involved in other genomic aberrations such as trisomy 12, del(6q), trisomy 2p, trisomy 19, or trisomy 8q, are unknown.
Array-based genomic technologies allow a genome-wide screening for genetic lesions. The use of matrix-comparative genomic hybridization in CLL led to the discovery of novel disease-related aberrant regions.14 The development of high-resolution genome-wide scanning technologies, such as single-nucleotide polymorphism (SNP)–array analysis, has enabled the detection of subtle copy number alterations (CNAs) and regions of copy-neutral loss-of-heterozygosity (CN-LOH), which is also termed uniparental disomy. Using these arrays, genetic alterations of key regulators in B-lymphoid development and cell cycle could be identified in B-progenitor pediatric acute lymphoblastic leukemia.15-17 CN-LOH is supposed to play a role in the pathogenesis of a variety of hematologic malignancies.18 So far, only a few SNP-array based studies have been performed in CLL19-23 and these studies were limited by heterogeneous patient cohorts and by the lack of intraindividual, nontumor reference DNA.
The objectives of the present study were to identify novel regions of recurring CNAs, to delineate minimally affected regions of recurrent lesions and CN-LOH, and to perform sequence analyses of candidate disease genes located in critical regions. We performed high-resolution SNP-array analysis on samples from 353 previously untreated CLL patients who were enrolled in the CLL8 treatment trial of the German CLL Study Group (GCLLSG).
Methods
Patients and samples
The present study included peripheral blood samples from 353 previously untreated CLL patients (Binet stage A, n = 16; stage B, n = 217; stage C, n = 119; and stage unknown, n = 1). All samples were taken at enrollment in the CLL8 multicenter trial for first-line treatment.4 Mononuclear cells were isolated by Ficoll density gradient centrifugation and an immunomagnetic tumor cell enrichment via CD19 (Midi MACS; Miltenyi Biotec) was done on all cases. A CD19− fraction of mononuclear cells with a tumor cell load of less than 5% was available for 144 samples as intraindividual normal reference DNA. Data on genomic aberrations [del(11)(q22.3), +12, del(13)(q14), del(17)(p13), and t(11;14)] was available for all but 2 samples. The IGHV and TP53 mutational status was available for 343 and 349 cases, respectively. Genetic characteristics are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Written informed consent and local ethics committee approval was obtained in accordance with the Declaration of Helsinki for all patients.
SNP-array analysis
Genomic DNA was extracted from frozen mononuclear cells of the CD19+ and CD19− fractions (Allprep DNA/RNA midi kit; QIAGEN) and hybridized to the genome-wide Human SNP Array Version 6.0 according to the manufacturer's protocol (Affymetrix). SNP genotype calls were generated by applying the birdseed algorithm in Genotyping Console Version 4.0 (Affymetrix) using at least 50 arrays in each analysis. After extraction of the raw data using dChipSNP,24 data were normalized using reference alignment.25 Aberrant copy number segments were detected by binary circular segmentation26 and LOH analysis was performed using dChipSNP. Segmentation was done pairwise for the 144 samples with intraindividual reference DNA. For samples lacking matched normal material, the segmentation was computed against a pool of 10 sex-matched reference samples. Resulting segments with a window of 5 consecutive markers and mean log2 ratios of > 0.2 and < −0.2 were visually inspected using dChipSNP to exclude inherited copy number variants and false calls because of experimental artifacts (eg, interbatch effects or noise). Lesions were cross-checked against public databases of copy number polymorphisms (http://www.genome.ucsc.edu/). Lesions occurring in a subclone with a clone size less than 25% were revised using the aroma.affymetrix software package for an exact determination of segment boundaries.27 When defining the number of CNAs per patient, discontinuous lesions on one chromosome were counted as one aberration as long as their segments were valuated with approximately the same mean log2 ratio. Biallelic deletions were counted as 2 aberrations if deletion sizes differed on the 2 alleles. If candidate genes were identified only by unpaired analyzed samples, an exploratory comparison of the mean log2 ratio in tumor and reference DNA samples was performed. According to prior experience, CN-LOH was considered to be a true lesion when containing homozygous SNPs in at least 20 consecutive markers with less than 10% intervening or conflicting calls.28 CN-LOH in unpaired analyzed patients was also required to have a size larger than 10 Mb and to have coincidence with a proven tumor-specific CN-LOH or CNA. Size position and location of genes were identified with the UCSC Genome Browser (assembly March 2006, NCBI36/hg18; http://www.genome.ucsc.edu/). For the presence of chromothripsis, data were analyzed as described previously and was inferred in cases in whom at least 10 switches between 2 or more copy number states were apparent on an individual chromosome.29 Microarray raw data were made publicly available at the Gene Expression Omnibus (access no. GSE36908).
FISH
Based on availability of material, a subset of newly identified recurrent CNAs was validated using FISH, as described previously.1 DNA clones were ordered at http://www.imagenes-bio.de, and the following chromosomal locations were analyzed (respective clones in parentheses): 2p16.1-p15 (RP11-17D23; RP11-81L13; RP11-89J7), 19p13.2 (RP11-197O4; RP11-841N21; RP11-79F15), 8q24.21(yeast artificial chromosome clone 935A12), 15q15.1 (RP11-380D11), 17p13.3 (RP11-380H7), 3p21.31 (RP11-88B8), 1p35.3 (RP11-261P19), 18p11.23 (RP11-789C17), 10q24.32 (RP11-18I14). To rule out cryptic translocations of the NFKB2 gene at 10q24, FISH analysis was performed with 2 probes flanking the locus (RP11-107I14 and RP11-541N10).
Mutational analysis
TP53 gene mutational analysis was performed as described previously.30 Based on the SNP-array data, additional DNA sequence analyses were performed for ATRIP, RPA2, and MGA. The entire coding region of each gene was amplified using intron-exon flanking primer pairs and subjected to direct sequencing in accordance with standard protocols. Primers were selected by Primer3 Input Version 0.4.0 (http://frodo.wi.mit.edu/primer3/) and are listed in supplemental Table 2.
Statistical analysis
Distributions of patient subgroups defined by aberrations were tested for significance by a Wilcoxon rank-sum test. To investigate the association between recurrent genomic aberrations and outcome, estimates of progression-free survival (PFS) and overall survival (OS) were computed using the Kaplan-Meier method. Comparisons were performed using the exact log-rank test and significance was set at P < .05.
Results
Frequency of CNAs
High-resolution genome-wide copy number analysis was performed on CD19-enriched PBMCs from 353 previously untreated CLL patients (supplemental Tables 3 and 4). Intraindividual reference DNA from CD19− fractions was available for paired analysis in 144 cases (supplemental Figure 1). Paired analysis revealed a mean of 1.81 somatically acquired CNAs per case. In unpaired analysis (209 samples), the mean number of CNAs was higher (2.18 CNAs per case) even after cross-checking against public databases of copy number polymorphisms. Therefore, data from paired analysis were used to identify new genomic regions affected by CNAs and/or uniparental disomy, whereas data from unpaired analyzed cases were used only to determine the frequencies of these aberrations and to delineate critical regions of recurrent lesions.
