Abstract
NF-E2–related factor 2 (Nrf2) transcription factor regulates a range of cytoprotective transcriptional responses, preventing further cellular injury by removing biochemical damage and renewing tissue. Here we show that acute myeloid leukemia (AML) cells possess greater constitutive nuclear levels of Nrf2 than normal control CD34+ cells because of an imbalance between mRNA expression levels of Nrf2 and its inhibitor Keap1 but not through their somatic mutation. Elevated Nrf2 was reduced by NF-κB inhibitors. Using promoter assays, ChIP and siRNA knockdown, we demonstrated NF-κB subunits p50 and p65 induce transcription of Nrf2 in AML cells at a specific promoter κB-site and that long-term lentiviral miRNA-knockdown of Nrf2 significantly reduced clonogenicity of AML patient cells and improved their chemotherapeutic responsiveness. Normal physiologic Nrf2 protects cells from damage, but here we have exposed aberrant continuous nuclear activation of Nrf2 in AML that allows cell survival, even against cytotoxic chemotherapeutics. We show for the first time that Nrf2, an important regulator of several biologic processes involved in the progression of cancer, has abnormal NF-κB–driven constitutive expression in AML. Such a mechanism allows for a greater cytoprotective response in human AML cells and encourages their evasion of chemotherapy-induced cytotoxicity, which is necessary for improved clinical outcomes.
Introduction
Acute myeloid leukemia (AML) comprises a biologically heterogeneous group of disorders that occur as a consequence of a wide variety of genetic abnormalities in hematopoietic progenitors. In fact, hundreds of different genetic lesions have been described in AML, but despite this genetic heterogeneity, it appears these tumors share common programs of self-renewal and transformation downstream of leukemia-associated oncogenes. This finding suggests that mechanistically common therapeutic approaches to AML are likely to be possible, regardless of the identity of the driver oncogene involved, and argues for the presence of common mechanisms of leukemia cell survival in this group of patients.1
The NF-E2–related factor 2 (Nrf2) is 1 of the cancer cell survival pathways that is implicated in protecting cancer cells from apoptosis.2-5 Nrf2 functions to rapidly change the sensitivity of the cells environment to oxidants and electrophiles by stimulating the transcriptional activation of more than 100 cytoprotective and detoxification genes.6,7 Genes regulated by Nrf2 have the capacity to prevent damage to DNA, proteins, and lipids and assist in the generation of new tissue.8-10 Unlike in nonmalignant CD34+ hematopoietic cells, Nrf2 is present in the nucleus of most patients with AML, where its pathway is primed for immediate activation, leading to cytoprotective and detoxification gene up-regulation in response to chemotherapeutic drugs,2,11 and subsequently reduces apoptosis compared with AML cells with no nuclear Nrf2 expression.
The mechanism that controls Nrf2 expression and nuclear localization in AML is presently unknown. In non-AML cells, the inhibitor of Nrf2 (Keap1 or INrf2) mediates the ubiquitin-26S proteasome-mediated turnover of Nrf2. Exposure to oxidative and electrophilic stresses, such as reactive oxygen species (ROS) through the addition of chemotherapeutic drugs,12,13 impairs Keap1-mediated proteasomal degradation of Nrf2, causing its activation and translocation to the nucleus.14 Nrf2 then forms a complex with Maf proteins, which bind to the antioxidant response element (ARE) to mediate transcription of Nrf2-inducible genes. Another control element to the Nrf2 pathway is the transcriptional repressor Bach1, which can bind ARE enhancers blocking Nrf2 until naive cells are stimulated by pro-oxidants.15,16 Consequently, under normal cellular conditions Nrf2 is anticancerous because of induction of cytoprotective and detoxification genes that protect cells from electrophilic/oxidative damage. Paradoxically, however, these very same cytoprotective and detoxification genes that provide protection from cancer initiation enhance the resistance of cancer cells to chemotherapeutic drugs, and in this context Nrf2 is rapidly attracting a protumoral identity.2,4,17-19 Taken together. it is likely that AML cells acquire a growth advantage and chemoresistance via activation of Nrf2-dependent defense responses and suggests that the pathways that control constitutive nuclear Nrf2 expression in AML may be appropriate therapeutic targets. In this study we aimed to understand the mechanism by which constitutively active Nrf2 is present in AML and its role in protecting AML cells against current chemotherapeutic drugs.
Methods
Materials
The AML-derived cell lines U937, HL-60, and THP-1 were obtained from the European Collection of Cell Cultures. Anti-NF-κB antibodies p50 and p65 and U0126 were purchased from Cell Signaling Technologies. All other antibodies were obtained from Santa Cruz Biotechnology. BAY 11-7082 (inhibitor of IκB phosphorylation) was procured from Calbiochem. Control, Nrf2, and p50 and p65 siRNA were purchased from Applied Biosystems. All other reagents were obtained from Sigma-Aldrich unless indicated.
Cell culture
Primary AML cells were obtained under local ethical approval (LREC ref. 07/H0310/146). For primary cell isolation, heparinized blood was collected from volunteers and human peripheral blood mononuclear cells (PBMCs) isolated by Histopaque (Sigma-Aldrich) density gradient centrifugation. PBMCs (4 × 106/mL) were incubated in complete medium for 2 hours at 37°C to allow adherence of monocytes. We obtained hematopoietic CD34+ cells from 2 sources, StemCell Technologies and volunteers. The positive selection of CD34+ cells was isolated from PBMCs with the use of a human CD34+ MicroBead selection kit (Miltenyi Biotec). For all CD34+ and primary monocyte experiments, at least 3 different donors were used to obtain the results presented in this article. AML samples that were < 80% blasts were purified using the CD34-positive selection kit (denoted by an asterisk in Table 1). Cell type was confirmed by microscopy and flow cytometry.
AML sample information
| Number . | Age, y . | Sex . | WHO diagnosis . | Cytogenetics . | % blasts . | Nrf2 mRNA (fold over CD34+) . | Nrf2 binding activity (fold over CD34+) . | NF-κB binding activity (fold over CD34+) . |
|---|---|---|---|---|---|---|---|---|
| AML#1 | 80 | M | AML without maturation | Normal | 95 | 10.04 | 6.00 | |
| AML#2 | 66 | F | Therapy-related myeloid neoplasm | Complex | 85 | 5.87 | 4.62 | |
| AML#3 | 61 | M | AML with myelodysplasia | Complex | 80 | 5.54 | 2.33 | 3.17 |
| AML#4 | 53 | M | Acute promyelocytic leukemia | t(15;17) | 95 | 0.68 | 1.16 | 1.26 |
| AML#5 | 77 | M | AML with myelodysplasia | Complex | 95 | 6.99 | 2.88 | 4.06 |
| AML#6 | 49 | M | AML with maturation | Normal | 80 | 6.24 | 3.26 | 4.65 |
| AML#7 | 39 | M | AML with maturation | Normal | 65* | 3.57 | 2.03 | 2.68 |
| AML#8 | 58 | M | Acute erythroid leukemia | Normal | 70 | 4.54 | 2.03 | 2.67 |
| AML#9 | 64 | M | AML with RUNX1-RUNX1T1 | t(8;21) | 85 | 0.88 | 1.09 | 1.98 |
| AML#10 | 92 | F | AML with myelodysplasia | Not available | 70 | 4.28 | 4.29 | |
| AML#11 | 57 | M | AML with myelodysplasia | Normal | 55 | 3.05 | 1.69 | 1.55 |
| AML#12 | 82 | F | AML with myelodysplasia | Deletion 13 | 85 | 2.96 | 2.10 | 1.99 |
| AML#13 | 46 | F | AML with maturation | +4,+8, t(9;22) | 70* | 4.97 | 2.27 | 3.05 |
| AML#14 | 40 | M | Acute promyelocytic leukemia | t(15;17) | 95 | 4.42 | 2.93 | 1.71 |
| AML#15 | 66 | F | AML without maturation | t(2;12) | 65* | 0.96 | 1.36 | 1.38 |
| AML#16 | 78 | M | AML with myelodysplasia | Not available | 85 | 1.13 | 0.93 | 0.89 |
| AML#17 | 57 | M | AML with minimal differentiation | Not available | 95 | 4.75 | 2.12 | 2.81 |
| AML#18 | 27 | M | AML with RUNX1-RUNX1T1 | t(8;21) | 60* | 6.42 | 3.86 | 5.64 |
| AML#19 | 28 | F | Acute myelomonocytic leukemia | Normal | 70 | 11.97 | 3.24 | 4.62 |
| AML#20 | 51 | F | AML with maturation | Normal | 75 | 6.85 | 3.56 | 5.15 |
| AML#21 | 77 | M | AML without maturation | Normal | 90 | 2.36 | 1.90 | |
| AML#22 | 85 | F | AML not otherwise specified | Not available | 75* | 4.31 | 2.32 | 3.14 |
| Number . | Age, y . | Sex . | WHO diagnosis . | Cytogenetics . | % blasts . | Nrf2 mRNA (fold over CD34+) . | Nrf2 binding activity (fold over CD34+) . | NF-κB binding activity (fold over CD34+) . |
|---|---|---|---|---|---|---|---|---|
| AML#1 | 80 | M | AML without maturation | Normal | 95 | 10.04 | 6.00 | |
| AML#2 | 66 | F | Therapy-related myeloid neoplasm | Complex | 85 | 5.87 | 4.62 | |
| AML#3 | 61 | M | AML with myelodysplasia | Complex | 80 | 5.54 | 2.33 | 3.17 |
| AML#4 | 53 | M | Acute promyelocytic leukemia | t(15;17) | 95 | 0.68 | 1.16 | 1.26 |
| AML#5 | 77 | M | AML with myelodysplasia | Complex | 95 | 6.99 | 2.88 | 4.06 |
| AML#6 | 49 | M | AML with maturation | Normal | 80 | 6.24 | 3.26 | 4.65 |
| AML#7 | 39 | M | AML with maturation | Normal | 65* | 3.57 | 2.03 | 2.68 |
| AML#8 | 58 | M | Acute erythroid leukemia | Normal | 70 | 4.54 | 2.03 | 2.67 |
| AML#9 | 64 | M | AML with RUNX1-RUNX1T1 | t(8;21) | 85 | 0.88 | 1.09 | 1.98 |
| AML#10 | 92 | F | AML with myelodysplasia | Not available | 70 | 4.28 | 4.29 | |
| AML#11 | 57 | M | AML with myelodysplasia | Normal | 55 | 3.05 | 1.69 | 1.55 |
| AML#12 | 82 | F | AML with myelodysplasia | Deletion 13 | 85 | 2.96 | 2.10 | 1.99 |
| AML#13 | 46 | F | AML with maturation | +4,+8, t(9;22) | 70* | 4.97 | 2.27 | 3.05 |
| AML#14 | 40 | M | Acute promyelocytic leukemia | t(15;17) | 95 | 4.42 | 2.93 | 1.71 |
| AML#15 | 66 | F | AML without maturation | t(2;12) | 65* | 0.96 | 1.36 | 1.38 |
| AML#16 | 78 | M | AML with myelodysplasia | Not available | 85 | 1.13 | 0.93 | 0.89 |
| AML#17 | 57 | M | AML with minimal differentiation | Not available | 95 | 4.75 | 2.12 | 2.81 |
| AML#18 | 27 | M | AML with RUNX1-RUNX1T1 | t(8;21) | 60* | 6.42 | 3.86 | 5.64 |
| AML#19 | 28 | F | Acute myelomonocytic leukemia | Normal | 70 | 11.97 | 3.24 | 4.62 |
| AML#20 | 51 | F | AML with maturation | Normal | 75 | 6.85 | 3.56 | 5.15 |
| AML#21 | 77 | M | AML without maturation | Normal | 90 | 2.36 | 1.90 | |
| AML#22 | 85 | F | AML not otherwise specified | Not available | 75* | 4.31 | 2.32 | 3.14 |
This table defines the nature of the AML disease, including WHO diagnosis and cytogenetics. Percent blast denotes the percentage of AML blasts after purification with the use of density gradient centrifugation and in some instances CD34+ positive selection. NF-κB and Nrf2 binding is indicated as a fold increase above levels from CD34+ cells. Nrf2 mRNA status indicates a marked difference in the levels of Nrf2 mRNA in AML samples compared with CD34+ control cells, as shown in Figure 1D.
AML indicates acute myeloid leukemia; Nrf2, NF-E2–related factor 2; and WHO, World Health Organization.
Samples that have been CD34+ positive selected.
RNA extraction and real-time PCR
Total RNA was extracted from 5 × 105 cells by use of the Nucleic acid PrepStation from Applied Biosystems, according to the manufacturer's instructions. Reverse transcription was performed using the RNA PCR core kit (Applied Biosystems). Real-time PCR primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Nrf2 were purchased from Invitrogen. Relative quantitative real-time PCR used SYBR green technology (Roche) on cDNA generated from the reverse transcription of purified RNA. After preamplification (95°C for 2 minutes), the PCRs were amplified for 45 cycles (95°C for 15 seconds and 60°C for 10 seconds and 72°C for 10 seconds) on a 384-well LightCycler 480 (Roche). Each mRNA expression was normalized against GAPDH mRNA expression via use of the standard curve method.
Genomic DNA extraction and sequencing
Genomic DNA was extracted from blood samples with the GenElute DNA miniprep kit (Sigma-Aldrich); we then performed PCR by using multiple primers to flank the Nrf2 and Keap1 exonic regions (supplemental Figure 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Product sequencing was performed by Dundee DNA Sequencing & Services, with 100% match between the AML samples, CD34+ HSC tested, and the GenBank gene sequence.
DNA binding assays
NF-κB DNA binding was measured with the NF-κB p65 transcription factor ELISA kit (Panomics). Nrf2 DNA binding was measured with the Nrf2 DNA binding activity Kit (TransAM kits; Active Motif).
Virus construction and infection
MicroRNA sequence miRNA-Nrf2 (5′-TTAATGAGTTCACTGTCAACT-3′) targeting human Nrf2 was selected with Invitrogen Block-iT RNAi Designer software (www.invitrogen.com/rnai), and plasmid pcDNATM6.2-GW/EmGFP-miR-neg (Invitrogen) was used as source for the negative control. MicroRNA-encoding sequences were cloned into Block-iT Pol II miR-RNAi vector (Invitrogen) and then EmGFP/pre-miRNA fragments were subcloned into the BamHI/XhoI site in the LNT/SffvMCS plasmid (kind gift from Penny Powell, University of East Anglia, Norfolk, United Kingdom). MicroRNA-encoding viruses were produced in 293T cells as described previously20 by the use of packaging plasmids pCMVΔR8.91 (expressing gag-pol) and pMD.G (expressing VSV-G; kindly provided by Dr Ariberto Fassati, University College London, Longon, United Kingdom). Lentiviral stocks were concentrated by the use of Lenti-XTM Concentrator (Clontech), and titers were obtained with Lenti-XTM qRT-PCR Titration kit (Clontech). For transduction, AML and control cells were plated onto 12-well plates at 5 × 104 cells/well. Cells were infected with lentiviral stocks at an MOI of 10 in presence of polybrene. Transduced cells were analyzed by flow cytometry (Accuri), real-time PCR (Roche), and Western blotting.
Promoter assays
To generate the Nrf2 promoter construct [a], a DNA fragment containing 1.5 kb of the human Nrf2 promoter region was amplified from genomic DNA with PCR and specific primers 5′-ATGAGCTGTGGACCGTGTGTT-3′ reverse primer and 5′-TGGGCGTTGATTGCTATAGTC-3′ forward primer. The fragment was cloned into the PGL4 basic plasmid (Promega). To generate mutated κB1 (construct [c]) and κB2 (construct [b]) constructs, the PCR primers used were mutκB2 5′-CGCGCGGGCTGAGCTTCCGAACCAACCCCACCCGCG-3′ and mutκB1 5′-CCAGAGCTGGGAGAAAAACGGTCTACCCAAGAGAACCTCTTCCCAC-3′. These mutations were introduced with the QuikChange XL Site-Directed Mutagenesis Kit (Agilent).
ChIP assays
THP-1 cells were untreated or treated with BAY 11-7082 for 8 hours before the cells were fixed with 1% formaldehyde in medium for 10 minutes at room temperature. The sonication conditions were optimized to determine generation of DNA fragments between 300 and 600 bp in length. Chromatin was immunoprecipitated with IgG (Sigma-Aldrich), anti-p65, and anti-p50 (New England Biolabs). The association of p50 and p65 was measured by RT-PCR on immunoprecipitated chromatin by use of the following primers spanning the κB1 site at -820 5′-TGCACTCGGTAATCGGCTACA-3′ (forward) and 5′-GGGGAGCTAACGGAGACCT-3′ (reverse) and κB2 site at -220 5′-ACTCCCACGTGTCTCCATTC-3′ (forward) and 5′-CGATTACAGCATGTTGTGGTATT-3′ (reverse).
ChIP-seq (ENCODE)
These data were generated by the laboratory of Michael Snyder using the human lymphoblastoid cell line GM12891. The following antibody was used for NF-κB (sc-372; Santa Cruz Biotechnology). Data were accessed at http://genome.ucsc.edu/ENCODE/.
Western immunoblotting and flow cytometry
SDS-PAGE and Western analyses were performed as described previously. To summarize, whole cell lysates were extracted by use of the radioimmunoprecipitation assay buffer method and SDS-PAGE separation performed. Nuclear extracts were prepared as previously described.3,21 Protein was transferred to nitrocellulose and Western blot analysis performed with the indicated antisera according to their manufacturer's guidelines. A dichlorofluorescein (DCF) assay was used to determine cellular ROS generation; fluorescence was analyzed by Accuri flow cytometry as described.22
Transfections
We transfected AML cells and control cells (106/well) with Amaxa Nucleofector Technology by using equivalent molar concentrations of the siRNA (to yield a final concentrations of 30nM).23 A total of 0.5 μg of PGL4 reporter and pRL-CMV control constructs were cotransfected into THP-1. Transfected cells were incubated for 24 hours before the indicated treatments. For reporter assay, cells were treated with Dual-Luciferase Reporter Assay System (Promega).
Clonogenic methylcellulose assays
Control cell, AML cell lines, and primary AML cells (1 × 103 to 5 × 104 cells) were plated in methylcellulose medium (R&D systems) and colonies were visualized, measured, and counted after 10 days.
Proliferation/death assays
Cell number was measured by incubation with MTS one-solution assay reagent (Promega) at 37°C for 1 hour before reading absorbance in quadruplicate at 490 nm.
Statistical analyses
Student t test was performed to assess statistical significance from controls unless otherwise stated. Results with P < .05 or P < .01 were considered statistically significant. Results represent the mean ± SEM of 3 independent experiments. For Western blotting experiments, data are representative of 3 independent experiments.
Results
High Nrf2 protein in human AML cells is because of elevated Nrf2 RNA levels
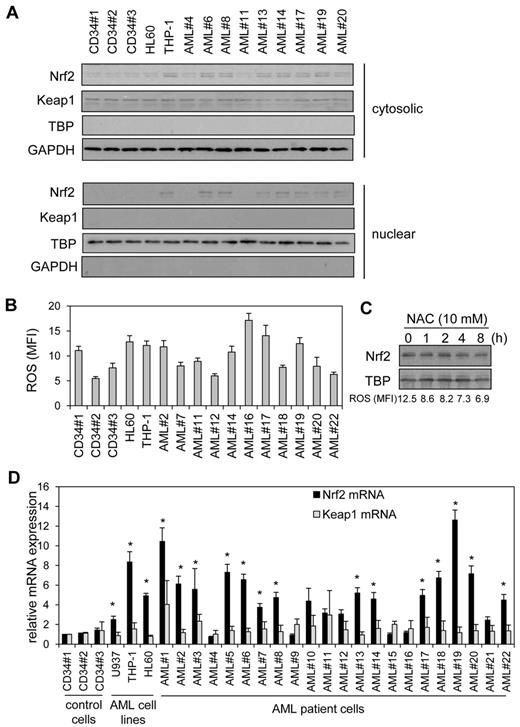
Recently we showed that Nrf2 was constitutively active in human AML cells.2,3 Here we wanted to understand the mechanism by which Nrf2 was constitutively active in these cells. Figure 1A shows that in human AML primary cells and cell lines, Nrf2 protein was high in 8 of 11 AML samples tested. There was no correlation between Keap1 and Nrf2 protein levels in AML (supplemental Figure 1), suggesting that aberrant activation of Nrf2 is responsible for its nuclear localization in AML. Because mutations in Keap1 and/or Nrf2 have been identified previously in other cancers, we sought to determine whether Keap1and/or Nrf2 somatic mutations were responsible for the elevated Nrf2 protein in AML by using a combination of PCR and DNA sequencing in our primary AML samples. However, we found no mutations in the 21 of 22 primary AML samples tested (supplemental Figure 2) with 100% match to CD34+ control cells and PubMed sequence, suggesting Nrf2 expression in AML is induced by an alternative mechanism.
High Nrf2 protein in human AML cells is because of elevated Nrf2 RNA levels. (A) Nuclear extracts were obtained from human AML samples and control cells, extracts were separated by SDS-PAGE, and Western blot analysis was conducted for Nrf2 and Keap1 protein levels. Nuclear blots were reprobed with TATA binding protein (TBP) to confirm sample loading and cytosolic blots with GAPDH. (B) AML cells and control cells washed with PBS and incubation with 10μM H2DCFDA for 15 minutes. Cells were then assessed for H2DCFDA oxidation by the use of flow cytometry. (C) THP-1 cells were treated with NAC for up to 8 hours. Nuclear extracts were obtained and analyzed for Nrf2 expression. Nuclear blots were reprobed with TBP to confirm equal sample loading. The numbers under the Western blot indicate ROS levels in NAC-treated THP-1 cells. (D) RNA was extracted from AML cells and control cells and Nrf2 and Keap1 mRNA was measured with real-time PCR. mRNA expression was normalized to GAPDH mRNA levels. *Statistical significance (P < .05) between Nrf2 and Keap1 mRNA levels where indicated.
High Nrf2 protein in human AML cells is because of elevated Nrf2 RNA levels. (A) Nuclear extracts were obtained from human AML samples and control cells, extracts were separated by SDS-PAGE, and Western blot analysis was conducted for Nrf2 and Keap1 protein levels. Nuclear blots were reprobed with TATA binding protein (TBP) to confirm sample loading and cytosolic blots with GAPDH. (B) AML cells and control cells washed with PBS and incubation with 10μM H2DCFDA for 15 minutes. Cells were then assessed for H2DCFDA oxidation by the use of flow cytometry. (C) THP-1 cells were treated with NAC for up to 8 hours. Nuclear extracts were obtained and analyzed for Nrf2 expression. Nuclear blots were reprobed with TBP to confirm equal sample loading. The numbers under the Western blot indicate ROS levels in NAC-treated THP-1 cells. (D) RNA was extracted from AML cells and control cells and Nrf2 and Keap1 mRNA was measured with real-time PCR. mRNA expression was normalized to GAPDH mRNA levels. *Statistical significance (P < .05) between Nrf2 and Keap1 mRNA levels where indicated.
Because oxidative stress induces nuclear Nrf2 expression by deactivating Keap1, we sought to determine whether elevated ROS levels could be responsible for the nuclear localization of Nrf2 observed in AML. In the AML cells, there was no relationship between high ROS levels and high nuclear Nrf2 (Figure 1A-B and supplemental Figure 1B). Furthermore, use of the ROS quencher N-acetyl cysteine (NAC), which successfully sequesters endogenous ROS in AML (supplemental Figure 3), had no effect on nuclear Nrf2 levels (Figure 1C). Interestingly, although Keap1 appears unable to restrain high Nrf2 levels in AML, it seemed to remain functional as nuclear Nrf2 increases in response to H2O2 (supplemental Figure 3D). Taken together, this excludes oxidative stress as the mechanistic cause of high nuclear Nrf2 in resting human AML cells.
We next examined the RNA profile of Nrf2 in AML to determine whether this was the cause of aberrant Nrf2 protein expression in AML. Figure 1D shows that there was an increase in Nrf2 mRNA in AML samples and cell lines in correlation with the presence of nuclear Nrf2 protein observed in Figure 1A (supplemental Figure 1A). We also show that 15 of 22 AML samples tested had significantly increased Nrf2 mRNA levels compared with Keap1 mRNA. Moreover, there was no correlation between high Nrf2 mRNA levels and Keap1 mRNA expression in AML (Figure 1D and Supplementary Figure 1C). Nrf2 target gene mRNA levels also were elevated compared with CD34+ control cells in high Nrf2-expressing AML cells (supplemental Figure 4). Therefore, high Nrf2 protein in human AML cells is because of elevated Nrf2 RNA levels.
NF-κB regulates Nrf2 overexpression in human AML
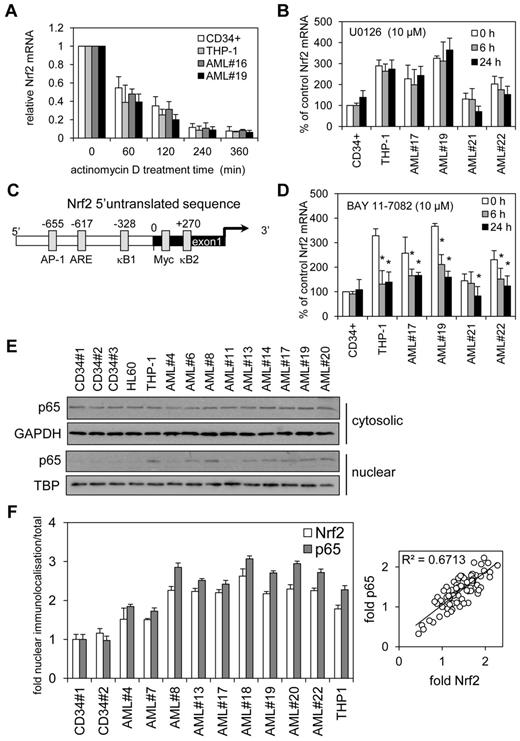
Next we wanted to understand the cause of the aberrant mRNA expression in AML. We identified 2 potential causes, aberrant transcription or increased RNA stability. To determine whether increased Nrf2 RNA stability was resulting in high Nrf2 protein, we analyzed THP-1, AML cells, and control cells with the transcription inhibitor actinomycin D (Act D) during a 6-hour time-course. Act D had no significant effect on the rate of decay of Nrf2 mRNA between control and AML cells (Figure 2A). Moreover, Nrf2 protein levels between THP-1 and CD34+ cells showed no difference in decay in response to treatment with Act D for 6 hours (data not shown). These results indicate that an increase in Nrf2 mRNA in human AML is through transcriptional processes rather than effects on RNA stability.
NF-κB regulates Nrf2 overexpression in human AML. (A) AML and control cells were treated with Act D (5 μg/mL) for up to 6 hours. RNA was extracted and examined for Nrf2 mRNA expression by real-time PCR. mRNA expression was normalized to GAPDH mRNA levels. (B) AML and control cells were treated for 6 and 24 hours with U0126 (10μM) and RNA was extracted and examined for Nrf2 mRNA expression by real-time PCR. mRNA expression was normalized to GAPDH mRNA levels. Data presented as percent of control. (C) Schematic representation of the Nrf2 promoter. (D) AML and control cells were treated for 6 and 24 hours with BAY 11-7082 (10μM) and RNA was extracted and examined for Nrf2 mRNA expression by real-time PCR. mRNA expression was normalized to GAPDH mRNA levels. Data are presented as percent of control. *P < .05 between the different treatment groups. (E) Nuclear and cytosolic Western blots from Figure 1A were reprobed for p65 expression. (F) Analysis of p65 and Nrf2 subcellular localization in AML cell nucleus. Data were calculated as a percentage of total localization and is expressed as fold increase compared with healthy CD34+ cell levels. Values indicate the mean ± SEM from at least 5 cells per sample (statistical significance *P ≤ .05 compared with an average of both CD34+ samples). Relative fold p65 versus fold Nrf2 scatter plot indicates a correlation with a slope of 1.
NF-κB regulates Nrf2 overexpression in human AML. (A) AML and control cells were treated with Act D (5 μg/mL) for up to 6 hours. RNA was extracted and examined for Nrf2 mRNA expression by real-time PCR. mRNA expression was normalized to GAPDH mRNA levels. (B) AML and control cells were treated for 6 and 24 hours with U0126 (10μM) and RNA was extracted and examined for Nrf2 mRNA expression by real-time PCR. mRNA expression was normalized to GAPDH mRNA levels. Data presented as percent of control. (C) Schematic representation of the Nrf2 promoter. (D) AML and control cells were treated for 6 and 24 hours with BAY 11-7082 (10μM) and RNA was extracted and examined for Nrf2 mRNA expression by real-time PCR. mRNA expression was normalized to GAPDH mRNA levels. Data are presented as percent of control. *P < .05 between the different treatment groups. (E) Nuclear and cytosolic Western blots from Figure 1A were reprobed for p65 expression. (F) Analysis of p65 and Nrf2 subcellular localization in AML cell nucleus. Data were calculated as a percentage of total localization and is expressed as fold increase compared with healthy CD34+ cell levels. Values indicate the mean ± SEM from at least 5 cells per sample (statistical significance *P ≤ .05 compared with an average of both CD34+ samples). Relative fold p65 versus fold Nrf2 scatter plot indicates a correlation with a slope of 1.
To consider the possibility that mutated K-Ras acting through c-Jun and c-myc activation via Raf/MEK/ERK pathway might lead to an increase in Nrf2 transcription and elevated basal Nrf2 protein levels, as recently shown in pancreatic cancer,24 we sought to determine whether the MEK inhibitor U0126 could reduce the levels of Nrf2 in AML cells that had high basal Nrf2 mRNA expression. In AML, however, U0126 had no effect on Nrf2 mRNA expression (Figure 2B), despite significantly inhibiting p-ERK at 10μM at 2 and 4 hours after treatment (data not shown). Together with the fact that K-Ras mutations are rare in AML (circa 5% of AML),25 this finding suggests that this pathway is not responsible for the elevated Nrf2 transcription in this disease.
To understand the transcriptional processes by which Nrf2 mRNA is increased in human AML cells, we used transcription factor binding site analysis programs (www.genomatix.de/online_help/help_matinspector/matinspector_help.html) to analyze the Nrf2 5′-promoter sequence for transcription control sites. These analyses identified 2 potential κB-binding sites within the Nrf2 promoter and exon1 sequence (Figure 2C) highlighting NF-κB as the potential controller of Nrf2 expression in AML. Other transcription factor binding sites also are included,24,26 and the DNA sequence for this region is shown in supplemental Figure 2B. We used inhibitor of IκB phosphorylation, BAY 11-7082 to test the hypothesis that NF-κB inhibition would reduce Nrf2 expression in AML. Nrf2 mRNA expression was reduced in AML in response to BAY 11-7082 (Figure 2D). BAY-11-7082 had no effect on either AML or control nonmalignant CD34+ cell viability (data not shown).
To further test whether NF-κB is responsible for the high expression of Nrf2 in AML we examined the subcellular protein expression of the NF-κB subunit p65. Western blots from Figure 1A were reprobed for p65 expression and showed that p65 was increased in the nucleus of AML cells that also tested positive for Nrf2 protein expression (Figure 2E and supplemental Figure 1F). Moreover, when we examined subcellular localization of p65 and Nrf2 in AML nuclei by immunocytochemistry (supplemental Figure 5), the analysis showed both nuclear protein levels to correlate in a 1:1 manner (Figure 2F). We also showed that both nuclear localization of p65 and Nrf2 can be inhibited by BAY 11-7082 and increased in response to lipopolysaccharide (LPS; supplemental Figure 6). Together, these data suggest that NF-κB is responsible for the aberrant expression of Nrf2 in human AML.
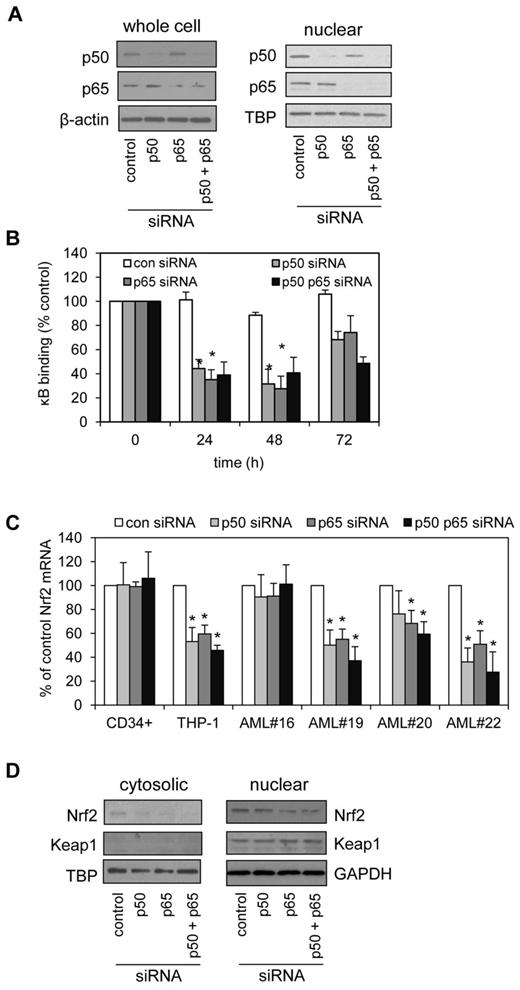
To further clarify the role of the NF-κB subunits p50 and p65 in the regulation of Nrf2 in human AML, we used siRNA knockdown to silence p50 and p65 in THP-1 cells. siRNA constructs, effective at silencing p50 or p65, individually and in combination (Figure 3A) were also able to inhibit the binding of p50 and p65 to the κB sites of THP-1 cells in a κB-binding assay (Figure 3B). Importantly, both p50 and p65 were necessary for maintaining high Nrf2 RNA expression that is seen in AML cell line and in 4 AML samples tested (1 normal Nrf2 and 3 high Nrf2 expression; Figure 3C). In total we tested 6 AML samples (2 normal Nrf2 and 4 high Nrf2 expression). The 1 difference seen in the data is that p50 siRNA did not significantly inhibit Nrf2 mRNA expression in AML#20 (Figure 3C). We also examined cytosolic and nuclear Nrf2 and Keap1 protein expression in response to p50 and p65 siRNA knockdown in THP-1 cells (Figure 3D).
Blocking NF-κB restores elevated Nrf2 levels to normal. (A) THP-1 cells were transfected with 30nM of control, p50 or p65 siRNA, or p50 and p65 combined and incubated for 48 hours. Extracts were separated by SDS-PAGE and Western blot analysis was conducted for p50 and p65 total protein or nuclear protein levels. Blots were reprobed for β-actin and TBP to confirm equal sample loading. (B) THP-1 cells were transfected with 30nM of control, p50 or p65-siRNA, or p50 and p65 combined and incubated for 24, 48, and 72 hours, then examined for the nuclear binding of p65 and p50 to specific κB sites. (C) AML cells were transfected with 30 nM of control, p50, or p65-siRNA, or p50 and p65 combined and incubated for 24 hours. RNA was extracted and examined for Nrf2 mRNA expression by real-time PCR. Data presented as percent of control. *P < .05 between the different treatment groups. (D) THP-1 cells were transfected with 30nM of control, p50 or p65-siRNA, or p50 and p65 combined and incubated for 48 hours. Nuclear and cytosolic extracts were separated by SDS-PAGE and Western blot analysis was conducted for Nrf2 and Keap1 protein levels were measured.
Blocking NF-κB restores elevated Nrf2 levels to normal. (A) THP-1 cells were transfected with 30nM of control, p50 or p65 siRNA, or p50 and p65 combined and incubated for 48 hours. Extracts were separated by SDS-PAGE and Western blot analysis was conducted for p50 and p65 total protein or nuclear protein levels. Blots were reprobed for β-actin and TBP to confirm equal sample loading. (B) THP-1 cells were transfected with 30nM of control, p50 or p65-siRNA, or p50 and p65 combined and incubated for 24, 48, and 72 hours, then examined for the nuclear binding of p65 and p50 to specific κB sites. (C) AML cells were transfected with 30 nM of control, p50, or p65-siRNA, or p50 and p65 combined and incubated for 24 hours. RNA was extracted and examined for Nrf2 mRNA expression by real-time PCR. Data presented as percent of control. *P < .05 between the different treatment groups. (D) THP-1 cells were transfected with 30nM of control, p50 or p65-siRNA, or p50 and p65 combined and incubated for 48 hours. Nuclear and cytosolic extracts were separated by SDS-PAGE and Western blot analysis was conducted for Nrf2 and Keap1 protein levels were measured.
Understanding the regulation of Nrf2 by NF-κB
To determine whether the κB-binding sites within the Nrf2 promoter sequence are important in regulating Nrf2 in AML, we created κB-deletion mutant reporter constructs of the human Nrf2 promoter (Figure 4A). To examine the functional role of these mutants, THP-1 cells were transfected with each individual construct and assayed in the presence of NF-κB inhibition (BAY 11-7082) and NF-κB activation (TNF-α). The construct with mutated κB2 site (construct [b]) showed decreased basal levels of promoter activity compared with wild-type Nrf2 promoter construct [a]. However, the construct with mutated κB1 (construct [c]) showed similar promoter activity to those seen with the wild-type Nrf2 promoter construct [a] (Figure 4B). As a positive control we treated transfected cells with BAY 11-7082 and TNF alone. Figure 4B shows a decrease in promoter activity in wild-type Nrf2 promoter construct [a] and the mutated κB1 construct [c] but not mutated κB2 in response to BAY 11-7082, and an increase in promoter activity in wild-type Nrf2 promoter construct [a] and the mutated κB1 construct [c] but not mutated κB2 construct [b] when treated with TNF (Figure 4B). These results demonstrate the κB2 site, located at +270 upstream of the transcription start site, is responsible for the increased expression of Nrf2 seen in AML cells.
Understanding the regulation of Nrf2 by NF-κB. (A) Schematic presentation of the Nrf2 promoter constructs that were created for this study, being either wild-type [a], κB2 site–deleted [b], or κB1 site–deleted [c] constructs. (B) THP-1 cells were transiently transfected with 0.5 μg of each promoter construct shown in panel A and pRL-CMV for normalization of transfection efficiency. Cell extracts were harvested, and luciferase assays were performed. Values are the means ± SD, n = 4. *P < .01 of deleted κB against untreated Nrf2 wild-type control. TNF (10 mg/mL) treatment acted as a positive control to activate NF-κB and BAY 11-7082 (10μM) to inhibit NF-κB. #P < .01 of TNF treated or BAY 11-7082 treated against untreated controls. (C) ChIP analysis of the Nrf2 promoter using antibodies against p50 and p65. Normal rabbit IgG was used as a control. THP-1 cells were left untreated or treated with BAY 11-7082 for 8 hours. Real-time PCR was performed in triplicate on immunoprecipitated DNA and input DNA. Data presented as percent of input. *P < .05 between the different treatment groups. (D) ChIP-seq data from the ENCODE Consortium demonstrates NF-κB subunit p65 binding to the Nrf2 promoter. NF-κB binding occurs in exon1 of Nrf2 and overlaps with sites of RNA pol II binding. Black bars denote positive signal above background.
Understanding the regulation of Nrf2 by NF-κB. (A) Schematic presentation of the Nrf2 promoter constructs that were created for this study, being either wild-type [a], κB2 site–deleted [b], or κB1 site–deleted [c] constructs. (B) THP-1 cells were transiently transfected with 0.5 μg of each promoter construct shown in panel A and pRL-CMV for normalization of transfection efficiency. Cell extracts were harvested, and luciferase assays were performed. Values are the means ± SD, n = 4. *P < .01 of deleted κB against untreated Nrf2 wild-type control. TNF (10 mg/mL) treatment acted as a positive control to activate NF-κB and BAY 11-7082 (10μM) to inhibit NF-κB. #P < .01 of TNF treated or BAY 11-7082 treated against untreated controls. (C) ChIP analysis of the Nrf2 promoter using antibodies against p50 and p65. Normal rabbit IgG was used as a control. THP-1 cells were left untreated or treated with BAY 11-7082 for 8 hours. Real-time PCR was performed in triplicate on immunoprecipitated DNA and input DNA. Data presented as percent of input. *P < .05 between the different treatment groups. (D) ChIP-seq data from the ENCODE Consortium demonstrates NF-κB subunit p65 binding to the Nrf2 promoter. NF-κB binding occurs in exon1 of Nrf2 and overlaps with sites of RNA pol II binding. Black bars denote positive signal above background.
To determine the in vivo relevance of p50 and p65 in regulating Nrf2 expression, we evaluated the recruitment of p50 and p65 to the Nrf2 promoter on untreated cells and cells treated with BAY 11-7082 by ChIP assay (Figure 4C). Analysis of untreated and BAY 11-7082–treated THP-1 cells was performed with the corresponding antibodies followed by PCR with specific primers amplifying the κB1 or κB2 regions. Recruitment of p50 and p65 were markedly enhanced to the κB2 site, but not the κB1 site, on the Nrf2 promoter in untreated THP-1 cells. In the presence of BAY 11-7082, recruitment of p50 and p65 to the Nrf2 promoter was inhibited, also confirming that NF-κB controls aberrant Nrf2 expression in vivo. Finally, ChIP-seq revealed that the transcriptional start site of the Nrf2 locus is a direct binding target of NF-κB subunit p65 when lymphoblastoid cell line GM12891 was activated by TNF (Figure 4D).27
Does NF-κB regulate Nrf2 in normal blood cells?
If the promoter of Nrf2 does contain an authentic κB binding site and AML cells do not have genetic aberrance in the Nrf2 pathway, then activation of the NF-κB pathway should enhance Nrf2 mRNA in normal cells. Previously we have shown that LPS can induce Nrf2 mRNA in THP-1 cells by up to 4-fold.28 To determine whether this is regulated by NF-κB, we stimulated human monocytes and THP-1 cells with LPS with and without BAY 11-7082 treatment and demonstrate that LPS can induce Nrf2 mRNA in human monocytes. Moreover, treatment with BAY-11-7082 confirms that LPS up-regulates Nrf2 transcription through NF-κB activation and induces Nrf2 and NF-κB nuclear localization (supplemental Figure 7).
Knockdown of Nrf2 enhances chemotherapy-induced apoptosis in AML
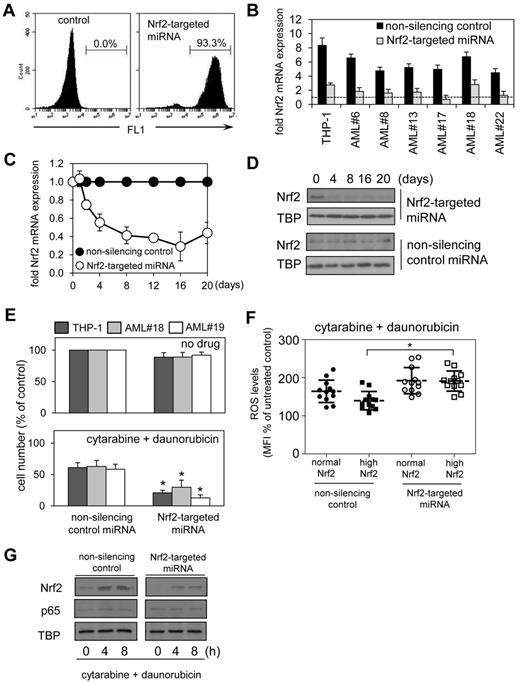
To assess the functional significance of aberrant Nrf2 expression in AML, we used lentivirus-mediated delivery for long-term Nrf2 knockdown by using targeted artificial microRNA (Nrf2-targeted miRNA) and visualization of infected cells by a GFP tag. The artificial GFP-tagged miRNA to target Nrf2 knockdown was constructed for lentivirus-mediated infection. This construct induced Nrf2 knockdown confirmed for up to 20 days by visualization of infected cells and assessment of the role of Nrf2 in long-term clonogenic assays. The construct was specifically designed to reduce levels of Nrf2 mRNA by 40%-75% (and not > 75%) in AML cells infected with this construct, which better reflects physiologic levels of Nrf2 mRNA compared with control cells, providing a more realistic model of Nrf2 knock-down in AML (Figure 5B). The Nrf2-targeted miRNA construct initially was validated in the THP-1 cell line; we observed a 94% transfection efficiency (Figure 5A) with the transfection efficiency of primary AML cells ranging from 55% to 80%, with knockdown of Nrf2 mRNA (Figure 5C) and Nrf2 protein (Figure 5D) efficiently maintained over at least 20 days. In all the AML samples tested, knockdown of Nrf2 expression by Nrf2-targeted miRNA had no observed effect on cell proliferation rates in both high Nrf2-expressing tumors (AML#19) or normal Nrf2-expressing tumors (AML#18) compared with nonsilenced control cells (Figure 5E).
Silencing Nrf2 enhances chemotherapy-induced apoptosis in AML. THP-1 cells were transduced with Nrf2-targeted miRNA lentiviral constructs. (A) The percentage of gated GFP-positive cells was measured with flow cytometry. (B). RNA was extracted from AML cells transduced with Nrf2-targeted and nonsilencing miRNA control constructs and examined for Nrf2 expression by real-time PCR compared with CD34+ control cells (dashed line; C) RNA was extracted from THP-1 cells transduced with Nrf2-targeted and nonsilencing miRNA control constructs and examined for Nrf2 expression by real-time PCR at the indicated times. mRNA expression was normalized to GAPDH mRNA levels. (D) Protein extracts were also obtained and Western blot analysis was conducted for nuclear Nrf2 protein levels. (E) AML cells were transduced with either Nrf2-targeted miRNA or nonsilencing control miRNA construct for 4 days, then treated with 0.5μM cytarabine and 0.2μM daunorubicin for 24 hours. Cell number was assessed by MTS assay. In all panels values indicate the mean ± SEM from 3 independent experiments. *Statistical significance of P < .05 between the different treatment groups. (F) AML cells and control cells were transduced with Nrf2-targeted miRNA and control miRNA constructs and treated with 0.5μM cytarabine and 0.2μM daunorubicin. Cells were washed with PBS and incubated with 10μM of H2DCFDA for 15 minutes. Cells were then assessed for H2DCFDA oxidation using flow cytometry. *Statistical significance of P < .05 by 2-way ANOVA analysis. (G) Protein extracts also were obtained, and Western blot analysis was conducted for Nrf2 and p65 protein levels.
Silencing Nrf2 enhances chemotherapy-induced apoptosis in AML. THP-1 cells were transduced with Nrf2-targeted miRNA lentiviral constructs. (A) The percentage of gated GFP-positive cells was measured with flow cytometry. (B). RNA was extracted from AML cells transduced with Nrf2-targeted and nonsilencing miRNA control constructs and examined for Nrf2 expression by real-time PCR compared with CD34+ control cells (dashed line; C) RNA was extracted from THP-1 cells transduced with Nrf2-targeted and nonsilencing miRNA control constructs and examined for Nrf2 expression by real-time PCR at the indicated times. mRNA expression was normalized to GAPDH mRNA levels. (D) Protein extracts were also obtained and Western blot analysis was conducted for nuclear Nrf2 protein levels. (E) AML cells were transduced with either Nrf2-targeted miRNA or nonsilencing control miRNA construct for 4 days, then treated with 0.5μM cytarabine and 0.2μM daunorubicin for 24 hours. Cell number was assessed by MTS assay. In all panels values indicate the mean ± SEM from 3 independent experiments. *Statistical significance of P < .05 between the different treatment groups. (F) AML cells and control cells were transduced with Nrf2-targeted miRNA and control miRNA constructs and treated with 0.5μM cytarabine and 0.2μM daunorubicin. Cells were washed with PBS and incubated with 10μM of H2DCFDA for 15 minutes. Cells were then assessed for H2DCFDA oxidation using flow cytometry. *Statistical significance of P < .05 by 2-way ANOVA analysis. (G) Protein extracts also were obtained, and Western blot analysis was conducted for Nrf2 and p65 protein levels.
These Nrf2 knockdown experiments were then repeated with the addition of 2 frontline chemotherapeutic agents; cytarabine and daunorubicin. AML cell lines and patient samples were treated with cytarabine and daunorubicin in combination with and without Nrf2-targeted miRNA for 24 hours. In combination these drugs induced cell death in AML cells. However, stable Nrf2 knockdown in these AML cells significantly augmented the observed cell death in vitro compared with any cytotoxicity observed in control miRNA-treated cells (Figure 5E). To determine the role of high Nrf2 levels in regulating ROS in response to cytarabine and daunorubicin, we examined the levels of ROS in AML cells transduced with either control or Nrf2-targeted miRNA (Figure 5F). The results show that ROS are increased to levels observed in normal Nrf2 AML cells in response to cytarabine and daunorubicin when Nrf2 is silenced (Figure 5F). We also observe that Nrf2 but not p65 is induced in cytarabine and daunorubicin treated THP-1 cells (both control and targeted Nrf2-miRNA (Figure 5G). These results demonstrate that high Nrf2-expressing AML cells have a distinct advantage when regulating ROS levels in response to chemotherapy.
Reduced colony formation of AML cells in response to chemotherapy after knockdown of Nrf2
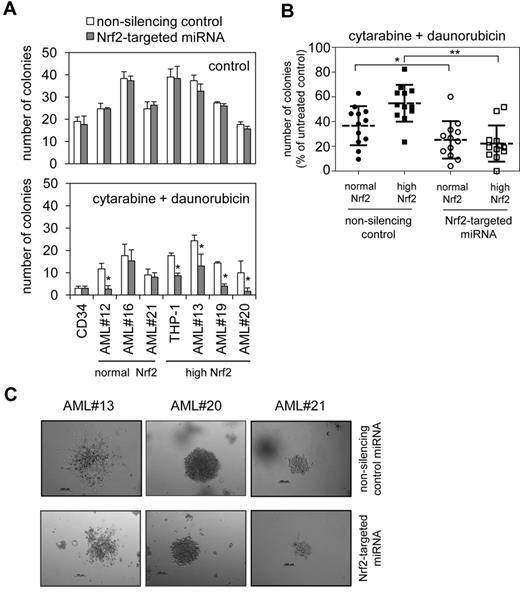
AML cells transduced with Nrf2-targeted miRNA were examined in clonogenic assays to assess the influence of Nrf2 knockdown. In THP-1 cells and 3 high Nrf2-expressing AML samples, and CD34+ cells and 3 normal-expressing samples, Nrf2 knockdown resulted in no significant reduction in the actual number of colonies that were formed (Figure 6A). However, the addition of cytarabine and daunorubicin chemotherapeutics showed that Nrf2 knockdown resulted in a significantly augmented chemotherapy-induced reduction in colony formation in the high-Nrf2 expressing AML cells (Figure 6A-B). Comparison with normal-Nrf2 expressing AML cells (AML#16 and AML#21) showed that Nrf2 knockdown in these cells did not change the effects of the chemotherapeutic drugs. However, we did observe a clear effect on AML#12, which is considered a normal Nrf2-expressing AML cell.
Reduced colony formation of AML cells after knockdown of Nrf2. (A) AML cells and control cells were transduced with Nrf2-targeted miRNA and control miRNA constructs and colony forming assays were performed to show the number of colonies in response to combined treatment with 0.5μM cytarabine and 0.2μM daunorubicin. In all panels values indicate the mean ± SD from 3 independent experiments. *Statistical significance of P < .05 between the different treatment groups when Student t test was used. (B) AML cells treated with 0.5μM cytarabine and 0.2μM daunorubicin from panel A expressed as number of colonies as a percentage of untreated AML colonies. *P < .05 and **P < .01 by 2-way ANOVA analysis. (C) Transmission light microscopic examination of colonies derived from AML cells and control cells infected with Nrf2-targeted or control miRNA construct.
Reduced colony formation of AML cells after knockdown of Nrf2. (A) AML cells and control cells were transduced with Nrf2-targeted miRNA and control miRNA constructs and colony forming assays were performed to show the number of colonies in response to combined treatment with 0.5μM cytarabine and 0.2μM daunorubicin. In all panels values indicate the mean ± SD from 3 independent experiments. *Statistical significance of P < .05 between the different treatment groups when Student t test was used. (B) AML cells treated with 0.5μM cytarabine and 0.2μM daunorubicin from panel A expressed as number of colonies as a percentage of untreated AML colonies. *P < .05 and **P < .01 by 2-way ANOVA analysis. (C) Transmission light microscopic examination of colonies derived from AML cells and control cells infected with Nrf2-targeted or control miRNA construct.
One explanation for this is that AML#12 has slightly elevated levels of Nrf2 binding and RNA, as shown in Table 1; however, these levels were not considered significant from our analysis. This finding suggests that there must be a threshold level of Nrf2 activity within AML cells that is below significance levels from our statistical tests. Representative colonies formed were viewed by transmission light microscopy. In this assay we examined colonies derived from AML cells and control cells infected with Nrf2-targeted or control miRNA construct (Figure 6C and supplemental Figure 8). Finally, in silencing Nrf2, we also were able to reduce AML cell colony formation in response to cytarabine or daunorubicin alone and in response to proteasome inhibitor bortezomib alone (supplemental Figure 9).
Discussion
In this study we have shown that the basic leucine zipper transcription factor Nrf2 is highly expressed in human AML cells. High expression of Nrf2 in human cancer cells is well characterized, with DNA mutations in its inhibitor, Keap1, or in Nrf2 itself, having previously been reported as causal in several classes of tumor. In this study, however, we show that in human AML high Nrf2 is not linked to mutations within Keap1 or in Nrf2 but is in fact the result of an aberrant up-regulation of Nrf2 by NF-κB; a mechanism not previously described in any other tumor type. Furthermore, we found a direct correlation between high basal nuclear levels of NF-κB and Nrf2 transcription factors. Moreover, analysis of primary AML cells with clonogenic assays and cell death assays established that deregulated Nrf2 has a functional significance, insomuch as this transcription factor can protect AML cells from the cytotoxic effects of frontline chemotherapy drugs daunorubicin and cytarabine. Taken together this leads us to hypothesize that a specific knowledge of Nrf2 regulation is required to enable the exploitation of this pathway therapeutically in different tumors.
Nrf2 activity is controlled primarily through posttranscriptional regulation. Less is known about the transcriptional regulation of Nrf2 and members of this family. Our current finding that NF-κB controls the inducible levels of Nrf2 mRNA in some AML samples is confirmed by siRNA knockdown of NF-κB subunits p50 and p65 (Figure 3C). These results show that Nrf2 mRNA expression is not inhibited by lack of p50/p65 in samples expressing normal levels of Nrf2 (AML#16) but is reduced in samples expressing high levels of Nrf2 (AML#19, AML#20 and AML#22). These results highlight the absolute requirement for p50 and p65 in maintaining Nrf2 overexpression in AML.
We now believe that Nrf2 mRNA transcription can be regulated by NF-κB in normal cells, as well as cancer cells, under the stimulation of LPS (supplemental Figure 7); moreover, LPS also can induce a cellular environment, which includes elevated ROS that activates Nrf2 and drives Nrf2-regulated gene expression.28 However, the induction of Nrf2 mRNA in response to LPS is small, which might suggest that other factors are working in tandem with NF-κB.
Recently, dysregulation of Nrf2 has been uncovered in cancer cells,29-31 and somatic mutations found in the Nrf2-Keap1 axis are considered the main reason such dysregulation of Nrf2.29,30 In this study we completed mutational analysis of all Nrf2 and Keap1 exonic coding regions and found no mutations in these genes. However, we cannot rule out mutations in Nrf2-associated regulatory genes, such as MAF or Cul3ligase. It is also interesting to note that polymorphisms have been identified in the Nrf2 promoter that can affect basal and inducible Nrf2 levels.26,31,32 Moreover, several different transcription factors that have been shown to regulate Nrf2 include ARE, AP-1, and Myc sites.24,26 Here we report that NF-κB can regulate Nrf2 expression in AML, leading to enhanced activation of Nrf2-dependent antioxidant defense responses. Similarly, DeNicola et al (2011) have described oncogenic signaling as an alternative mechanism to activate Nrf2 transcription during tumorigenesis in human pancreatic cells.24 Thus, constitutively elevated Nrf2 activity in cancer cells can occur through 2 distinct mechanisms: diminished Nrf2 turnover caused by mutations; and aberrant control of Nrf2 mRNA levels as we see here.
It is of note that 3 AML samples (AML#9, #12, #21) displayed normal Nrf2 levels despite constitutive NF-κB activation. It is unclear why these samples are different; however, we can speculate that the biologic variation and heterogeneity normally observed within AML may well be a factor. One possible explanation for these observations is the mechanism by which NF-κB is constitutively activated varies in different AML subtypes. This can be both independent of and dependent on the AML subtype. For example, mutated C/EBPα, which defines a subgroup of acute myeloid leukemia (approximately 10% of all AML), has the capacity to displace HDACs from NF-κB p50:p50 homodimers in unstimulated cells to activate NF-κB target genes.33,34 Other mechanisms of activity include the phosphorylation of ataxia telangiectasia mutated, a nuclear kinase that is required for NF-κB activation and has been shown to be active in AML,35 and constitutively active PI3-kinase/Akt, which activates downstream NF-κB in AML.36 Neither of these studies show a link to specific AML mutations or subtypes. These explanations suggest that the mechanism for constitutive NF-κB activation in AML is diverse; however, the frequency with which we observe NF-κB activation alludes to a more fundamental role in AML. Further work to delineate which mechanism of NF-κB activation is responsible for induced transcription of Nrf2 is underway, which offers the possibility of paving the way for the development of combination therapies more effective in the treatment of AML.
AML is a genetically and phenotypically heterogeneous disease. However, data support the existence of a subpopulation of rare leukemic stem cells that are responsible for initiation and maintenance of the disease in most, if not all, AML subtypes.33 Here we examined the sensitivity to chemotherapy of AML cells and normal noncancerous control cells transduced with either Nrf2-targeted miRNA or control miRNA. We showed in all cell types a slight trend (in some cases significant) for reduced colony formation in Nrf2 knockdown cells when exposed to chemotherapeutic drugs. Nrf2 activity induces the expression of a plethora of genes that could influence sensitivity toward chemotherapeutics, including HO-1, NQO1 glutathione rate-limiting enzymes (GCLM and GCLC), and multidrug resistant proteins.37 Nrf2 recently has been shown to regulate the expression of miRNA, which also may have a bearing on chemotherapy resistance in human AML.38 The concept that Nrf2, or indeed the absence of Nrf2, is able to interfere with cell proliferation is not unusual; for example, Reddy et al (2008) have shown that Nrf2-deficient epithelial cells have impaired cell-cycle progression, mainly at a G2/M-phase arrest.39 They also demonstrated that glutathione synthesis was impaired in Nrf2-deficient cells, with GSH supplementation restoring normal cell-cycle progression. Similarly, we have shown recently that the glutathione rate-limiting enzymes GCLM and GCLC are regulated by Nrf2 in human AML cells.2 Taken together, these results suggest that Nrf2 influences AML cell sensitivity to chemotherapeutic drugs.
In conclusion, our results demonstrate that high levels of nuclear Nrf2 expression are regulated by high levels of nuclear NF-κB in AML cells. These findings also support the hypothesis that aberrant expression of Nrf2 contributes to the malignant and drug-resistant nature of AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Professor Richard Ball, Norfolk and Norwich University Hospitals Human Tissue Bank for sample storage, Dr Penny Powell (UEA) and Dr Ariberto Fassati (UCL) for lentiviral reagents, and Mr John Wood (UCL) for advice on statistical analyses.
This work was supported by Leukemia Lymphoma Research, the AICR, The Big C, and the Flexibility and Sustainability Funding from the National Institutes of Health Research.
Authorship
Contribution: S.A.R., L.Z., M.Y.M., and D.J.M. designed the research and analyzed data; S.A.R., K.M.B., and D.J.M. wrote the paper; K.M.B. contributed vital new reagents; and S.A.R., L.Z., M.Y.M., and N.M.S. performed the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Professor David J. MacEwan, School of Pharmacy, University of East Anglia, Norwich, NR4 7TJ, United Kingdom; e-mail: d.macewan@uea.ac.uk.

![Figure 4. Understanding the regulation of Nrf2 by NF-κB. (A) Schematic presentation of the Nrf2 promoter constructs that were created for this study, being either wild-type [a], κB2 site–deleted [b], or κB1 site–deleted [c] constructs. (B) THP-1 cells were transiently transfected with 0.5 μg of each promoter construct shown in panel A and pRL-CMV for normalization of transfection efficiency. Cell extracts were harvested, and luciferase assays were performed. Values are the means ± SD, n = 4. *P < .01 of deleted κB against untreated Nrf2 wild-type control. TNF (10 mg/mL) treatment acted as a positive control to activate NF-κB and BAY 11-7082 (10μM) to inhibit NF-κB. #P < .01 of TNF treated or BAY 11-7082 treated against untreated controls. (C) ChIP analysis of the Nrf2 promoter using antibodies against p50 and p65. Normal rabbit IgG was used as a control. THP-1 cells were left untreated or treated with BAY 11-7082 for 8 hours. Real-time PCR was performed in triplicate on immunoprecipitated DNA and input DNA. Data presented as percent of input. *P < .05 between the different treatment groups. (D) ChIP-seq data from the ENCODE Consortium demonstrates NF-κB subunit p65 binding to the Nrf2 promoter. NF-κB binding occurs in exon1 of Nrf2 and overlaps with sites of RNA pol II binding. Black bars denote positive signal above background.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/26/10.1182_blood-2012-04-422121/4/m_zh89991200180004.jpeg?Expires=1767737516&Signature=1h3Sm14kRPfLXFabgFFGGoKMuRSoeJ9dOT0Xfsn66mn1AWcx0f-T9rpEK9NsOVqPkXnbtcKIRFng7LF1IPMV5PgxScv6Mx~XlyP-GCxFYeOMvZxLOATzu72IoGrK4vtYZ8nAOR4mVjSU7QDB6UOe3HXZT9c7a91J5AqxrErSFRXJzKZzP3Ai2vGQ8wzorYMNEr0U~6XvQkAX4AUdP-28XRviDuUMJB7ioWqrfh3KXUSZO3X68sIPPcwinXbSK5pB4EKwaCTB5v4dbnUiYkzPInwuhdRlbD~lsv8BGvoXmqpntXvix3YFEnknPqtSsTqmQCntJCYNy8Jp4l5inyqc9g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal