Abstract
Aberrations in the p53 tumor suppressor pathway are associated with hematologic malignancies. p53-dependent cell cycle control, senescence, and apoptosis functions are actively involved in maintaining hematopoietic homeostasis under normal and stress conditions. Whereas loss of p53 function promotes leukemia and lymphoma development in humans and mice, increased p53 activity inhibits hematopoietic stem cell function and results in myelodysplasia. Thus, exquisite regulation of p53 activity is critical for homeostasis. Most of our understanding of p53 function in hematopoiesis is derived from genetically engineered mice. Here we summarize some of these models, the various mechanisms that disrupt the regulation of p53 activity, and their relevance to human disease.
Introduction
The tumor suppressor p53 is a ubiquitously expressed transcription factor maintained in an inactive, latent form in most cell types. p53 is stabilized and activated in response to cellular stressors, such as DNA damage, hypoxia, and oncogene activation.1 Activated p53 triggers a multifaceted transcriptional program, which, depending on the cell type and damage signal, initiates cell cycle arrest, senescence, or apoptotic pathways.1-3 As p53 activation determines the fate of the cell, it is very tightly regulated in vivo. Multiple biologic inhibitors have been identified that regulate p53 stability and activity.4 Mdm2 and Mdm4 (also known as Mdmx) are homologous proteins and the primary negative regulators of p53 activity and stability under both normal and stress conditions.5 Both these proteins bind p53 and inactivate it by masking its transactivation domain.6-8 In addition, Mdm2 also functions as an E3-ubiquitin ligase that targets p53 and promotes its degradation by the 26S proteosomal machinery.9-11
The mouse is commonly used in conjunction with ionizing radiation (IR) to probe p53 activation in vivo. IR acts as a prototypical DNA damage insult causing double-strand DNA breaks that induce p53 activity.12 IR induced p53-dependent acute pathologies can be observed in different tissues such as the CNS, hair follicles, hematopoietic system (HP), gastrointestinal tract, and germ cells.13 However, long-term effects of increased p53 activity are most obvious in the HP and gastrointestinal systems, both of which are highly regenerative systems that typically collapse in a p53- and IR dose-dependent manner. Whole-body radiation, typically around 8 Gy, obliterates the hematopoietic system, whereas higher dosages are required to collapse the gastrointestinal system in a wild-type mouse.13 Loss of cellularity of these tissues is the primary cause of IR-mediated demise in these animals.
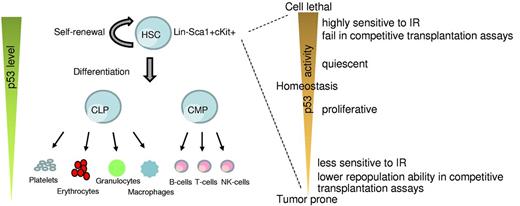
The HP system is most vulnerable to DNA damage because of its highly proliferative nature.14 It is composed of multiple hematopoietic organs (fetal liver, bone marrow, thymus, and spleen), which provide the microenvironment required for self-renewal and multilineage differentiation of hematopoietic stem cells (HSCs).15-17 HSCs are multipotent cells that have the potential to self-renew and also give rise to progenitor cells, namely, common myeloid progenitors and common lymphoid progenitors, which subsequently differentiate to mature hematopoietic cells of myeloid and lymphoid lineages (Figure 1).18 These abilities of HSCs are critical for maintaining the lifelong supply of hematopoietic cell types in the bone marrow and peripheral blood. Under normal conditions, HSCs are maintained in a quiescent state, and only a small fraction of cells enter the cell cycle to give rise to lineage-specific progenitors, necessary for replenishing the daily need of circulating blood cells.19 Under stress conditions, HSCs readily enter the cell cycle and maintain their cell pool by cell-renewal. Mouse HSCs lack the expression of cell surface molecule for lineage-specific mature cells (Lin−) but express stem cell antigen 1 (Sca1+) and c-Kit (Kit+, CD117), a transmembrane tyrosine kinase that serves as a receptor for stem cell factor. These HSCs can be identified by flow cytometry as the Lin−Sca1+Kit+ (LSK) population.20,21 Whereas LSK defines the basic marker set used to identify the HSCs, additional markers, such as CD34, CD38, CD48, CD150, and Flt3, are also used in conjunction to further refine HSCs.22-26 Some investigators also use rhodamine staining or Hoechst dye efflux side population to identify the stem cell population.26
Role of p53 in HSCs. A simplistic model depicting changes in p53 levels during differentiation of HSCs into the lineage components (left). p53 activity levels influence HSC functionality (right).
Role of p53 in HSCs. A simplistic model depicting changes in p53 levels during differentiation of HSCs into the lineage components (left). p53 activity levels influence HSC functionality (right).
Most of our understanding about p53 regulation and stress response comes from a series of genetically engineered mouse models generated during the past 2 decades. In this review, we discuss models that disrupt the p53 pathway that expand our understanding of p53-mediated stress and tissue-dependent cell fate decisions in hematopoiesis.
p53 deregulation in hematopoietic tumors
p53 regulates a range of physiologic functions, including ageing, development, cell metabolism, differentiation, fertility, skin pigmentation, stem cell function, and tissue homeostasis.27 Given its pleiotropic activity, it comes as no surprise that disruption of the p53 pathway is a common denominator in many malignancies. Indeed, > 50% of human tumors carry a mutation in TP53, and many other tumors carrying wild-type TP53 alleles exhibit attenuated p53 activity by various mechanisms.28,29 Germline mutations of TP53 cause Li-Fraumeni syndrome, a genetic disorder characterized by a dramatically increased incidence of many kinds of tumors that include sarcomas, breast cancers, leukemia, and lymphomas.30,31 Genetically engineered mice that lack p53 or express a mutant form of p53 (p53R172H, p53R172P, p53R270H) are prone to early onset of tumor development.32-36 A high percentage of these tumors are lymphomas, a tumor of hematopoietic T- or B-cell origin.33-36 Furthermore, p53 gene dosage impacts tumor latency as mice heterozygous for these mutations develop tumors slightly later than homozygous mutant mice.33-35
Surprisingly, in contrast to solid tumors, hematologic malignancies present a rather low incidence of genetic alterations in TP53 (10%-20%).37,38 TP53 mutations/deletions have been reported in chronic lymphocytic leukemia (CLL), marginal zone lymphoma, follicular lymphoma, and diffuse large B-cell lymphoma.39 Nonetheless, aberrations in TP53 correlate with an inferior clinical outcome in hematologic cancers.40,41 For instance, in patients with CLL, deletion of the short arm of chromosome 17 (TP53 maps to band 17p13.1), is remarkably predictive of poor outcome with standard chemo-immunotherapeutic regimens.42 Loss of TP53 also acts as a prognostic marker for acute myeloid leukemia.43
These data suggest that other mechanisms besides mutations could be involved in deregulation of the p53 pathway. Indeed, attenuation of p53 function by increased levels of its inhibitors Mdm2 and Mdm4 results in tumors of hematologic origin in mice.44,45 In humans, overexpression of MDM2 has been associated with CLL, follicular lymphoma, diffuse large B-cell lymphoma, marginal zone lymphoma, acute myeloid leukemia, mantle cell lymphoma, Burkitt lymphoma, acute lymphocytic leukemia, multiple myeloma, and plasma cell leukemia.39,46 A single nucleotide polymorphism (SNP) in MDM2 (SNP309), which increases the transcription of MDM2 from the P2 promoter, has been linked to many different hematologic malignancies, including CLL and other B-cell hematologic tumors.47
The genomic region at chromosome 1q harboring the p53− regulator MDM4 (also known as HDMX) is also amplified in several types of cancer.48 MDM4 amplifications have been recently described in a high percentage of patients with myeloproliferative neoplasm during their transformation to acute myeloid leukemia.49 In this context, 1q gains (ie, MDM4 amplifications) are mutually exclusive with TP53 mutations, a phenomenon also observed in solid tumors.50 Overexpression of MDM4 has also been reported in CLL, which responds poorly to the MDM2 inhibitor Nutlin, implicating MDM4 as an important p53 inhibitor in CLL.51 Altogether, these data highlight the importance of dampening p53 levels in transformation of the hematopoietic system.
p53 loss impairs HSC function
In the hematopoietic system, p53 is primarily expressed in HSCs and regulates their quiescence and self-renewal.52 This is critical for preserving the lifelong pool of HSCs that sustain the highly regenerative hematopoietic system.52 Genetically engineered mice that lack p53 have a 2- to 3-fold increase in their HSC pool.52-55 This is probably the result of the higher rate of cellular proliferation of p53-deficient HSCs.52 Interestingly, though, these highly proliferative p53-deficient HSCs lack the equivalent repopulation capability compared with their wild-type counterparts (Figure 1).52-55 This implies that the absence of p53 compromises the functional fitness of HSCs. The engraftment potential of HSCs, a mark of their fitness, is also reduced with age and correlates with decreasing p53 activity in mice.56,57 On the other hand, total bone marrow cells from p53-null mice easily outgrow wild-type bone marrow cells in competitive bone marrow transplantation assays,52-55 indicating that either the progenitor cells or external factors originating in the bone marrow modulate HSC function. Of note, the short-term engraftment advantage of p53−/− bone marrow cells comes at a price as the recipient mice typically die much earlier because of hematologic tumors. Unregulated proliferation of HSCs in the absence of p53 also makes them prone to accumulate mutations. The early tumorigenesis and death of p53−/− mice or mice implanted with p53−/− bone marrow cells precludes any long-term analyses of HSC composition on life span of the animal.58
Under nonstress conditions, p53 functions as an antioxidant and lowers reactive oxygen species (ROS) levels, thereby protecting HSCs from DNA damage and mutations.59 This function of p53 could explain the increased incidences of hematologic tumors in p53+/− and p53−/− mice.
These data suggest that, under certain conditions, transient inhibition of p53 function could potentially be used to promote proliferation of HSCs and accelerate the recovery of bone marrow without impairing the “stemness” of these cells. Indeed, inhibition of p53 by pifithrins (which specifically inhibit p53 activity) stimulates HSC proliferation in both in vitro and in vivo settings.60 This also correlates with the finding that suppression of p53 function facilitates hematopoietic reconstitution after cytotoxic therapy.61 Taken together, these data indicate that p53 plays a cytoprotective role in HSCs by preventing their entry into the cell-cycle and maintaining their quiescence.
Damage-induced p53 activity is detrimental for hematopoiesis
Induction of p53 activity in response to DNA damage also impairs hematopoiesis. DNA damage results in phosphorylation of p53 by ATM and Chk2 kinases and subsequent activation.67-69 Histopathologic analyses and bone marrow transplantation assays confirm that mice with enhanced p53 activity (via various genetic modifications) die of sublethal doses of IR exclusively because of hematopoietic failure.70 Furthermore, unirradiated HSCs outcompete HSCs that are treated with low-dose IR in transplantation assays.71 This is probably the result of the increased p53 activity, impairing the cellular fitness of irradiated stem and progenitor cells of the hematopoietic system. In agreement, loss of p53 protects HSCs and progenitor cells from IR damage in p53-null mice,72 possibly because radiation confers a selective advantage for long-term expansion of p53-deficient HSCs and progenitor cells.71,73 Treatment of irradiated mice with pifithrin-α, a drug that inhibits p53 activity, also nullifies the effects of radiation on the hematopoietic system.60
Damage-induced p53 activates downstream targets involved in both apoptosis and cell cycle arrest, Nonetheless, the apoptotic pathway is perceived as predominant in hematopoietic organs.52 Meticulous studies show that the spleen, thymus, and bone marrow (major hematopoietic organs) in p53-null mice are refractory to p53 dose-dependent DNA damage.74 Thymocytes from wild-type mice undergo apoptosis after IR, whereas p53-null thymocytes remain completely resistant and p53 heterozygous thymocytes show an intermediate apoptotic response to DNA damage (Table 1).75,76 Irradiation also accelerates lymphomagenesis in p53-deficient mice. Remarkably, the most dramatic increase in lymphoma incidence is seen in p53+/− mice.58,77,78 The prevailing view that acute p53 apoptotic response to IR protects against tumorigenesis by eliminating cells with excessive DNA damage was challenged by 2 recent studies. Concurrent restoration of p53 activity in a mouse model with radiation did not protect against tumor onset, whereas restoration 8 days after IR was effective in tumor suppression. Similarly, conditional deletion of p53 4 weeks after IR resulted in lymphoma rates comparable with disrupting p53 before IR.79,80 These data suggest that p53 activity after IR is more important for tumor protection.
Summary of mouse models with distinct hematopoietic phenotypes
| Mice/alleles . | p53 activity, % . | Mdm2 dosage, % . | Mdm4 dosage, % . | Phenotype . | References . |
|---|---|---|---|---|---|
| Wt | 100 | 100 | 100 | Normal | — |
| p53+/− | 50 | 100 | 100 | Tumor prone after IR | 58 |
| p53−/− | 0 | 100 | 100 | Tumor prone, increases in the number and proliferation of HSCs, radioresistant | 33,34,53,58 |
| p53R172P | ∼ 50 | 100 | 100 | Tumor prone, undergo cell cycle arrest but not apoptosis, radioresistant | 32 |
| p53R172H | 0 | 100 | 100 | Transcriptionaly null, tumor prone, radioresistant | 35,36 |
| p53S18A | < 100 | 100 | 100 | Compromised apoptosis in thymocytes | 139 |
| p53S23A | < 100 | 100 | 100 | Homozygous mice develop B-cell lineage tumors, decreased apoptosis in thymocytes | 140 |
| p53S18AS23A | < 100 | 100 | 100 | Tumor prone, no apoptosis after IR in thymocytes | 141 |
| p53T21AT23A | > 100 | 100 | 100 | Small, lymphopenic, low HSCs, aging phenotype, radiosensitive | 81 |
| p537KR | > 100 | 100 | 100 | Higher p53 activity after IR, low HSCs at baseline, radiosensitive | 82 |
| Super p53 | > 100 | 100 | 100 | Radiosensitive | 83 |
| Mdm2+/− | > 100 | 50 | 100 | Radiosensitive, delayed lymphomagenesis in Eμ-Myc background | 70,91 |
| Mdm2−/− | > 100 | 0 | 100 | Embryo lethal because of enhanced apoptosis | 87,88 |
| Mdm2Puro/Δ7-12 | > 100 | ∼30 | 100 | Small, lymphopenic, hypocellular spleen, thymus and bone marrow; radiosensitive | 84 |
| Mdm2−/−p53P/R172P/R172P | ∼ 50 | 0 | 100 | Died at weaning of hematopoietic defects | 110,112 |
| Mdm2Tg | < 100 | > 100 | 100 | Tumor prone, more sarcomas | 44 |
| Mdm2SNP309G | < 100 | > 100 | 100 | Early-onset tumors | 94 |
| Mdmx−/− | > 100 | 100 | 0 | Embryo lethal because of proliferation defects | 95,96 |
| Mdmx+/− | 100 | 100 | 50 | Radiosensitive, delayed lymphomagenesis in Eμ-Myc background | 70 |
| Mdm4+/ΔRING | 100 | 100 | 100 | Homozygous mice lethal, radioresistant | 98 |
| Mdmx3SA | 100 | 100 | 100 | Radioresistant at 10 Gy | 97 |
| Mdm2+/−Mdm4+/− | > 100 | 50 | 50 | Lethal by weaning, hematopoietic defects | 70 |
| Mdm2+/−Mdm4+/−p53+/− | 50 | 50 | 50 | developmentally normal | 70 |
| p21−/− | 100 | 100 | 100 | Normal, late tumors, radioresistant in C57BL/6 background, sensitive in mixed strain background | 103,104 |
| Puma−/− | 100 | 100 | 100 | Normal, radioresistant | 142,143 |
| Mice/alleles . | p53 activity, % . | Mdm2 dosage, % . | Mdm4 dosage, % . | Phenotype . | References . |
|---|---|---|---|---|---|
| Wt | 100 | 100 | 100 | Normal | — |
| p53+/− | 50 | 100 | 100 | Tumor prone after IR | 58 |
| p53−/− | 0 | 100 | 100 | Tumor prone, increases in the number and proliferation of HSCs, radioresistant | 33,34,53,58 |
| p53R172P | ∼ 50 | 100 | 100 | Tumor prone, undergo cell cycle arrest but not apoptosis, radioresistant | 32 |
| p53R172H | 0 | 100 | 100 | Transcriptionaly null, tumor prone, radioresistant | 35,36 |
| p53S18A | < 100 | 100 | 100 | Compromised apoptosis in thymocytes | 139 |
| p53S23A | < 100 | 100 | 100 | Homozygous mice develop B-cell lineage tumors, decreased apoptosis in thymocytes | 140 |
| p53S18AS23A | < 100 | 100 | 100 | Tumor prone, no apoptosis after IR in thymocytes | 141 |
| p53T21AT23A | > 100 | 100 | 100 | Small, lymphopenic, low HSCs, aging phenotype, radiosensitive | 81 |
| p537KR | > 100 | 100 | 100 | Higher p53 activity after IR, low HSCs at baseline, radiosensitive | 82 |
| Super p53 | > 100 | 100 | 100 | Radiosensitive | 83 |
| Mdm2+/− | > 100 | 50 | 100 | Radiosensitive, delayed lymphomagenesis in Eμ-Myc background | 70,91 |
| Mdm2−/− | > 100 | 0 | 100 | Embryo lethal because of enhanced apoptosis | 87,88 |
| Mdm2Puro/Δ7-12 | > 100 | ∼30 | 100 | Small, lymphopenic, hypocellular spleen, thymus and bone marrow; radiosensitive | 84 |
| Mdm2−/−p53P/R172P/R172P | ∼ 50 | 0 | 100 | Died at weaning of hematopoietic defects | 110,112 |
| Mdm2Tg | < 100 | > 100 | 100 | Tumor prone, more sarcomas | 44 |
| Mdm2SNP309G | < 100 | > 100 | 100 | Early-onset tumors | 94 |
| Mdmx−/− | > 100 | 100 | 0 | Embryo lethal because of proliferation defects | 95,96 |
| Mdmx+/− | 100 | 100 | 50 | Radiosensitive, delayed lymphomagenesis in Eμ-Myc background | 70 |
| Mdm4+/ΔRING | 100 | 100 | 100 | Homozygous mice lethal, radioresistant | 98 |
| Mdmx3SA | 100 | 100 | 100 | Radioresistant at 10 Gy | 97 |
| Mdm2+/−Mdm4+/− | > 100 | 50 | 50 | Lethal by weaning, hematopoietic defects | 70 |
| Mdm2+/−Mdm4+/−p53+/− | 50 | 50 | 50 | developmentally normal | 70 |
| p21−/− | 100 | 100 | 100 | Normal, late tumors, radioresistant in C57BL/6 background, sensitive in mixed strain background | 103,104 |
| Puma−/− | 100 | 100 | 100 | Normal, radioresistant | 142,143 |
— indicates not applicable.
Overall, p53 response to genotoxic stressors perturbs hematopoiesis by promoting apoptosis and altering the dynamics of HSCs and progenitor cell proliferation.
Constitutive p53 activity impairs hematopoietic homeostasis
Increase in p53 activity above basal levels is detrimental to long-term HSC fitness (Figure 1). p53 activity is characterized by the phosphorylation of amino terminal serine and threonine residues. Transgenic mice in which the p53 phosphorylation sites at threonine-21 and serine-23 are mutated to aspartic acid, a phospho-mimicking amino acid, have constitutive p53 activation and exhibit an ageing phenotype that correlates with depletion of adult stem cells in multiple tissues, including the bone marrow (Table 1).81 Similarly, another knock-in mouse model (p53-7KR) in which 7 C-terminal lysine residues, targets of Mdm2 ubiquitination, are mutated has constitutive p53 activity accompanied by a drastic depletion of the HSC population.82 HSCs from these mice also do not measure up to the repopulation potential of wild-type cells in transplantation assays. Other mouse models of high basal p53 activity, such as Super p53 mice (which express additional copies of p53), and mice haploinsufficient for p53 inhibitors Mdm2 and Mdm4 or mice with even less Mdm2 levels (see below), are overtly radiosensitive and exhibit hematopoietic defects in response to low levels of IR exposure (Table 1).70,82-84 Thus, an increase in basal p53 level impairs the stemness of HSCs, thereby sensitizing mice to lower doses of IR and results in HP failure.
Aberrations in p53 regulators have major implications for hematopoiesis
Mouse models show that aberrations in the expression level of Mdm2 and Mdm4 modulate p53 activity and impact the hematopoietic system. Decreased expression of either of these 2 inhibitors leads to activation of p53 and associated anomalies in the hematopoietic system (Table 1).70,84
Mdm2 is required during primitive erythropoiesis to inhibit p53-mediated apoptosis.86 Genetically engineered mice with homozygous deletion of Mdm2 die prematurely at E3.5 because of lethal activation of p53.87,88 Recently Evan's group89 used a novel strategy to bypass this limitation and probed Mdm2 role in p53 regulation during hematopoiesis. They used a mouse model in which p53 activity could be reversibly toggled between inactive and active stages.89 Restoration of p53 activity in an Mdm2-null adult mouse led to an almost complete ablation of radio-sensitive tissues that included the bone marrow, thymus, spleen, small intestine, and colon via apoptosis and resulted in lethality. Consistent with the induction of bone marrow aplasia, analyses of peripheral blood also revealed a dramatic reduction in all hematopoietic lineages. Thus, inhibition of p53 activity by Mdm2 is required for HP viability.
Mice heterozygous for Mdm2 are normal and do not exhibit any apparent phenotypic abnormalities.87,88 They do, however, exhibit hyperpigmentation of foot pads, a characteristic of enhanced p53 activity.90 This enhancement of p53 activity is probably the reason that unirradiated BM cells from Mdm2+/− mice fail to compete with wild-type BM cells in competitive transplantation assays.71 In addition, Mdm2+/− mice are radiosensitive and die of sublethal IR dosage predominantly because of p53-mediated disruption of the hematopoietic system.70 Haploinsufficiency of Mdm2 also inhibits lymphomagenesis probably through up-regulation of p53 function as Eμ-myc Mdm2+/− mice exhibit a delayed lymphoma onset compared with Eμ-myc mice.91 These data suggest that a small increase in p53 activity is tolerated but compromises the ability to cope with stress.
Reduction of Mdm2 to < 50% of normal levels results in even more distinctive hematopoietic abnormalities in mice. Mice expressing hypomorphic and null alleles of Mdm2 encoding only 30% of normal Mdm2 levels (Mdm2Puro/Δ7-12) are small and have abnormally low levels of lymphocytes, red cells, neutrophils, and platelets in peripheral blood (Table 1).84,92,93 The spleen, thymus, and bone marrow of these mice also show a decrease in the number of lymphoid cells and are hypocellular. This correlates with increased p53 activity and subsequent apoptosis of hematopoietic cells in these organs. This HP defect is completely dependent on p53 as it disappears in a p53-null background.84
Even more subtle fluctuations in Mdm2 levels impact p53 activity. Data from human studies suggest a correlation of a single nucleotide polymorphism in the MDM2 promoter, SNP309G, with early onset of lymphoma and leukemogenesis.47 Our laboratory has generated mouse models for the corresponding human MDM2 SNP309 polymorphism. Mice homozygous for the Mdm2SNP309G allele express slightly elevated Mdm2 levels probably because of a more efficient SP1 binding site at the P2-MDM2 promoter.94 Mdm2SNP309G/G mice exhibit lower levels of apoptosis in thymus than Mdm2SNP309T/T mice in response to 1 Gy IR.94 Mdm2SNP309G/G mice also die of early-onset tumors, most of which are lymphomas. The impact of this polymorphism on HSCs function remains to be investigated.
Altogether, these data suggest that Mdm2 dosage manifests p53-mediated hematologic anomalies (Table 1).84,92 Mdm2 levels inversely correlate with p53 levels/activity. Decreased Mdm2 levels recapitulate conditions of high p53 activation, which over time impairs the cellular fitness of stem cells and leads to hematologic aberrations. Down-modulation of Mdm2 levels or disruption of Mdm2-p53 interaction could be used therapeutically to activate p53 in hematologic tumors with wild-type p53. Given that p53 is very quickly activated to lethal limits in the absence of Mdm2, such strategies have to be transient and reversible to preclude any side effects. Of note, the effect of transient activation of p53 in the HSC population has not been examined.
Mdm4, the other critical but relatively weaker inhibitor of p53 compared with Mdm2, is required during the rapidly growing phase of definitive erythropoiesis.86 Mdm4-null mice exhibit p53-dependent proliferation defects and lethality during embryogenesis (Table 1).95,96 Mice heterozygous for Mdm4 are normal but overtly radiosensitive.70 Similar to Mdm2+/− mice, irradiated Mdm4+/− mice die exclusively because of bone marrow aplasia.70 Importantly, p53 loss rescues this radiolethal phenotype. In addition, similar to Mdm2, haploinsufficiency of Mdm4 delays lymphoma onset in Eμ-myc mice.70
Conversely, mice encoding a stable form of Mdm4 (named Mdmx-3SA), which is not targeted by Mdm2 for degradation, are radioresistant and can survive 10 Gy IR (Table 1).97 This is probably because of irreversible p53 binding and, thus, inhibition of p53 activity. We also observe a similar radioresistant phenotype in mice with an Mdm4 allele that lacks the sequence encoding the RING domain necessary for Mdm4-Mdm2 interaction.98 RING domain-deficient Mdm4ΔRING is more stable than Mdm4, resulting in less p53 activity. Thus, heterozygous Mdm4+/ΔRING mice survive 6 Gy IR, unlike Mdm4+/− mice, and do not die of hematopoietic defects. These data validate the role of Mdm4 in dampening p53 activity during hematopoietic homeostasis and in response to DNA damage.
Mdm4 has also been tested as a therapeutic target for p53 activation.99 Restoration of p53 in Mdm4−/− adult tissues induces p53-target genes and causes widespread apoptosis in radiosensitive tissues. Most of the damage is limited to the lymphoid organs and the bone marrow.99 Interestingly, unlike Mdm2, the p53-dependent pathologies induced in Mdm4-deficient tissues are milder and fully reversible, implying that short-term inhibition of Mdm4 for p53 activation could be a more beneficial strategy with less toxicity in hematologic malignancies.
Compound haploinsufficiency of both Mdm2 and Mdm4 in a double-heterozygous mouse results in a much more severe phenotype.70 At mid-gestation, the Mdm2+/−Mdm4+/− embryos show characteristics of aplastic anemia and are smaller than wild-type embryos. Some of the embryos also have exencephaly associated with cleft palate or other types of neural tube closure defects. At birth, some double-heterozygous pups survive and are smaller than their wild-type littermates. Histologic examination of tissue sections from these pups show marked hypoplasia and atrophy in the bone marrow, spleen, and thymus along with decreased extramedullary hematopoiesis in the liver. Complete peripheral blood count and bone marrow pathology indicate systemic and severe depletion of all blood cells in these double-heterozygous animals. Importantly, deletion of a single p53 allele reverts all these defects, suggesting a dose-dependent equilibrium between p53 and its inhibitors.70 These experiments highlight the exquisite balance between p53 and Mdm proteins in maintaining hematopoietic homeostasis. The hematopoietic compartment contains highly proliferative cells that are exquisitely sensitive to Mdm2 and Mdm4 dosage and the resultant p53 activity.
In summary, loss of p53 function provides radioprotection and promotes HSC proliferation together with an increase in hematologic tumors. On the other hand, enhanced p53 activity inhibits HSC proliferation and imparts radiosensitivity (Figure 1). Altogether, these results indicate that p53 plays a critical role in regulating hematopoietic cell functions under both normal and stressed conditions.
Role of p53 downstream targets in hematopoiesis
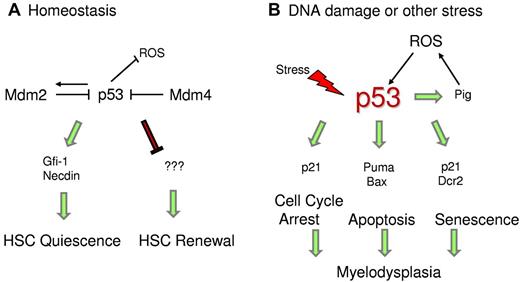
p53-target genes involved in both apoptotic (eg, Puma) and cell cycle arrest (eg, p21) pathways are significantly induced in the bone marrow after IR (Figure 2). The role of p53 target proapoptotic genes Puma and Bax in modulating the function of hematopoietic stem and progenitor cells in response to high-dose irradiation is already established.100-102 Mice lacking either of these genes do not exhibit radiation-induced lethality (Table 1). Indeed, deletion of Puma or Bax rescues the lethality caused by p53 hyper-activity in sensitive mouse models.81,102 HSCs from Puma-null mice are highly quiescent and along with the progenitor cells are also more efficient in DNA-damage repair.100,101 This provides further evidence in support of dominant role of apoptotic genes in hematopoietic degeneration. In agreement, mice with compromised p53 apoptotic activity (p53R172P retain activation of cell cycle genes) do not undergo HP failure after DNA damage (V.P., unpublished observations, November 2011). These data suggest that inhibition of p53 apoptotic activity or down modulation of specific targets could be used as a potential strategy to protect HSCs from the cytotoxic and myelosuppressive effects of radio/chemotherapy.
p53 regulates HSC quiescence. (A) Under homeostatic conditions, p53 regulates HSC quiescence mediated by Gfi-1 and Necdin and maintains HSC self-renewal by an unknown mechanism. (B) Under stress conditions, enhanced p53 activity promotes transcription of downstream targets, which trigger cell cycle arrest, senescence, or apoptosis. p53 also transactivates Pig genes, which induce ROS levels and promote p53 stability.
p53 regulates HSC quiescence. (A) Under homeostatic conditions, p53 regulates HSC quiescence mediated by Gfi-1 and Necdin and maintains HSC self-renewal by an unknown mechanism. (B) Under stress conditions, enhanced p53 activity promotes transcription of downstream targets, which trigger cell cycle arrest, senescence, or apoptosis. p53 also transactivates Pig genes, which induce ROS levels and promote p53 stability.
Still unclear is the role of p21-mediated cell cycle arrest in response to radiation. In a mixed strain background, loss of p21 affected hematopoietic recovery after IR as the HSC pool was completely depleted because of uncontrolled cell cycling, leading to lethality (Table 1).103 However, in the C57Bl/6 background, no such fatality or HSC exhaustion was observed.104 In our hands, we also did not observe an increased radio-susceptibility in p21 null mice on a C57Bl/6 background after 6 Gy IR (V.P., unpublished observations, October 2010). In addition, it was recently shown that deletion of a single p21 allele could partially rescue the radio-lethality of p53-7KR mice after 6 Gy IR.82 p53-7KR mice display a differential transcriptional activity with higher induction of p21 in the bone marrow over apoptotic genes. These data further emphasize that, under certain conditions, p21 contributes to hematopoietic sensitivity by controlling transition through the G1/S boundary. However, as mentioned in the previous paragraph, blocking apoptosis completely rescues the HSC defects in wild-type and p53 hyperactive mice, supporting it as the predominant pathway involved.
Studies are ongoing to identify p53 downstream targets that are specifically involved in maintaining HSC quiescence. Gfi-1 (growth factor independent-1) and Necdin are 2 direct p53 targets that have been implicated in HSC quiescence under steady-state conditions (Figure 2).52 Gfi-1 is a SNAG-domain containing zinc-finger transcriptional repressor that restricts HSC proliferation and preserves their functional integrity.105 In in vivo competitive assays, Gfi-1-null HSCs demonstrate excessive cycling and impaired self-renewal.106,107 The other target, Necdin, is a growth-suppressing protein that is highly expressed in HSCs.52,108 Down-regulation of Necdin diminishes HSC quiescence, whereas its up-regulation increases HSC quiescence.52 Recent mouse studies have also confirmed that Necdin-null adult HSCs are less quiescent and more proliferative than normal.109 Other targets modulated by p53 during differentiation of HSCs remain to be identified.
p53515C/515C mice as a model for apoptosis-independent activities of p53 in hematopoiesis
Previously, our laboratory has generated a knock-in mouse (p53515C) that expresses a mutant form of p53 protein, p53R172P, which is defective for apoptosis but retains partial cell cycle arrest and senescence functions.32 Homozygosity of the p53515C allele partially rescues the early embryonic lethality of Mdm2-null mice.110 Mdm2−/−p53515C/515C mice are born at expected Mendelian ratio with normal cellularity of all major hematopoietic organs at birth. However, soon after, these organs undergo progressive degradation, which culminates in complete arrest of hematopoiesis. By day P10-P14, the spleens and thymi lose all lymphoid cells, and the bone marrow becomes completely aplastic, resulting in death of these young pups. These observations indicate that, in the absence of Mdm2, increase in p53R172P activity causes rapid and progressive loss of hematopoietic stem and progenitor cells postpartum. These results are in congruence with a report that p53 is maintained at low levels during embryonic development but increases a few days after birth.111 Overall, these data highlight the importance of p53-dependent cell cycle arrest in hematopoiesis.
To gain further insights into the mechanism causing the HP defect, we investigated the stress signals that instigate increase in p53 levels and activity during prenatal and postnatal development.112 We detected higher ROS levels in the bone marrow of Mdm2−/−p53515C/515C mice compared with controls. Previous studies have shown that p53 transactivates a set of ROS-inducing genes dubbed p53-induced genes (PIG) that lead to cell death.113 In agreement, we observed higher mRNA levels of some of the Pig genes in BM cells of Mdm2−/−p53515C/515C mice. Thus, a positive feedback loop is established wherein p53R172P transactivates Pig genes, which induce ROS levels and in turn further stabilize p53. Importantly, antioxidant treatment of Mdm2−/−p53515C/515C postnatal pups with N-acetyl cysteine in vivo rescued the hematopoietic defect. These data uphold the pro-oxidant role of p53 under stress conditions, perhaps as a possible signal to stabilize p53, in contrast to its antioxidant role in nonstress conditions (Figure 2).59 Stress because of the absence of Mdm2 induces ROS and stabilizes/activates p53R172P, leading to an increase in cell cycle arrest and senescence functions.112 This is further corroborated by elevated mRNA levels of p53 targets p21 and Dcr2 and an independent senescence marker p15 in the BM cells.114 Furthermore, senescence-associated βgal activity is also higher in the postnatal BMs. The hematopoietic defect in these mice is HSC-specific as bone marrow cells from these mice fail to rescue lethally irradiated wild-type mice in BM transplantation assays. Importantly, these defects are p53-dependent.
Extrapolation of these data implicate Mdm2 as a critical regulator of p53 activity in hematopoietic tissues when ROS levels increase. This regulation is critical for stem cell survival and differentiation. The increased ROS levels observed in patients with myeloid malignancies115 may also be p53-dependent. As such, antioxidant treatment could be a therapeutic strategy for these patients. Furthermore, the data also show that, in addition to apoptosis, other p53 downstream pathways, such as cell cycle arrest and senescence, are also involved in depletion of HSCs and progenitors cells.
TP53 in ribosomopathies: Diamond Blackfan anemia and 5q− syndrome
Ribosomal stress resulting from impaired nascent ribosome biogenesis or defects in ribosome function is another mechanism by which the p53 pathway is activated and commonly associates with the hematopoietic phenotype observed in several bone marrow failure syndromes.116 Ribosomal proteins function primarily as structural components of the 40S and 60S ribosomal subunits. Mutations in individual ribosomal proteins disrupt the assembly of the protein into the ribosomal subunits.117-121 As a result, unengaged ribosomal proteins remain free to bind and block Mdm2, leading to p53 accumulation and activation of a cell cycle arrest response in the erythroid lineage.122-124 This ultimately results in an erythroid failure phenotype that characterizes several genetic syndromes, such as Diamond-Blackfan anemia (DBA).117,125 DBA is an autosomal dominant syndrome characterized by aplasia of the erythroid lineage that is associated with growth retardation and limb, cardiac and craniofacial malformations with a very wide range of severity in all its phenotypic manifestations.126 Patients with DBA exhibit severe macrocytic anemia with reticulocytopenia and aplasia of red blood cell precursors in the context of an otherwise normocellular bone marrow.127 The absence of red blood cell precursors is the result of impaired proliferation and increased apoptosis at the erythrocyte burst-forming unit stage. Knockdown of RPS19 (the ribosomal protein most frequently involved in DBA) in mouse fetal liver cells induces increased levels of p53 and p21 with decreased proliferation and delay of the G1/S phase of the cell cycle.128 A similar erythroid phenotype in RPS19-deficient zebrafish is alleviated on suppression of p53.129 In human bone marrow–derived CD34+ cells in which the expression of RPS19 or RPS14 is down-modulated, the subsequent erythroid phenotype is completely rescued on treatment with the p53 inhibitor pifithrin-α. Conversely, nutlin-3, a compound that activates p53 through inhibition of Mdm2, selectively impairs erythropoiesis.130 Mice homozygous for RPS19 loss are not viable, whereas heterozygous RPS19 mice are born but exhibit no DBA phenotype.131,132 Interestingly, an inducible mouse model of mutated RPS19 (R62W mutation) completely recapitulates a DBA phenotype (growth retardation, anemia, and reduced erythroid progenitors and circulating red blood cells), suggesting a dominant negative role for RPS19 mutations.133 Induction of the mutant allele reduces the expression levels of RPS19 to 10% of normal endogenous levels. Ubiquitous expression of the mutant transgene results in lethality in early embryogenesis, but inducible expression at later time points induces ribosomal RNA-processing defects in bone marrow cells that phenocopy DBA. McGowan and Mason also identified a knock-in allele of RPS19 in a forward genetic screen in mice for defects in skin color.134 The RPS19DSK/+ mice are characterized by dark tail, ears, and feet, a white belly spot, growth retardation, and mild macrocytic anemia. Intriguingly, deletion of p53 from the bone marrow mitigated the skin and anemia phenotypes. Incidentally, dark skin phenotype are also observed in mice haploinsufficient for Mdm2 (Mdm2 heterozygous or Mdm2 hypomorphic alleles) or Mdm4 (Mdm4 heterozygosity).70,90 These data indicate that p53 function is critical for the phenotypic manifestations of human ribosomapathies.
Since the initial description of RPS19 mutations as the cause of DBA,117 mutations in RPS24, RPS17, and RPL35A have also been implicated in DBA.120,121,135 Recent studies have determined that mutations in ribosomal genes RPS19, RPS24, RPS17, RPS10, RPS26, RPS7, RPL5, RPL11, and RPL35A account for ∼ 50% of the known genetic alterations in affected persons.116,126 Defective ribosome biogenesis has been linked to many other congenital syndromes, including Schwachman-Diamond syndrome, X-linked dyskeratosis congenita, cartilage hair hypoplasia, Treacher Collins syndrome, and the 5q− syndrome.116 Importantly, all these clinical phenotypes, when induced in genetically engineered mouse models, can be rescued by crossing such models to p53-null mice, indicating their dependence on hyperactivation of the p53 pathway.134
A prime example is the 5q− syndrome, a clonal disorder characterized by macrocytic anemia with erythroid hypoplasia and increased platelet counts with dysplastic hypolobulated megakaryocytes, typically affecting women.136 The striking overlapping features with DBA lead to the identification of haploinsufficiency of RPS14 as responsible for this phenotype. RPS14 was identified in an RNA interference screen of each gene within the commonly deleted region as responsible for the 5q− syndrome.137 A mouse model in which coordinate deletion of loci syntenic with the common deleted region of the 5q− syndrome (including RPS14 haploinsufficiency) recapitulates the erythroid phenotype characteristic of 5q− syndrome.138 Intercrossing this mouse with p53-deficient mice completely rescues the progenitor cell defect, restoring all hematopoietic bone marrow populations, again implicating a p53-dependent mechanism in the pathophysiology of this disorder induced by RPS14 alterations.138 Thus, increased p53 activity caused by ribosomal dysfunction is critical in the evolution of myelodysplastic syndromes, again emphasizing the need to squelch p53 activity.
Perspectives and conclusions
Studies using genetically modified mice have provided invaluable insights regarding the regulation of the p53-Mdm axis under physiologic conditions as well as under stress (oncogenic or otherwise) in a variety of human diseases. This information is critical from a translational viewpoint as pharmacologic agents that impinge on the p53 pathway are starting to be tested in clinical trials. For instance, p53-reactivating therapies are being tested for treatment of leukemia and other hematologic diseases with wild-type but deregulated p53 function. One such strategy involves the disruption of the p53-Mdm2 interaction using synthetic inhibitors, such as Nutlin. However, based on the results from the mouse models, caution is warranted when treating patients with such drugs as too much p53 activity could also impart toxicity to healthy hematopoietic cells as well as other normal tissues and cause depletion of HSCs and other toxicities. Furthermore, caution should be exercised when agents of that kind are used in patients whose malignancies express mutant forms of p53 (such as the p53R172H or p53R172P model) as their use in such context might lead to stabilization of mutant p53 with potentially catastrophic consequences. On the other hand, a transient disruption of the p53-Mdm2 interaction could be used to achieve myelosuppression before bone marrow transplantation while minimizing the use of chemotherapeutic agents in conditioning regimens, thus avoiding the toxicity associated with them. Inhibitors of Mdm4 are currently under development as well and could also be used for the same indications.
The knowledge gained regarding p53 regulation in hematopoiesis can be translated to the treatment of leukemia. Leukemogenesis is the result of uncontrolled proliferation and abnormal differentiation of leukemia stem cells. These cells share multiple features with normal HSCs, such as quiescence, and are also regulated by p53. It is plausible that alterations in the p53 pathway that sensitize normal HSCs to apoptosis or regulate their quiescence may produce similar effects in leukemia stem cells. Although the therapeutic window may be too narrow, in patients where the p53 pathway is still intact, disrupting the p53-mdm2 interaction could be conducive to apoptosis and eradication of leukemia stem cells.
The availability of mouse models that recapitulate human disease further our mechanistic insight into disease manifestation and provide a unique opportunity to test strategies targeting p53 activity in a preclinical setting. However, mouse models do have their own set of limitations. Most of the models used in laboratory carry a germline deletion/mutation of p53 pathway component, which differs from the subtle somatic mutations observed in human tumors. The genetic background of mouse models also influences the tumor spectrum. Differences in the mouse and human stroma could affect the tumor initiation, progression, and treatment profiles. Moreover, the common strategy of using whole-body radiation response for testing the fitness of the hematopoietic system presents a scenario unlikely ever to be recapitulated in human patients. Nonetheless, the benefits of using these models to test hypotheses far outweigh the negatives.
In conclusion, mouse models with alterations in the p53 pathway have been invaluable in shedding light as to the role of p53 in hematopoiesis. During steady state, basal p53 activity regulates HSC quiescence and self-renewal. Lack of p53 activity promotes HSC proliferation, which eventually leads to accumulation of DNA damage and tumor development. Excessive p53 activation, on the other hand, adversely affects cellularity of key organs and promotes loss of hematopoietic progenitors. Therefore, fine-tuned regulation of p53 activity is critical to maintain homeostasis and recovery after tissue damage, particularly in tissues with high cellular turnover, such as the bone marrow. A better understanding of the p53 regulatory mechanisms is critical to harness p53 activity for therapeutic purposes in cancer and other diseases.
Acknowledgments
The authors thank Dr James Jackson, MD Anderson Cancer Center, for critical reading of the manuscript.
This work was supported by the National Institutes of Health (grant CA47296, G.L.). V.P. is supported by the Brown Foundation.
National Institutes of Health
Authorship
Contribution: V.P., A.Q.-C., and G.L. wrote different sections of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guillermina Lozano, Department of Genetics, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: gglozano@mdanderson.org.