Among the 144 cases with paired intraindividual reference DNA, CNAs were identified in 127 cases (88%); 56 (39%) cases had 1 CNA, 33 (23%) cases had 2 CNAs, 23 (16%) cases had 3 CNAs, and only 15 (10%) cases had more than 3 CNAs, with a maximum of 6 CNAs per case. In total, 261 tumor-specific CNAs were identified. Deletions were far more common than gains, with 1.44 losses versus 0.38 gains per case. No significant difference in the number of CNAs per case was seen between mutated and unmutated IGHV status (1.85 vs 1.80 CNAs per case), whereas according to the hierarchical model of genetic alterations, an increased number of CNAs was found in genetic categories associated with inferior survival. Cases with the high-risk genomic changes del(11)(q22.3), del(17)(p13) detected by FISH, and/or TP53 mutation (n = 48) had a significantly higher number of CNAs than cases without these lesions (n = 96): 2.61 versus 1.40 CNAs per case (P < .01; Table 1). Ten of the 15 cases with more than 3 CNAs were associated with del(11)(q22.3), del(17)(p13), and/or TP53 mutations.
Mean number of CNAs per patient detected by paired SNP array analysis of 144 CLL patients
| . | All CNAs . | CNAs detected beyond FISH analysis . |
|---|---|---|
| All patients (n = 144) | 1.81 | 0.72 |
| IGHV mutated (n = 61) | 1.85 | 0.70 |
| IGHV unmutated (n = 79) | 1.80 | 0.75 |
| No aberration (n = 28) | 0.86 | 0.86 |
| del(13)(q14) as sole abnormality (n = 49) | 1.49 | 0.31 |
| trisomy 12 (n = 19) | 2.11 | 0.84 |
| del(11)(q22.3) (n = 33) | 2.48 | 0.91 |
| del(17)(p13)/TP53 mutated (n = 15) | 2.80 | 1.27 |
| Low-risk patients (n = 96) | 1.40 | 0.55 |
| High-risk patients (n = 48) | 2.61 | 1.06 |
| . | All CNAs . | CNAs detected beyond FISH analysis . |
|---|---|---|
| All patients (n = 144) | 1.81 | 0.72 |
| IGHV mutated (n = 61) | 1.85 | 0.70 |
| IGHV unmutated (n = 79) | 1.80 | 0.75 |
| No aberration (n = 28) | 0.86 | 0.86 |
| del(13)(q14) as sole abnormality (n = 49) | 1.49 | 0.31 |
| trisomy 12 (n = 19) | 2.11 | 0.84 |
| del(11)(q22.3) (n = 33) | 2.48 | 0.91 |
| del(17)(p13)/TP53 mutated (n = 15) | 2.80 | 1.27 |
| Low-risk patients (n = 96) | 1.40 | 0.55 |
| High-risk patients (n = 48) | 2.61 | 1.06 |
Genomic aberrations are grouped according to the hierarchical model.1 High-risk is defined by the presence of del(11)(q22.3), del(17)(p13), and/or TP53 mutation; low-risk by the absence of these alterations.
Comparison of data from FISH and SNP-array analyses
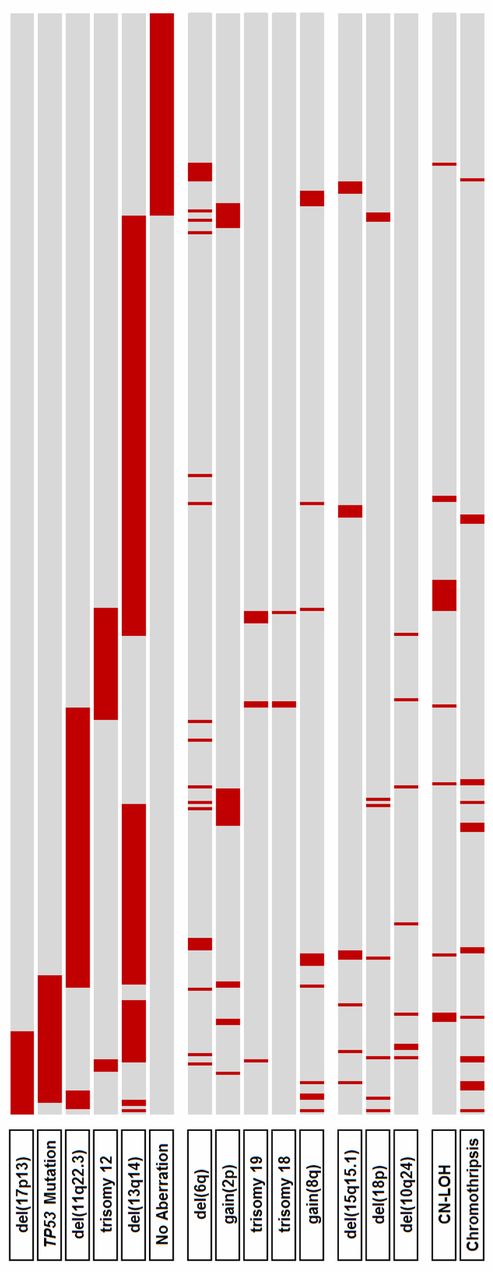
A total of 159 of the 261 (61%) CNAs detected by paired analysis were already known based on routine FISH analyses performed for del(11)(q22.3), +12, del(13)(q14), and del(17)(p13). Only 102 additional CNAs were identified. Among others, these CNAs included genomic lesions previously known in CLL, such as del(6q), +8q, +2p, +19, and +18. The majority of lesions detected beyond the FISH panel were found in cases with high-risk genomic changes: 1.06 CNAs per case versus 0.55 in cases without.
Combining data from paired and unpaired analyses, all genomic aberrations previously identified by FISH analysis were also detected by SNP-array analysis (supplemental Figure 2). Few additional lesions were detected: del(11)(q22.3) in 3 cases and del(13)(q14) in 2 cases. Three deletions were too small to be detected by the FISH probe [del(11)(q22.3) in case number CLL075 and case number CLL237up; del(13)(q14) in case number CLL275up]. The other 2 deletions were subclonal lesions in cases with low tumor burden presumably not detectable before tumor cell enrichment [del(11)(q22.3) in case number CLL131 and del(13)(q14) in case number CLL140].
Identification of novel CNAs
Novel recurrent aberrations occurred at a low frequency (Table 2). The following regions were affected in at least 2 paired cases analyzed (frequencies based on paired and unpaired analyses): del(15)(q15.1) (4%; clustering around position 39 800 000 Mb with the smallest lesion of 70.48 kb lying within the MAX gene associated [MGA] gene), del(10)(q24) (2%; clustering around position 104 500 000 Mb close to the NFKB2 locus), del(14)(q24.3) (2%; minimally deleted region [MDR] 462 kb), del(1)(q23.3) (1%; MDR 445 kb), del(1)(q42.12) (1%; MDR 635 kb), del(3)(p21.31) (1%; MDR 1.256 Mb), del(6)(p22.1) (1%; MDR 221 kb), gain(6)(p25.3) 1%; minimally amplified region 1.391 Mb), del(9)(q13-q21.11) (1%; MDR 648 kb), del(14)(q13.2) (1%; MDR 1.076 Mb). A less stringent analysis taking into consideration regions that proved to be tumor specific in at least 1 case revealed additional recurrent aberrations: del(18)(pter-p11.22) (3%; MDR 9.470 Mb), del(4)(p16) (2%; MDR 434 kb), del(8)(p12) (1%; MDR 91 kb), and del(20)(pter-p12.2) (1%; MDR 11.605 Mb; supplemental Table 5). All deletions analyzed by FISH could be confirmed (supplemental Table 6).
New recurrent copy number alterations
| Genomic lesion . | Paired patients, n . | Unpaired patients, n . | Frequency within cohort . | Start . | Stop . | Segment length . | Genes . |
|---|---|---|---|---|---|---|---|
| del(15)(q15.1) | 1* | 7† | 4% | 39 745 774 | 39 816 254 | 70 480 | MGA |
| del(10)(q24) | 2* | 4* | 2% | 104 146 465 | 105 040 096 | 893 631 | 19 genes (eg, NFKB2) |
| del(14)(q24.3) | 2 | 3‡ | 2% | 75 152 557 | 75 614 215 | 461 658 | FLVCR2, C14orf1, TTLL5, BCYRN1, TGFB3, C14orf179 |
| del(1)(q23.3) | 2 | 1 | 1% | 158 997 196 | 159 441 807 | 444 611 | 19 genes |
| del(1)(q42.12) | 2 | 1 | 1% | 223 854 863 | 224 490 349 | 635 486 | 12 genes |
| del(3)(p21.31) | 2 | 2 | 1% | 47 565 887 | 49 821 882 | 1 255 995 | 58 genes (eg, ATRIP) |
| del(6)(p22.1) | 3 | 2 | 1% | 26 121 993 | 26 343 281 | 221 288 | Histone cluster, HFE |
| gain(6)(p25.3) | 2 | 1 | 1% | 94 649 | 1 485 307 | 1 390 658 | DUSP22, IRF4, EXOC2, HUS1B, LOC285768, FOXQ1, FOXF2 |
| del(9)(q13-q21.11) | 2 | 3 | 1% | 70 139 836 | 70 788 159 | 648 323 | PGM5, C9orf71, PIP5K1B |
| del(14)(q13.2) | 2 | 1% | 34 107 436 | 35 183 612 | 1 076 176 | 11 genes (eg, NFKBIA) |
| Genomic lesion . | Paired patients, n . | Unpaired patients, n . | Frequency within cohort . | Start . | Stop . | Segment length . | Genes . |
|---|---|---|---|---|---|---|---|
| del(15)(q15.1) | 1* | 7† | 4% | 39 745 774 | 39 816 254 | 70 480 | MGA |
| del(10)(q24) | 2* | 4* | 2% | 104 146 465 | 105 040 096 | 893 631 | 19 genes (eg, NFKB2) |
| del(14)(q24.3) | 2 | 3‡ | 2% | 75 152 557 | 75 614 215 | 461 658 | FLVCR2, C14orf1, TTLL5, BCYRN1, TGFB3, C14orf179 |
| del(1)(q23.3) | 2 | 1 | 1% | 158 997 196 | 159 441 807 | 444 611 | 19 genes |
| del(1)(q42.12) | 2 | 1 | 1% | 223 854 863 | 224 490 349 | 635 486 | 12 genes |
| del(3)(p21.31) | 2 | 2 | 1% | 47 565 887 | 49 821 882 | 1 255 995 | 58 genes (eg, ATRIP) |
| del(6)(p22.1) | 3 | 2 | 1% | 26 121 993 | 26 343 281 | 221 288 | Histone cluster, HFE |
| gain(6)(p25.3) | 2 | 1 | 1% | 94 649 | 1 485 307 | 1 390 658 | DUSP22, IRF4, EXOC2, HUS1B, LOC285768, FOXQ1, FOXF2 |
| del(9)(q13-q21.11) | 2 | 3 | 1% | 70 139 836 | 70 788 159 | 648 323 | PGM5, C9orf71, PIP5K1B |
| del(14)(q13.2) | 2 | 1% | 34 107 436 | 35 183 612 | 1 076 176 | 11 genes (eg, NFKBIA) |
Loss in close vicinity in 1 patient.
Losses in close vicinity in 5 different patients.
Losses in close vicinity in 2 different patients.
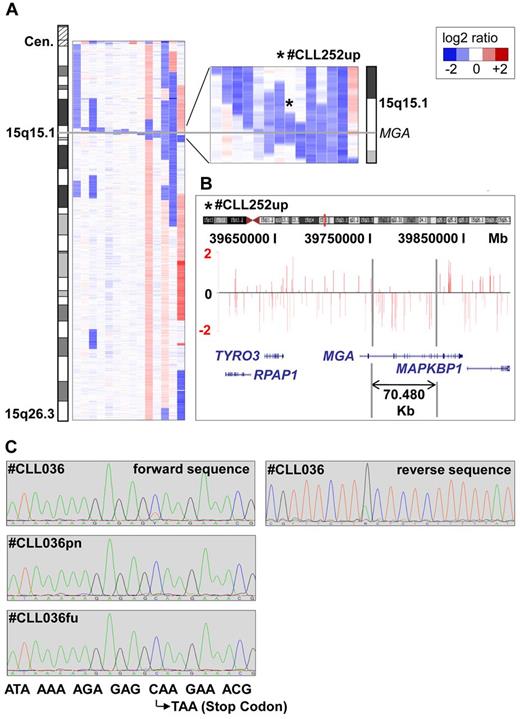
Among the novel CNAs, losses in 15q15.1 were most common, with 14 of 353 (4%) cases being affected (Figure 1A-C). The smallest lesion was 70.48 kb in size (patient number CLL252up), proved to be tumor specific, and was located within the MGA gene (Figure 1B and supplemental Figure 3). Altogether, 8 of the 14 deletions contained the MGA locus. The others were in the vicinity of MGA, either centromeric (n = 5) or telomeric (n = 1), with a maximum distance of 1.373 Mb. Mutational analysis of all coding exons of MGA in 59 CLL samples, including 9 patients with del(15)(q15.1) and 50 randomly chosen patients without the deletion, revealed a tumor-specific point mutation in exon 17 leading to a premature stop codon in 1 case without del(15)(q15.1) (case number CLL036). In a follow-up sample taken 9 months after enrollment, this mutation in MGA was no longer detectable (Figure 1C), whereas the proportion of the 17p-deleted clone rose from 30% to 95%. Exploratory survival analysis showed no difference in PFS and OS between patients with and without del(15)(q15.1) (supplemental Figure 4).
Microdeletions in band 15q15.1 involving the MGA locus. (A) Inferred log2-ratio of SNP copy number data on chromosome 15q (median smoothing with a window of 5 consecutive markers; losses are indicated in blue, gains in red). Eight deletions are covering the MGA locus, the other 5 are located in close vicinity to a maximum distance of 1.373 Mb. (B) Raw log-ratio, chromosome 15 (case number CLL252up) displayed with the UCSC genome browser (hg18). Red bars represent the determined log2 ratio of single probe sets sorted by their physical position along the chromosome. Shown is the smallest deletion on 15q15.1 within the MGA locus, 70.48 kb in size. (C) Forward and reverse sequence analysis of exon 17 revealing a subclonal nucleotide exchange C → T in position 6143 that codes for a premature stop codon in case number CLL036. This sequence variation was not detectable in the nontumor reference sample (case number CLL036pn) and in a follow-up sample taken after treatment (case number CLL036fu).
Microdeletions in band 15q15.1 involving the MGA locus. (A) Inferred log2-ratio of SNP copy number data on chromosome 15q (median smoothing with a window of 5 consecutive markers; losses are indicated in blue, gains in red). Eight deletions are covering the MGA locus, the other 5 are located in close vicinity to a maximum distance of 1.373 Mb. (B) Raw log-ratio, chromosome 15 (case number CLL252up) displayed with the UCSC genome browser (hg18). Red bars represent the determined log2 ratio of single probe sets sorted by their physical position along the chromosome. Shown is the smallest deletion on 15q15.1 within the MGA locus, 70.48 kb in size. (C) Forward and reverse sequence analysis of exon 17 revealing a subclonal nucleotide exchange C → T in position 6143 that codes for a premature stop codon in case number CLL036. This sequence variation was not detectable in the nontumor reference sample (case number CLL036pn) and in a follow-up sample taken after treatment (case number CLL036fu).
Del(10)(q24) was found in 8 cases (2%) and clustered around the NFKB2 locus (supplemental Figure 5). A commonly deleted region could not be defined. The smallest deletion was 511.5 kb in size (case number CLL323up). Cryptic translocations could be ruled out by FISH analysis using deletion flanking probes.
Deletion 18p was observed in 9 of 353 (3%) cases. In 6 of the 9 cases, this deletion was associated with high-risk genomic changes and 7 cases had an unmutated IGHV status. On average, cases had 3 other aberrations (range, 1-5) and a concomitant trisomy 2p was found in 5 of 9 cases. A concomitant del(17)(p13) was present in 3 cases.
Identification of CN-LOH
Within the 144 paired-analyzed cases, 39 cases of CN-LOH were identified. However, only 9 of them proved to be tumor specific as determined by comparison with nontumor control DNA. Whereas tumor-specific CN-LOH had a median size of 48.4 Mb (range, 2.5-199.3), CN-LOHs also present in nonmalignant tissue (required minimal size of 5 Mb) were smaller on average, with a median size of 12.1 Mb (range, 5.1-122). Therefore, in unpaired analyses, CN-LOH was only taken into consideration when it had a size of at least 10 Mb and when it occurred in a region previously shown to be altered tumor specifically by recurrent CNA or CN-LOH. This led to 20 CN-LOHs within the whole cohort (Table 3). CN-LOH was identified on 13q in 10 cases, on 17p in 3 cases, and on 11q in 2 cases. All cases with CN-LOH on 13q harbored a focal biallelic del(13)(q14) with exactly the same deletion boundaries on the 2 alleles (supplemental Figure 6). All cases with CN-LOH on 17p carried homozygous TP53 mutations. On 11q, both CN-LOHs were accompanied by a monoallelic deletion encompassing the ATM locus. One CN-LOH was identified on 12q21.33-qter (case number CLL058). Outside of the previously described critical regions, only 3 CN-LOHs were found on chromosome 1p (n = 1; case number CLL037), 3p (n = 1; case number CLL032), and 22q (n = 1; case number CLL094).
Copy-neutral loss of heterozygosity
| Chromosome . | Probe ID . | Start . | Stop . | Length, mb . | Cytoband . | IGHV mutational status . | FISH . | TP53 mutation . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutated . | Unmutated . | Normal . | Del(13q) . | +12 . | Del(11q) . | Del(17p) . | |||||||
| 1 | CLL037 | 554 484 | 49 046 634 | 48 492 150 | pter-p33 | X | X | ||||||
| 3 | CLL032 | 41 866 | 199 340 830 | 199 298 964 | pter-qter | X | X | ||||||
| 11 | CLL032 | 188 260 | 2 707 910 | 2 519 650 | p15.5 | X | X | ||||||
| CLL175up | 65 369 924 | 134 449 982 | 69 080 058 | q13.1-qter | X | X | NA | ||||||
| CLL243up | 71 431 230 | 134 449 982 | 63 018 752 | q13.4-qter | X | X | X | ||||||
| 12 | CLL058 | 89 090 531 | 132 287 718 | 43 197 187 | q21.33-qter | X | X | NA | |||||
| 13 | CLL041 | 26 496 372 | 114 125 098 | 87 628 726 | q12.13-qter | X | X bidel | ||||||
| CLL070 | 18 876 771 | 114 125 098 | 95 248 327 | q12.11-qter | X | X bidel | |||||||
| CLL110 | 19 273 074 | 114 125 098 | 94 852 024 | q12.11-qter | X | X bidel | |||||||
| CLL211up | 45 637 626 | 114 125 098 | 68 487 472 | q14.12-qter | X | X bidel | |||||||
| CLL213up | 20 194 267 | 114 125 098 | 93 930 831 | q12.11-qter | X | X bidel | |||||||
| CLL234up | 18 833 721 | 114 125 098 | 95 291 377 | q12.11-qter | X | X bidel | |||||||
| CLL235up | 21 684 026 | 114 125 098 | 92 441 072 | q12.11-qter | X | X bidel | |||||||
| CLL288up | 17 963 628 | 114 125 098 | 96 161 470 | q11-qter | X | X bidel | |||||||
| CLL315up | 44 533 321 | 114 125 098 | 69 591 777 | q14.12-qter | X | X bidel | |||||||
| CLL353up | 26 698 826 | 114 125 098 | 87 426 272 | q12.13-qter | X | X bidel | X | ||||||
| 17 | CLL067 | 6689 | 13 997 853 | 13 991 164 | pter-p12 | X | X | X | |||||
| CLL185up | 6689 | 17 985 823 | 17 979 134 | pter-p11.2 | X | X | X | ||||||
| CLL244up | 6689 | 20 649 281 | 20 642 592 | pter-p11.2 | X | X | X | ||||||
| 22 | CLL094 | 15 909 583 | 49 565 872 | 33 656 289 | q11.1-qter | X | X | ||||||
| Chromosome . | Probe ID . | Start . | Stop . | Length, mb . | Cytoband . | IGHV mutational status . | FISH . | TP53 mutation . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutated . | Unmutated . | Normal . | Del(13q) . | +12 . | Del(11q) . | Del(17p) . | |||||||
| 1 | CLL037 | 554 484 | 49 046 634 | 48 492 150 | pter-p33 | X | X | ||||||
| 3 | CLL032 | 41 866 | 199 340 830 | 199 298 964 | pter-qter | X | X | ||||||
| 11 | CLL032 | 188 260 | 2 707 910 | 2 519 650 | p15.5 | X | X | ||||||
| CLL175up | 65 369 924 | 134 449 982 | 69 080 058 | q13.1-qter | X | X | NA | ||||||
| CLL243up | 71 431 230 | 134 449 982 | 63 018 752 | q13.4-qter | X | X | X | ||||||
| 12 | CLL058 | 89 090 531 | 132 287 718 | 43 197 187 | q21.33-qter | X | X | NA | |||||
| 13 | CLL041 | 26 496 372 | 114 125 098 | 87 628 726 | q12.13-qter | X | X bidel | ||||||
| CLL070 | 18 876 771 | 114 125 098 | 95 248 327 | q12.11-qter | X | X bidel | |||||||
| CLL110 | 19 273 074 | 114 125 098 | 94 852 024 | q12.11-qter | X | X bidel | |||||||
| CLL211up | 45 637 626 | 114 125 098 | 68 487 472 | q14.12-qter | X | X bidel | |||||||
| CLL213up | 20 194 267 | 114 125 098 | 93 930 831 | q12.11-qter | X | X bidel | |||||||
| CLL234up | 18 833 721 | 114 125 098 | 95 291 377 | q12.11-qter | X | X bidel | |||||||
| CLL235up | 21 684 026 | 114 125 098 | 92 441 072 | q12.11-qter | X | X bidel | |||||||
| CLL288up | 17 963 628 | 114 125 098 | 96 161 470 | q11-qter | X | X bidel | |||||||
| CLL315up | 44 533 321 | 114 125 098 | 69 591 777 | q14.12-qter | X | X bidel | |||||||
| CLL353up | 26 698 826 | 114 125 098 | 87 426 272 | q12.13-qter | X | X bidel | X | ||||||
| 17 | CLL067 | 6689 | 13 997 853 | 13 991 164 | pter-p12 | X | X | X | |||||
| CLL185up | 6689 | 17 985 823 | 17 979 134 | pter-p11.2 | X | X | X | ||||||
| CLL244up | 6689 | 20 649 281 | 20 642 592 | pter-p11.2 | X | X | X | ||||||
| 22 | CLL094 | 15 909 583 | 49 565 872 | 33 656 289 | q11.1-qter | X | X | ||||||
To be considered as a true tumor-specific CN-LOH, defects had to have homozygous SNPs in at least 20 consecutive markers containing less than 10% intervening or conflicting calls. In unpaired (up) analyzed patients, regions of CN-LOH had to be larger than 10 Mb and had to coincide with a proven tumor-specific CN-LOH or CNA.
NA indicates not applicable.
The genomic regions on 1p and 3p with CN-LOHs were also affected by deletions. Del(3)(p21.31) was found in 4 cases (case numbers CLL022, CLL034, CLL321up, and CLL327up) and an MDR of 537 kb could be identified. Del(1)(p35.1) was found in 2 cases (case numbers CLL117 and CLL180up). The resulting MDRs contained members of the ataxia-telangiectasia mutated and Rad 3-related (ATR) pathway: the replication protein A2 (RPA2) gene on 1p35.3 and the ATR interacting protein (ATRIP) gene on 3p21.31. RPA2 and ATRIP mutational analyses were performed in a subset of 48 patients including those with CN-LOHs and deletions, but no mutations were detected.
Delineating critical regions of known recurrent CNAs
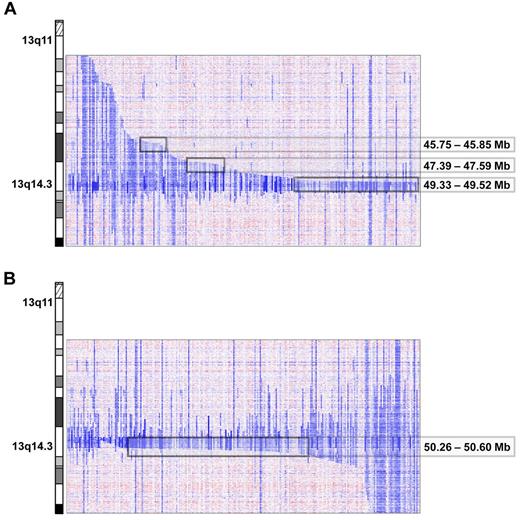
Del(13)(q14) were found in 216 of 353 (61%) cases. The deletion sizes were very heterogeneous, ranging from 267.042 kb to several mega base pairs. A total of 21 cases had discontinuous lesions, of which one had the breakpoints located within the critical region (case number CLL339up, supplemental Figure 7). By deletion mapping, we identified 3 proximal and one distal deletion breakpoint cluster. The major proximal breakpoint cluster (185 kb in size containing breakpoints of 76 deletions) was located between 49.33 and 49.52 Mb and contained the genes C13orf1, RFP2, KCNRG, and DLEU2. Two minor deletion breakpoint clusters centromeric of RB1 were identified between 47.39 and 47.59 Mb (200 kb in size; n = 13 patients) and between 45.75 and 45.85 Mb (87 kb in size; n = 9 patients; Figure 2A). These proximal breakpoint clusters were located in regions with abundance of repetitive DNA sequences, in particular short interspersed nuclear elements. The breakpoint cluster region telomeric of the 13q14 MDR was located between 50.26 and 50.6 Mb (331 kb in size; 118 deletion breakpoints); the genomic segment between this breakpoint cluster and the MDR region only contains the DLEU7 gene (Figure 2B and supplemental Figure 8).
Proximal and distal breakpoint clusters on chromosome 13q. Inferred log2 ratio of copy number data on 13q of all cases carrying a 13q deletion. Three proximal (A) and 1 distal (B) breakpoint cluster at the chromosomal positions indicated were identified visually after arranging the cases by their centromeric and telomeric breakpoints.
Proximal and distal breakpoint clusters on chromosome 13q. Inferred log2 ratio of copy number data on 13q of all cases carrying a 13q deletion. Three proximal (A) and 1 distal (B) breakpoint cluster at the chromosomal positions indicated were identified visually after arranging the cases by their centromeric and telomeric breakpoints.
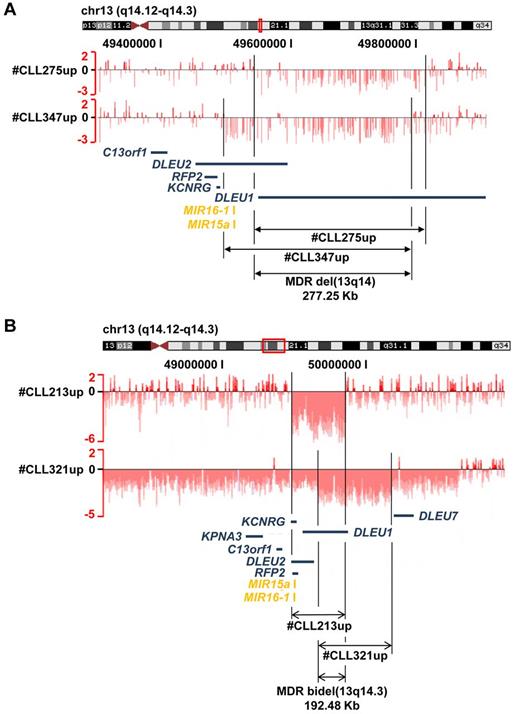
The MDR in 13q14 was 244.861 kb in size and was defined by 2 cases (case numbers CLL275up and CLL347up). The MDR did not encompass the 2 miRNAs mir16-1 and mir15a, but DLEU1 and the first 2 exons of DLEU2 (Figure 3A). Remarkably, the minimal region of biallelic deletion defined by 2 cases (case numbers CLL321up and CLL213up) was 192.477 kb in size and encompassed only DLEU1, not the 2 miRNAs or DLEU2 (Figure 3B). Biallelic 13q14 deletions were found in 36 cases. In 26 cases, deletions on the 2 alleles had different sizes. The remaining 10 cases had exactly the same deletion boundaries and were focal losses within large CN-LOH (supplemental Figure 6). Biallelic deletions were mostly small, with a median size of 1.162 Mb (range, 369 kb to 2.742 Mb).
Minimal consensus regions of monoallelic and biallelic del(13)(q14). Raw log2 ratio (chromosome 13; samples from case numbers CLL275up, CLL347up, CLL213, and CLL321) blotted to the UCSC genome browser (hg18). The minimal monoallelic (A) deleted region encompasses DLEU1 and the first exons of DLEU2, but not the 2 miRNAs, whereas the minimal biallelic (B) deleted region encompasses only DLEU1.
Minimal consensus regions of monoallelic and biallelic del(13)(q14). Raw log2 ratio (chromosome 13; samples from case numbers CLL275up, CLL347up, CLL213, and CLL321) blotted to the UCSC genome browser (hg18). The minimal monoallelic (A) deleted region encompasses DLEU1 and the first exons of DLEU2, but not the 2 miRNAs, whereas the minimal biallelic (B) deleted region encompasses only DLEU1.
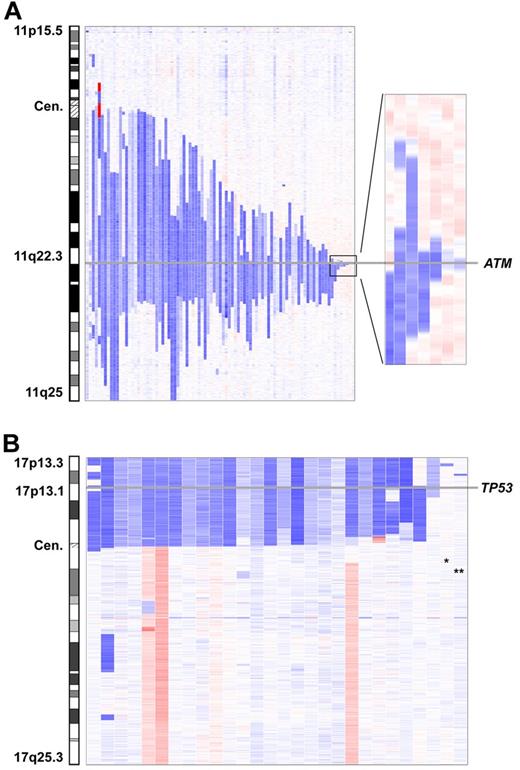
Del(11)(q22.3) were found in 96 of 353 (27%) cases. The deletion sizes were very heterogeneous and no breakpoint clusters were found. The smallest deletion was 78.507 kb in size (case number CLL237up) and was located within the ATM gene; the ATM locus was affected by all 11q deletions (Figure 4A). The BIRC3 locus was concomitantly deleted in 59 of the 96 cases.
Minimally deleted region of del(11)(q22.3) and del(17p13). (A) The smallest deletion in 11q22.3 has a size of 78.5 kb and is located within the ATM locus. (B) Involvement of the TP53 locus in all 17p-deleted patients except 2. The 2 deletions not encompassing TP53 are small deletions on 17p13.3 (*) and 17p13.2 (**).
Minimally deleted region of del(11)(q22.3) and del(17p13). (A) The smallest deletion in 11q22.3 has a size of 78.5 kb and is located within the ATM locus. (B) Involvement of the TP53 locus in all 17p-deleted patients except 2. The 2 deletions not encompassing TP53 are small deletions on 17p13.3 (*) and 17p13.2 (**).
Trisomy 12 was found in 40 of 353 (11%) cases. In addition, partial gains on chromosome 12 were found in 4 unpaired analyzed cases (case numbers CLL260up, CLL269up, CLL308up, and CLL314up). In an exploratory comparison of the mean log2 ratio in the tumor and the intraindividual reference DNA sample, only the gain in case number CLL308up appeared to be tumor specific (supplemental Figure 9). This gain was 13.7 Mb in size and located in 12q24.13-q24.32. The region coincided with a CN-LOH (43.2 Mb in size; case number CLL058).
Deletions on 17p were found in 29 of 353 (8%) cases. In 3 cases (case numbers CLL87, CLL260up, and CLL268up), the deletion likely resulted from the formation of an isochromosome 17q. Deletions usually encompassed the whole short arm (n = 21), with a breakpoint in the centromere (n = 10) or close to the centromere (n = 11). TP53 was part of the deletion in all cases except 2 and was found to be mutated in 85% on the remaining allele. The additional 2 cases had small deletions in 17p13.3 (case number CLL280up; 780 kb in size) and 17p13.2 (case number CLL065; 638 kb in size). Both deletions proved to be tumor specific and contained approximately 10 genes each. Candidate genes in 17p13.3 are the replication protein A1 (RPA1) and the MAX binding protein (MNT) genes (Figure 4B and supplemental Figure 10).
Gains on 2p were found in 25 of 353 cases (7%). The minimally gained region was defined by 2 cases (case numbers CLL311up and CLL062) and was located in 2p16.1-2p15. It had a size of 1.909 Mb and harbored 9 genes (Figure 5A). Gains on 2p were associated with high-risk genomic changes (Figure 6). However, we did not find an influence of gain(2p) on PFS or OS (supplemental Figure 11).
Minimally amplified region of gain(2p) and gain(8q). (A) The minimal region in bands 2p16.1-p15 is defined by 2 patients, has a size of 1.909 Mb, and harbors 9 genes. (B) Minimal consensus region of gain(8q) with focus on a 60-kb region in 8q24.21 located approximately 500 kb centromeric of the MYC locus.
Minimally amplified region of gain(2p) and gain(8q). (A) The minimal region in bands 2p16.1-p15 is defined by 2 patients, has a size of 1.909 Mb, and harbors 9 genes. (B) Minimal consensus region of gain(8q) with focus on a 60-kb region in 8q24.21 located approximately 500 kb centromeric of the MYC locus.
Distribution of genetic alterations. Primary categorization (top 6 lines) is according to the hierarchical model of genomic aberrations (with inclusion of TP53 mutational status).
Distribution of genetic alterations. Primary categorization (top 6 lines) is according to the hierarchical model of genomic aberrations (with inclusion of TP53 mutational status).
Deletions on 6q were found in 23 of 353 (7%) cases. Deletions were very heterogeneous and a MDR could not be identified. In 6q21, 1 potential candidate region with a size of 2.5 Mb was delineated that was affected in 80% of the 6q-deleted cases (supplemental Figure 12).
Gains in 8q24.21 were found in 16 of 353 (5%) cases. Eleven cases had long-range gains with a size larger than 15 Mb. The 5 remaining cases had small gains less than 500 kb, the smallest being 60.9 kb in size. The minimally gained region was defined by 2 cases (case numbers CLL023 and CLL120) and was located between 128.292.928 and 128.331.109 Mb, 486 kb proximal to the MYC locus (Figure 5B). Cases with gain(8q) had shorter PFS (P < .01), but there was no impact on OS (P = .37; supplemental Figure 13).
Six cases had trisomy 19. All cases had concomitant trisomy 12. In addition to these 6 cases, 2 additional cases with overlapping partial 19p gains were identified (case numbers CLL087 and CLL314up). The minimally gained region was located in 19p13.2 and had a size of 2.174 Mb containing approximately 40 genes (supplemental Figure 14).
Trisomy 18 was observed in 3 cases. All 3 cases also had trisomy 12 and trisomy 19. Partial gains on chromosome 18 were not found.
Chromothripsis
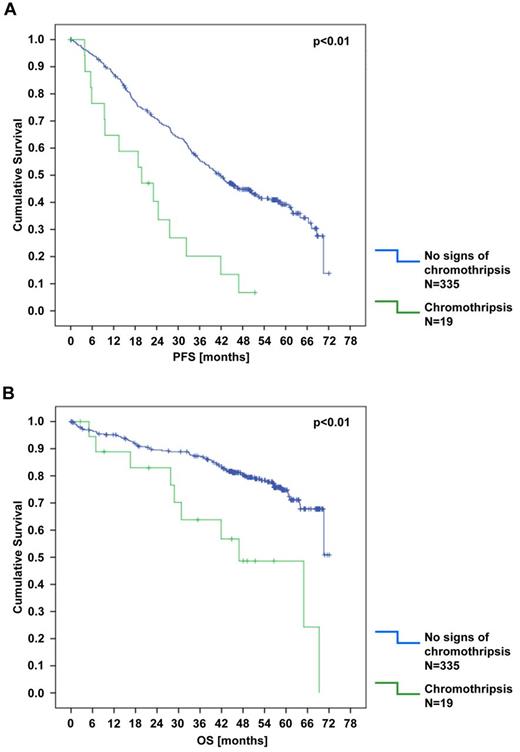
All samples were analyzed for the presence of chromothripsis, defined as the presence of at least 10 switches between 2 or 3 copy number states on an individual chromosome.29 Using these criteria, 7 cases showed chromothripsis (supplemental Figure 15). In 2 of the 7 cases, a second chromosome carried frequent CNAs that did not completely fulfill our criteria of chromothripsis (6 and 8 copy number changes). Furthermore, an additional 12 cases showed borderline signs of chromothripsis with at least 7 copy number oscillations on 1 or 2 chromosomes (supplemental Table 7). Of the 19 cases presumed to have chromothripsis, 74% showed unmutated IGHV status and 79% carried high-risk genomic aberrations (TP53 mutation in 6 of 19 cases). Patients with chromothripsis had inferior OS (P < .01) and PFS (P < .01; Figure 7).
Kaplan-Meier estimates for PFS and OS according to the presence of chromothripsis. (A) PFS. (B) OS. Included in the analysis were patients who showed clear and borderline signs of chromothripsis.
Kaplan-Meier estimates for PFS and OS according to the presence of chromothripsis. (A) PFS. (B) OS. Included in the analysis were patients who showed clear and borderline signs of chromothripsis.
Discussion
High-resolution SNP-array analysis of previously untreated CLL patients enrolled in the CLL8 trial revealed few somatic genetic alterations, with only 1.8 CNAs per case. Remarkably, two-thirds of the cases carried no aberrations beyond those already detected by the routine FISH panel covering only 4 genomic regions. In particular, this applied to CLL cases with a favorable risk profile according to the hierarchical model of genomic aberrations,1 with only 0.55 additional CNAs found per case. CLL cases with high-risk features, which were defined by del(11)(q22.3)/ATM, del(17)(p13)/TP53 by FISH and/or TP53 mutation, presented with an additional 1.06 CNAs per case, possibly because of secondary events resulting from defects in the DNA damage response associated with alterations of ATM and P53 proteins. Nonetheless, the low burden of CNAs, which is in contrast to the findings in B-lineage acute lymphoblastic leukemia,15 suggests that genomic instability is not a prominent feature of CLL. This is consistent with previously published data.20-23,31 Compared with these previous SNP-array studies on CLL, the number of CNAs was slightly lower in our cohort, possibly because of inclusion of only patients without prior therapy.
Similarly, tumor-specific CN-LOHs were detected in only 6% of cases, which is lower compared with other hematologic malignancies.18 In the present study, we found CN-LOHs mostly in regions affected by recurrent CNAs, 13q14.3, 17p13, and 11q22.3, resulting in biallelic deletions in 13q14.3 and homozygous TP53 mutations. The observation on 13q is consistent with previously published data in which focal biallelic deletions on 13q14 were found within larger CN-LOHs in few cases.19,20,32
Based on the high resolution and the large number of cases studied, we were able to further delineate critical regions and deletion boundaries of known CNAs. In the present study, we report a case with monoallelic 13q14 deletion located telomeric of the 2 miRNAs mir16-1 and mir15a, including only DLEU1 and the first 2 exons of DLEU2 (Figure 3A). This is consistent with previously published data.33 When taking into consideration the boundaries of biallelic deletions, we identified a case in whom the size of the deleted allele only included the DLEU1 locus (Figure 3B). Assuming that biallelic deletion on 13q has an impact on the pathogenesis of the disease, it is striking to find a minimal (consensus) biallelic deletion affecting only DLEU1.
Interestingly, both proximal and distal 13q deletion breakpoints showed clustering in confined genomic regions. The major proximal breakpoint cluster containing breakpoints of 76 deletions was located in a 185-kb genomic segment between RB1 and the MDR (Figure 2A). The major distal breakpoint cluster region encompassing 118 deletion breakpoints was located in a 331-kb segment just next to the MDR. Only the DLEU7 gene was located between the MDR and the breakpoint cluster (Figure 2B). This is particularly striking in light of a new mouse model of CLL in which codeletion of the DLEU7 homolog was shown to lead to higher disease penetrance and aggressiveness13 and a potential role of DLEU7 found in familial CLL.34 These major breakpoint clusters have been identified previously without further specification of these regions.22
The MDR of 11q deletions was clearly confined to the ATM locus, the smallest deletion of 78.5 kb being intragenic. Although few cases had discontinuous deletions along 11q, no other MDRs were identified on 11q. These data are consistent with previously published data and provide further evidence for a pathogenetic role of ATM in CLL.9 Rossi et al have recently described alterations affecting the BIRC3 locus.35 In our cohort, deletion of BIRC3 was found in approximately two-thirds of cases with 11q deletion, but only concomitantly to ATM deletions. Del(17p) affected TP53 in all cases except 2, with TP53 mutation of the second allele found in 23 of the 27 (85%) cases. The 2 small, tumor-specific 17p losses in 17p13.2 and 17p13.3 not affecting TP53 might hint at novel candidate genes (see below). No critical regions could be defined for trisomy 12 and 6q deletions. Further recurrent CNAs previously reported in CLL included gains on 2p (7%), 8q24 (5%), 19 (2%), and 18 (1%). Trisomy 2p was the most common aberration beyond those detected by the FISH probe panel. Compared with previously published data,19 the minimally amplified region could be further delineated and contained 9 genes, among them XPO1, which was recently reported to be recurrently mutated in CLL.36
In addition to known genetic lesions, in the present study, we were able to discover new lesions (Table 2). Deletions at 15q15.1 were most common, being found in 14 of 353 (4%) cases (Figure 1). The smallest deletion was 70.48 kb in size and was located within the MGA locus. In a whole-genome sequencing study of 4 CLL cases, MGA was found to be mutated in 1 case.36 We conducted MGA sequence analysis on 59 cases and revealed a somatic mutation in one case without del(15)(q15.1). Recurrent loss of one MGA allele and recurrent somatic mutations raise the possibility that this gene may function as a haploinsufficient tumor suppressor in CLL.
Interestingly, MGA is part of the network of MAX and MAX-interacting proteins that are involved in regulatory mechanisms for cell proliferation, differentiation, and apoptosis.37 We found that the genes MGA [del(15)(q15.1)] and MNT [del(17)(p13.3)], which encode for MAX-interacting proteins functioning as transcription factors after heterodimerization with MAX, as well as MYC [gain(8)(q24)] were recurrently affected by CNAs in CLL. Whereas MYC has an effect mostly in transcriptional activation promoting cell-cycle progression, apoptosis, and cellular transformation, the other MAX-interacting proteins such as MGA and MNT have been shown to function as corresponding transcriptional repressors.38,39 Remarkably, MNT has also been linked to T-cell lymphomagenesis.40,41 Recurrent gains on 8q have been described to occur at low frequency in CLL.1 In our cohort, we found gains on 8q24 in 5% of cases. Deregulated expression of MYC has been shown to be associated with a poor prognosis in CLL and other B-cell malignancies.42-45 However, in our study, not all 8q gains spanned the MYC locus; 5 small gains were located approximately 500 kb centromeric to the MYC locus. At present, it is unclear whether these focal gains have an influence on MYC expression. In the present study, gain(8)(q24) did not result in enhanced MYC expression (data not shown).
Finally, the integration of copy number and CN-LOH data suggested a potential role for the ATR pathway in CLL pathogenesis. We found 2 essential components of this pathway affected by CNAs: the ATR interacting protein (ATRIP; 3p21.31) and the replication protein A2 (RPA2; 1p35) were located within deletions that also coincided with tumor-specific CN-LOH. However, because the number of cases with copy number losses in these regions remained very low and no inactivating ATRIP and RPA2 mutations were identified (data not shown), a major role for the ATR pathway in CLL pathogenesis seems unlikely.
Chromothripsis was initially described in CLL as a new mechanism in tumorigenesis.46 In the present study, there was evidence of chromothripsis inferred by SNP profiles in few cases. In univariate analysis, patients with chromothripsis had inferior PFS and OS, but the majority also had unmutated IGHV (74%) and high-risk genomic aberrations (79%). One-third of the patients had a concomitant TP53 mutation. However, the association between chromothripsis and the TP53 mutation was not as strong as recently described for medulloblastoma.29
In summary, the results of the present study demonstrate a low frequency of genomic CNAs and CN-LOHs in previously untreated patients with CLL. Only few genetic lesions were detected beyond those already diagnosed by routine FISH analysis, and additional lesions were more commonly found in genetic high-risk CLL patients. Few novel lesions were detected, with deletions at 15q15.1/MGA being the most common. Recent studies applying next-generation sequencing have identified novel recurring gene mutations, for example, in NOTCH1 and SF3B1, that occur in a significant proportion of CLL cases.31,36,47-49 In the present study, the respective genomic regions, 9q34.3 and 2q33.1, were not affected by CNAs. Determining the role of these novel genetic changes will help in further elucidating CLL pathogenesis. Molecular studies on large cohorts of CLL patients will be necessary to determine which of the novel markers will become useful in patient management.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients who participated in the CLL8 trial; the investigators who treated patients and submitted samples; Myriam Mendila, Michael K. Wenger, and Jamie Wingate (F. Hoffmann-La Roche, Basel, Switzerland) for support; Sabrina Kless, Christina Galler, Doris Winter, and Karin Lanz for excellent technical assistance; and Silja Mack, Dr Anna-Maria Fink, Dr Kirsten Fischer, and Anne Westermann for data handling and analyses.
This work was supported by the Deutsche Forschungsgemeinschaft (grants STI 296/4-1 and SFB 1074/B2). Genetic reference diagnostics of CLL8 (CD19 enrichment, FISH, and IGHV and TP53 mutation analyses) were supported by F. Hoffmann-La Roche (Basel, Switzerland).
Authorship
Contribution: J.E. designed the research, performed the experiments, analyzed, collected, and interpreted the data, and wrote the manuscript; K.H., L.B., M.W.M.K., D.M., and J.R.D. designed the research and interpreted the data; F.M. performed the research; D.W., A.B., T.Z., A.G., and J.B. collected and analyzed the data; I.R., X.S., J.M., S.P., P.L., J.K., and R.B. analyzed the data; M.H. provided data; and S.S. and H.D. designed the research, collected and interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hartmut Döhner, MD, Department of Internal Medicine III, University Hospital of Ulm, Albert-Einstein-Allee 23, 89081 Ulm, Germany; e-mail: hartmut.doehner@uniklinik-ulm.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal