Abstract
Activated protein C (APC) exerts endothelial cytoprotective actions that require protease-activated receptor 1 (PAR1), whereas thrombin acting via PAR1 causes endothelial disruptive, proinflammatory actions. APC's activities, but not thrombin's, require PAR1 located in caveolae. PAR1 is a biased 7-transmembrane receptor because G proteins mediate thrombin's signaling, whereas β-arrestin 2 mediates APC's signaling. Here we elucidate novel mechanisms for APC's initiation of signaling. Biochemical studies of APC's protease specificity showed that APC cleaved PAR1 sequences at both Arg41 and Arg46. That PAR1 cleavage at Arg46 can occur on cells was supported by APC's cleavage of N-terminal-SEAP-tagged R41Q-PAR1 but not R41Q/R46Q-PAR1 mutants transfected into cells and by anti-PAR1 epitope mapping of APC-treated endothelial cells. A synthetic peptide composing PAR1 residues 47-66, TR47, stimulated protective signaling in endothelial cells as reflected in Akt and glycogen synthase kinase 3β phosphorylation, Ras-related C3 botulinum toxin substrate 1 activation, and barrier stabilization effects. In mice, the TR47 peptide reduced VEGF-induced vascular leakage. These in vitro and in vivo data imply that the novel PAR1 N-terminus beginning at residue Asn47, which is generated by APC cleavage at Arg46, mediates APC's cytoprotective signaling and that this unique APC-generated N-terminal peptide tail is a novel biased agonist for PAR1.
Introduction
Activated protein C (APC) is a homeostatic serine protease that provides beneficial effects via antithrombotic activities and also via cytoprotective actions that are based on its cell-signaling properties.1 Wild-type APC and cytoprotective-selective APC mutants reduce damage in vitro to cultured cells under stress and reduce mortality and organ injury in multiple in vivo murine injury models. Moreover, many reports, but not all studies, of APC's protective actions show that protease-activated receptor 1 (PAR1) and the endothelial protein C receptor (EPCR) are required for APC's protective actions.1-5 PAR1, a G protein coupled receptor (GPCR), which is also known as a 7-transmembrane receptor, is a primary thrombin receptor on human platelets, and thrombin-triggered signaling arises because of cleavage at the PAR1 canonical Arg41 site to generate the agonist N-terminal tethered ligand that begins with residue Ser42, which causes PAR1-initiated signaling.6-8 PAR1 as well as the other 3 known human and murine PARs are activated on many cell types by many proteases with a panoply of biologic activities.8,9
PAR1 mediates thrombin's proinflammatory and endothelial barrier disruptive actions.8,10 The conundrum then arose of how PAR1 signaling resulting from cleavage at Arg41 could mediate thrombin's proinflammatory and endothelial barrier disruptive actions as well as the opposite anti-inflammatory and barrier stabilizing effects of APC.10-12 Some insights came from demonstrations that membrane localization of PAR1 with EPCR in caveolae, or caveolin-1-rich microdomains, could influence PAR1-selective signaling and that APC-PAR1-dependent transactivation of other receptors, S1P1, PAR2, and Tie2, can arise.11,13-16 The selective nature of thrombin and APC for PAR1-dependent effects on endothelial barrier function is striking, as the former promotes Ras homolog gene family member A (RhoA) activation whereas the latter promotes Ras-related C3 botulinum toxin substrate 1 (Rac1) activation—RhoA promotes endothelial barrier leakage and Rac1 promotes barrier stabilization.10,11,17 Thrombin and APC are biased activators of PAR1 because thrombin requires G protein-dependent signaling for RhoA activation, whereas APC requires β-arrestin 2-dependent signaling for Rac1 activation.18 PAR1 can be allosterically influenced by retention of thrombin on activated PAR1 because of thrombin's binding to the hirudin-like sequence of residues 51-56 and by potential association of PAR1 with EPCR-protein C/APC, which appears to alter the functional selectivity of thrombin.19-23 PAR1 can be cleaved by MMP1 at Asp39 to generate an N-terminus differing from that due to cleavage at Arg41, but this alternative cleavage is not associated with remarkable differences in PAR1 activities.24 Nonetheless, these multiple insights, which show PAR1 to be a biased GPCR with multiple allosteric sites, fail to define a clear mechanism for biased activation of PAR1 by 2 different proteases, namely, thrombin and APC, with their remarkable differences in functional selectivity.
To explain the biased activation of PAR1 by APC, we hypothesized that PAR1-dependent signaling by APC involves a novel cleavage of the receptor's N-terminal domain, differing from that of thrombin, which then reveals a novel cryptic intramolecular pharmacophore that causes APC's cytoprotective, biased signaling. To test this hypothesis, we used mass spectroscopy, cells transfected with PAR1 constructs, synthetic peptides, cell-signaling assays, cytoprotective assays, and a murine vascular leakage injury model. The extensive in vitro and in vivo data in this report provide strong support for this hypothesis that provides a resolution for the thrombin-APC-PAR1-signaling conundrum.
Methods
Materials
SCH79797 (Tocris), thrombin receptor activating peptide that is a synthetic peptide (TFLLRNPNDK) resembling residues 42-51 of human PAR1 (TRAP; Anaspec), thrombin (ERL), and recombinant wild-type APC (Xigris, Eli Lilly) were from commercial sources. Mouse anti-ERK1/2 (3A7), rabbit anti-pThr202/Tyr204-ERK1/2 (197G2), mouse anti-Akt (40D4), rabbit anti-pSer473Akt (D9E), mouse anti–glycogen synthase kinase 3β (GSK3β; 3D10), and rabbit anti–pSer9-GSK3β (D3A4) were from Cell Signaling. Anti-PAR1 mouse monoclonal antibodies SPAN12b, ATAP2, and WEDE-15b (note the “b” denotes that this is a subcloned version of the original published antibody)25 were kindly provided by Dr L. Brass (University of Pennsylvania, Philadelphia, PA). Peptides (see supplemental Figure 2 for sequences; available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were synthesized and purified to > 95% purity (Biosynthesis).
Constructs
SEAP-PAR1 and wt-EPCR constructs were made as reported with a notable modification.26 Three Arg residues at positions 25 (−1 of mature PAR1), 27 (2), and 28 (3) were mutated to GQQ to prevent significant APC and thrombin-mediated cleavages at these sites in the N-terminal SEAP fusion constructs. The linker sequence now contains (… PGYSG-1A1QQP …) with the SEAP C-terminus underlined, the mature PAR1 N-terminus in italics and the GQQ mutations in bold. GQQ-SEAP-PAR1 is herein referred to as wt-SEAP-PAR1 and was used to introduce the R41Q, R46Q, and R41Q/R46Q mutations using Quickchange mutagenesis (Stratagene). SEAP-PAR1s, either in the presence or absence of wt-EPCR, were transfected into HEK-293 cells to obtain stable cells expressing the various SEAP-PAR1 constructs.
Cells
HEK-293 (ATCC) and EA.hy.926 (Dr C. J. S. Edgell, University of North Carolina, Chapel Hill, NC) were grown and maintained as described.26
PAR1 cleavage assays
Cleavage of the PAR1 synthetic peptide representing residues 33-62 of the PAR1 N-terminal tail by APC and thrombin was performed as described.26 All reactions contained 50μM TR33-62 peptide (supplemental Figure 2), 500nM APC or 10nM thrombin in HEPES-buffered saline (25mM HEPES, pH 7.4, 147mM NaCl, and 4mM KCl) with 4mM CaCl2 and 0.6mM MgCl2 unless stated otherwise. Molecular masses of isolated peptide fragments were determined by the Scripps Research Institute Mass Spectrometry and Metabolomics facility. Analysis of PAR1 cleavage by APC or thrombin using the SEAP-PAR1 cleavage reporters was done in 96-well plates as described.26 After correction for background activity in the absence of protease, values were expressed as percentage of the total SEAP activity present on the cells.
Mapping of the canonical and noncanonical cleavage site sensitive epitopes of anti-PAR1 antibodies
To determine the specific binding of antibodies WEDE15, ATAP2, and SPAN12 to PAR-1 peptides (TR24-41, TR24-46, TR42-66, TR47-66, and TR33-62), peptides were coated at 10μM on Maxisorp 96-well plates, followed by 2-hour incubation with TBS/3% (weight/volume) BSA to block the plates. Antibodies were diluted to 2 μg/mL in TBS/3% (weight/volume) BSA, and incubated for 1 hour at room temperature. Anti–mouse HRP-conjugated secondary antibody (Dako North America) was used to detect bound primary antibodies at A490nm.
On Cell Western (OCW) assays for anti-PAR1 antibodies
EA.hy.926 cells were grown in black clear-bottom 96-well plates (Corning) until confluent. Cells were washed and incubated at 37°C with thrombin (0.25nM) or APC (100nM) in HMM2 buffer (HBSS; Invitrogen) supplemented with 1.3mM CaCl2, 0.6mM MgCl2, and 0.1% endotoxin free BSA (Calbiochem) for 3 hours. After a quick wash with ice-cold HMM2, cells were fixed in methanol free 4% paraformaldehyde (Pierce Chemical) for 20 minutes at room temperature after which plates were blocked in Odyssey blocking buffer supplemented with 3% BSA overnight. Subsequently, plates were incubated with mouse anti-PAR1 antibodies SPAN12b, ATAP2, or WEDE-15b (all 10 μg/mL) in Odyssey blocking buffer, followed by 1/1000 biotinylated goat antimouse and 1/4000 IRDye 800CW Streptavidin (LI-COR) with 1/10 000 Draq5 (Biostatus). PAR1 on the cell surface was detected using the In Cell Western module of Odyssey Imager with Image Studio Software Version 2.0. Fluorescence signals were corrected for cell number and background staining, and signals were normalized to buffer only controls at the corresponding time point.
Western blotting
EA.hy.926 cells were grown in 6-cm dishes (3 × 106 cells per plate) for 48 hours and serum starved overnight. Peptides (50μM), APC (70nM), or thrombin (54nM) in serum-free media were added to the cells for different time points as indicated. In some experiments, cells were pretreated with SCH79797 (1μM) or vehicle control (DMSO) in serum-free media for 20 minutes. Cells were washed with cold Dulbecco PBS (Invitrogen), and cell lysates were made with 200 μL of 1.5% NP-40 lysis buffer containing 1 times protease and phosphatase inhibitor cocktail (Pierce Chemical). After clarification (30 minutes, 14 000 rpm), lysates (100 μg) were mixed with reducing SDS sample buffer (LI-COR) and separated on 12% SDS-PAGE (Bio-Rad). Proteins were transferred to Immobilon-FL PVDF membrane (Millipore), blocked with Odyssey Blocking Buffer (LI-COR), and incubated with a mouse-rabbit combination of pan and phospho-specific antibodies against either Akt, GSK3β and ERK1/2. Blots were developed with IRDye 680 and IRDye 800CW donkey antimouse or donkey antirabbit secondary antibodies (LI-COR) and scanned on the Odyssey Imager (LI-COR). Quantification of integrated fluorescence intensity (K counts) was done using Odyssey Application Version 3.0 software (LI-COR).
Rac1 activation
The pGEXTK-PAK1 70-117 construct encoding a GST fusion to p21-activated kinase (PAK-1)–binding domain was kindly made available by Dr J. Chernoff (Fox Chase Cancer Center, Philadelphia, PA; Addgene plasmid 12217). GST-PAK1was purified from transformed BL21 (DE3) Escherichia coli using B-Per lysis buffer (Pierce Chemical) with lysozyme, DNAse I, and Halt EDTA-free protease inhibitor cocktail (Pierce Chemical) on glutathione-agarose according to the manufacturer's recommendation. Pull-down of active Rac1 was performed as described.27 Briefly, endothelial EA.hy.926 cells (5 × 106 per plate) were grown in 100-mm dishes for 48 hours and serum starved overnight before addition of peptides (50μM) for 30 or 180 minutes. Lysates (2 mg) were mixed with GST-PAK1 glutathione-agarose (150 μg) and after washing active GTP-Rac1 was eluted from GST-PAK1 glutathione-agarose by boiling in reducing SDS sample buffer (LI-COR). Active GTP-Rac1 was resolved on 12% SDS-PAGE, transferred to Immobilon-FL PVDF membrane, and immunoblotted with a mouse anti-Rac1 antibody (BD Biosciences) and IRDye 800CW donkey anti–mouse secondary antibodies (LI-COR). Immunoblots were scanned on the Odyssey Imager (LI-COR). Quantification of integrated fluorescence intensity (K counts) was done using Odyssey Application Version 3.0 software (LI-COR).
Endothelial barrier protection
Permeability of endothelial cell barrier function was determined as described with minor modifications. Briefly, EA.hy.926 endothelial cells (5 × 104 cells/well) were grown on polycarbonate membrane Transwell inserts (Costar, 3-μm pore size, 12-mm diameter). When confluent, cells were incubated with APC (20nM), TR47 (50μM), or a synthetic peptide containing a scrambled, randomized sequence composed of amino acid residues 47-66 of human PAR1 (scrTR47; 50μM) for 4 hours in serum-free media with 0.1% BSA (fatty acid poor and endotoxin free fraction V, Calbiochem). After incubation with thrombin (10nM) for 10 minutes, the media in the inner chamber was replaced with complete medium containing 4% BSA and 0.67 mg/mL Evans blue. Endothelial cell permeability was determined by absorbance of Evans blue in the outer chamber at 650 nm. Permeability was expressed as the fold change in absorbance compared with that in the absence of thrombin (normalized to 1).
In vivo vascular permeability assay
The study was approved by the Institutional Animal Care and Use Committee of the Scripps Research Institute and complied with National Institutes of Health guidelines. SKH1-E hairless male (6-8 weeks old) were from Charles River Laboratories. Vascular permeability was determined using a VEGF-induced leakage model with some modifications.28,29 Briefly, 100 μL of a sterile-filtered solution containing 0.5% (weight/volume) Evans blue (Sigma-Aldrich) in 0.9% NaCl (Sigma-Aldrich) was injected in isoflurane-anesthetized mice. After 30 minutes, 50 μL of peptides (125 μg; TR47 or scrTR47) or PBS was injected intravenously in the retro-orbital sinus of mice anesthetized with ketamine-xylazine (100 and 10 mg/kg, respectively). After 5 minutes, mice received subcutaneously 15 μL of 75 ng/injection recombinant mouse VEGF165 (BioVision) in 0.1% BSA-PBS (3 sites on the right side of the abdomen) or vehicle (2 sites on the left side). After 30 minutes, mice were killed, photographed, and Evans blue extravasation in the skin was determined using the Odyssey Imager (LI-COR) in the 700-nm channel with 4-mm offset. Quantification of the intensity of the Evans blue dye signal was done using the Odyssey Application Software Version 3.0. A mean value for 3 data points (VEGF) or 2 data points (BSA; injection sites) was made for each mouse and normalized to the VEGF sites in PBS-injected mice. In total, 5 independent experiments were performed using a total of 23 mice (n = 11 for PBS, n = 6 for TR47, and n = 6 for scrTR47).
Statistical analysis
Statistical significance (P < .05) was determined using Student t test or 1-way ANOVA and Bonferroni multiple comparison posttests as appropriate (Prism Version 5.0 software, GraphPad Software).
Results
Identification of an APC-specific noncanonical cleavage in PAR1
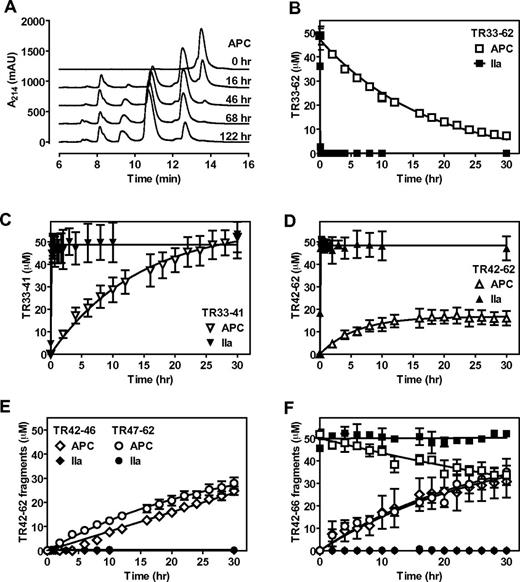
Previously, we have shown that APC cleaved a synthetic PAR1 N-terminal tail peptide (TR33-62), although proteolysis by APC was slow and required prolonged incubation with the peptide.26 It was noted at the time that additional proteolytic fragments were generated by APC that became apparent after 20 hours and that the appearance of these fragments were less obvious in the presence of EDTA. On additional characterization of TR33-62 cleavage by APC in the presence of CaCl2 and MgCl2, it became evident that initially APC generated 2 fragments (retention time 8.2 and 12.6 minutes compared with 13.8 minutes for the parental TR33-62 peptide; Table 1) that were identical to the 2 fragments generated by thrombin (Figure 1A). In time, APC, but not thrombin, generated 2 additional fragments (retention time 9.7 and 11.0 minutes; Table 1) that at least were partially derived from the initial 12.6 minutes cleavage fragment. The fact that no additional fragments were observed after more than 5 days of TR33-62 incubation with APC suggest that these second APC-derived fragments were derived from a specific cleavage rather than from nonspecific proteolysis. To identify the second APC cleavage site, peptide fragments were isolated and their molecular mass was determined by mass spectrometry (supplemental Figure 1). As anticipated, molecular mass of the initial APC and thrombin generated fragments (peak A, 1001 Da; and peak D, 2670 Da) corresponded to peptide fragments TR33-41 and TR42-62, respectively, consistent with cleavage at Arg41 (Table 1). The APC-specific fragments (peak B, 635 Da; and peak C, 2053 Da) were consistent with cleavage at Arg46 and the generation of TR42-46 and TR47-62 peptide fragments. Thrombin cleaved TR33-62 (Figure 1B) at Arg41 completely within 60 minutes with the concomitant generation of equimolar concentrations of TR33-41(Figure 1C) and TR42-62 (Figure 1D). In contrast, complete TR33-62 cleavage by APC required > 30 hours; and although TR33-41 was generated at the expected equimolar concentration (Figure 1C), TR42-62 never reached more than ∼ 40% of its potential maximum molar concentration (Figure 1D). Instead, similar rates were observed for generation of the TR42-62 fragment and generation of the TR42-46 and TR47-62 fragments derived from cleavage at Arg46 (Figure 1E). The formation of TR42-62 seemed not to be the rate-limiting step for the generation of TR42-46 and TR47-62 fragments as APC produced these fragments at approximately similar rates from a synthetic TR42-66 peptide (Figure 1F). Thrombin did not cleave the TR42-66 fragment. Thus, in a purified system, APC cleaved a synthetic PAR1 N-terminal tail peptide, TR33-62, at both Arg41 and Arg46, whereas thrombin cleaved TR33-62 only at Arg41 (Table 1).
PAR1 peptide fragments generated by APC or thrombin proteolysis
| Fragment . | Retention time, min . | Cleaved by . | Exp mass, Da . | Calculated mass, Da . | PAR1 peptide (33-62) . | Cleavage site . |
|---|---|---|---|---|---|---|
| A | 8.2 | IIa/APC | 1001 | 1000 | ATNATLDPR-41 | R41 |
| B | 9.7 | APC | 635 | 635 | 42-SFLLR-46 | R46 |
| C | 11.0 | APC | 2053 | 2053 | 47-NPNDKYEPFWEDEEKN | R46 |
| D | 12.6 | IIa/APC | 2670 | 2670 | 42-SFLLRNPNDKYEPFWEDEEKN | R41 |
| E | 13.8 | — | 3651 | 3652 | ATNATLDPRSFLLRNPNDKYEPFWEDEEKN | — |
| Fragment . | Retention time, min . | Cleaved by . | Exp mass, Da . | Calculated mass, Da . | PAR1 peptide (33-62) . | Cleavage site . |
|---|---|---|---|---|---|---|
| A | 8.2 | IIa/APC | 1001 | 1000 | ATNATLDPR-41 | R41 |
| B | 9.7 | APC | 635 | 635 | 42-SFLLR-46 | R46 |
| C | 11.0 | APC | 2053 | 2053 | 47-NPNDKYEPFWEDEEKN | R46 |
| D | 12.6 | IIa/APC | 2670 | 2670 | 42-SFLLRNPNDKYEPFWEDEEKN | R41 |
| E | 13.8 | — | 3651 | 3652 | ATNATLDPRSFLLRNPNDKYEPFWEDEEKN | — |
— indicates not applicable.
APC cleaves a synthetic PAR1 N-terminal peptide at Arg41 and Arg46. Cleavage of a PAR1 N-terminal tail peptide (TR33-62) by APC (500nM) or thrombin (10nM) in the presence of CaCl2 (4mM) and MgCl2 (0.6mM) and resolution of cleavage fragments by HPLC using C18 reverse phase chromatography. (A) Chromatograms of APC-mediated TR33-62 cleavage over time. (B) Time course of TR33-62 cleavage by APC (□) or thrombin (■). (C) Generation of the N-terminal cleavage fragment derived from cleavage at Arg41 (TR33-41) by APC (▿) or thrombin (▾). (D) Generation of the C-terminal cleavage fragment derived from cleavage at Arg41 (TR42-62) by APC (Δ) or thrombin (▴). (E) Generation of the N-terminal TR42-46 (♢, ♦) and C-terminal TR47-62 fragments (○, ●) derived from cleavage at Arg46 by APC (open symbols) or by thrombin (closed symbols). (F) Time courses for cleavage of peptide TR42-66 (□, ■) and generation of TR42-46 (♢, ♦) and TR47-66 (○, ●) cleavage fragments by APC (open symbols) or thrombin (closed symbols). Representative chromatograms (A) and (B-F) data points represent the mean ± SD (N ≥ 3).
APC cleaves a synthetic PAR1 N-terminal peptide at Arg41 and Arg46. Cleavage of a PAR1 N-terminal tail peptide (TR33-62) by APC (500nM) or thrombin (10nM) in the presence of CaCl2 (4mM) and MgCl2 (0.6mM) and resolution of cleavage fragments by HPLC using C18 reverse phase chromatography. (A) Chromatograms of APC-mediated TR33-62 cleavage over time. (B) Time course of TR33-62 cleavage by APC (□) or thrombin (■). (C) Generation of the N-terminal cleavage fragment derived from cleavage at Arg41 (TR33-41) by APC (▿) or thrombin (▾). (D) Generation of the C-terminal cleavage fragment derived from cleavage at Arg41 (TR42-62) by APC (Δ) or thrombin (▴). (E) Generation of the N-terminal TR42-46 (♢, ♦) and C-terminal TR47-62 fragments (○, ●) derived from cleavage at Arg46 by APC (open symbols) or by thrombin (closed symbols). (F) Time courses for cleavage of peptide TR42-66 (□, ■) and generation of TR42-46 (♢, ♦) and TR47-66 (○, ●) cleavage fragments by APC (open symbols) or thrombin (closed symbols). Representative chromatograms (A) and (B-F) data points represent the mean ± SD (N ≥ 3).
Cleavage of PAR1 at Arg46 by APC on cells
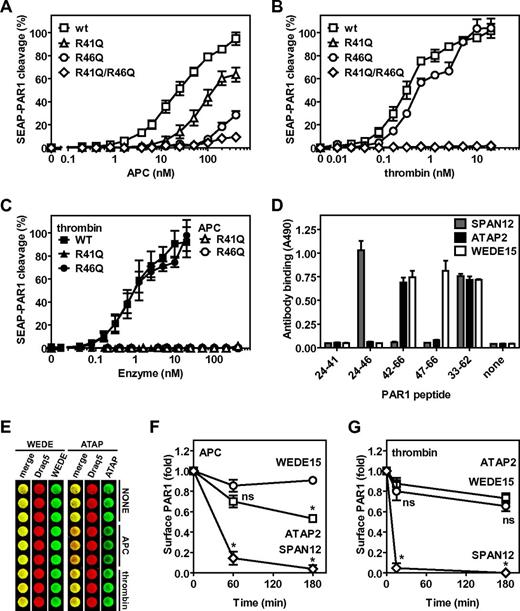
To determine whether APC cleaved PAR1 at Arg46 on cells, a SEAP-PAR1 fusion construct was used that readily detects PAR1 cleavage when transfected into HEK-293 cells by the release of alkaline phosphatase activity (SEAP) in the medium. Previously, we have used this system to demonstrate the important cofactor function of the EPCR for the cleavage of PAR1 by APC and the characterization of PAR1 cleavage by cytoprotective-selective and anticoagulant selective APC mutants.30 However, we discovered that such PAR1 reporter constructs are subject to potential artifacts when PAR1 cleavage site mutations at Arg41, Arg46, or both residues were made because thrombin and APC could still readily release SEAP into the media despite these cleavage site mutations, especially at higher concentrations of enzyme. We showed that Arg residues in the SEAP-PAR1 linker or either Arg27 or Arg28 (residues 2 and 3 of mature PAR1) were the sites for undesired cleavages because mutation of these 3 Arg residues to Gly/Gln/Gln abolished cleavage of R41Q-SEAP-PAR1 by thrombin. Thus, to avoid misleading or erroneous conclusions from SEAP-PAR1 cleavage studies that use constructs that have these 3 Arg residues intact, constructs lacking these 3 Arg residues are needed. Consistent with previous reports, APC cleaved GQQ-wt-SEAP-PAR1 (hereafter referred to as wild-type [wt]) in the presence of wt-EPCR at concentrations ranging between 1nM and 100nM that are typically required to convey PAR1-dependent cytoprotective effects on cells in vitro (Figure 2A). Half-maximal cleavage of wt-SEAP-PAR1 in the presence of wt-EPCR required 23nM APC compared with 0.31nM thrombin (Figures 2A-B). Mutation of the canonical thrombin cleavage site Arg41 to Gln resulted in a 5-fold shift of the APC dose-response curve (R41Q EC50 133nM), whereas mutation of noncanonical APC cleavage site in PAR1 at Arg46 almost completely ablated APC-mediated PAR1 cleavage (extrapolated R46Q EC50 961nM; Figure 2A). Thus, APC could cleave PAR1 on cells in the presence of EPCR at both Arg41 and Arg46. In contrast to APC, thrombin cleaved R46Q-SEAP-PAR1 like wt (R46Q EC50 0.66nM), whereas no cleavage by thrombin could be observed for R41Q-SEAP-PAR1, confirming that thrombin cleaved PAR1 at Arg41 and not at Arg46 (Figure 2B). No APC-mediated cleavage of PAR1 was observed when both Arg41 and Arg46 were mutated, indicating that Arg41 and Arg46 compose the only PAR1 cleavage sites for APC in the presence of EPCR (Figure 2A). EPCR was identified as an essential cofactor to APC for the cleavage and activation of PAR1 as no cleavage of R41Q-SEAP-PAR1 or R46Q-SEAP-PAR1 was observed in the absence of EPCR, indicating that cleavage at Arg46, like Arg41, by APC required EPCR (Figure 2C). Consistent with the results in the presence of EPCR, when EPCR was absent, thrombin cleaved wt-SEAP-PAR1 and R46Q-SEAP-PAR1 similarly, but no cleavage of R41Q-SEAP-PAR1 by thrombin could be observed (Figure 2C).
Noncanonical cleavage of PAR1 at Arg46 by APC on cells. Cleavage of wt-SEAP-PAR1 and SEAP-PAR1 cleavage site mutants by (A) APC and (B) thrombin in the presence of wt-EPCR and in the absence of EPCR (C). (□, ■) represent wt-SEAP-PAR1; (Δ, ▴), R41Q-SEAP-PAR1; (○, ●), R46Q-SEAP-PAR1; and (♢, ♦), R41Q/R46Q-SEAP-PAR1. (C) Closed symbols indicate thrombin; and open symbols, APC. (D) Mapping of the canonical and noncanonical cleavage site sensitive epitopes of anti-PAR1 antibodies SPAN12, ATAP2, and WEDE15 using synthetic peptides coated onto microtiter plate wells. Peptides represent the N-terminal (TR24-41) or C-terminal fragment (TR42-66) of PAR1 after cleavage at Arg41 or the N-terminal (TR24-47) or C-terminal fragment (TR47-66) of PAR1 after cleavage at Arg46 or the entire cleavage site region of the PAR1 N-terminal tail (TR33-62). (E) OCW of anti-PAR1 antibodies WEDE15 and ATAP2 on EA.hy.926 endothelial cells after incubation with control buffer (NONE), APC (100nM), or thrombin (0.25nM) for 3 hours. Cell-bound anti-PAR1 antibodies were detected with biotinylated goat anti–mouse secondary antibodies and IRDye 800CW streptavidin in the 800-nm channel of the Odyssey Imager (green). Draq5 was used for cell number normalization and detected in the 700-nm channel (red). Overlay of the Draq5 700 channel and anti-PAR1 800-nm channel is indicated in yellow (merge). The presence and loss of PAR1 epitopes on endothelial cell surfaces were determined using OCW quantification of cleavage site sensitive and insensitive PAR1 antibodies on EA.hy926 endothelial cells versus time for incubation of cells with (F) APC (100nM) or (G) thrombin (0.25nM). The cell surface was probed for PAR-1 with (○) WEDE15 (detecting all PAR1 regardless of cleavage at either Arg41 or Arg46), (□) ATAP2 (detecting uncleaved and PAR1 cleaved only at Arg41), or (♢) SPAN12 (detecting only uncleaved PAR1). (D-E) Representative experiment in triplicates. (A-C,F-G) Data points represent the mean ± SEM (N ≥ 3). (F-G) *P < .001, compared with WEDE15. ns indicates not significant.
Noncanonical cleavage of PAR1 at Arg46 by APC on cells. Cleavage of wt-SEAP-PAR1 and SEAP-PAR1 cleavage site mutants by (A) APC and (B) thrombin in the presence of wt-EPCR and in the absence of EPCR (C). (□, ■) represent wt-SEAP-PAR1; (Δ, ▴), R41Q-SEAP-PAR1; (○, ●), R46Q-SEAP-PAR1; and (♢, ♦), R41Q/R46Q-SEAP-PAR1. (C) Closed symbols indicate thrombin; and open symbols, APC. (D) Mapping of the canonical and noncanonical cleavage site sensitive epitopes of anti-PAR1 antibodies SPAN12, ATAP2, and WEDE15 using synthetic peptides coated onto microtiter plate wells. Peptides represent the N-terminal (TR24-41) or C-terminal fragment (TR42-66) of PAR1 after cleavage at Arg41 or the N-terminal (TR24-47) or C-terminal fragment (TR47-66) of PAR1 after cleavage at Arg46 or the entire cleavage site region of the PAR1 N-terminal tail (TR33-62). (E) OCW of anti-PAR1 antibodies WEDE15 and ATAP2 on EA.hy.926 endothelial cells after incubation with control buffer (NONE), APC (100nM), or thrombin (0.25nM) for 3 hours. Cell-bound anti-PAR1 antibodies were detected with biotinylated goat anti–mouse secondary antibodies and IRDye 800CW streptavidin in the 800-nm channel of the Odyssey Imager (green). Draq5 was used for cell number normalization and detected in the 700-nm channel (red). Overlay of the Draq5 700 channel and anti-PAR1 800-nm channel is indicated in yellow (merge). The presence and loss of PAR1 epitopes on endothelial cell surfaces were determined using OCW quantification of cleavage site sensitive and insensitive PAR1 antibodies on EA.hy926 endothelial cells versus time for incubation of cells with (F) APC (100nM) or (G) thrombin (0.25nM). The cell surface was probed for PAR-1 with (○) WEDE15 (detecting all PAR1 regardless of cleavage at either Arg41 or Arg46), (□) ATAP2 (detecting uncleaved and PAR1 cleaved only at Arg41), or (♢) SPAN12 (detecting only uncleaved PAR1). (D-E) Representative experiment in triplicates. (A-C,F-G) Data points represent the mean ± SEM (N ≥ 3). (F-G) *P < .001, compared with WEDE15. ns indicates not significant.
To determine the PAR1 proteolysis profile by APC on endothelial cells expressing endogenous PAR1 and EPCR, an OCW assay was set up using different anti-PAR1 antibodies whose epitopes are differentially affected by cleavage at either Arg41 or Arg46. SPAN12 is a cleavage-sensitive antibody (raised against residues 35-NATLDPRSFLLR-46)25 that bound to intact PAR1 (TR33-62) but not to PAR1 cleaved at either Arg41 (TR42-66) or Arg46 (TR47-66); this antibody recognized the N-terminal part of PAR1 after cleavage at Arg46 (TR24-46) but not at Arg41 (TR24-41; Figure 2D). The epitope of ATAP2 was mapped to residues 43-SFLLR-46 located just downstream of the cleavage site at residue Arg41.31 ATAP2 recognized intact PAR1 (TR33-62) and PAR1 cleaved at Arg41 (TR42-66) but not PAR1 cleaved at Arg46 (TR47-66) or the N-terminal part of PAR1 after cleavage at either Arg41 (TR24-41) or Arg46 (TR24-46; Figure 2D). WEDE15 (raised against residues 51-KYEPFWEDEEKNES-64)25 bound intact PAR1 (TR33-62) and cleaved PAR1 equally well regardless of whether cleavage occurred at Arg41 (TR42-66) or Arg46 (TR47-66; Figure 2D). Using a novel OCW assay, which proved to be a greatly improved cellular ELISA, allowed for careful normalization of the antibody-binding signal to the total cell number (Figure 2E). Incubation of EA.hy.926 endothelial cells with APC resulted in a rapid decease of SPAN12 reactivity (∼ 15% after 1 hour and < 5% after 3 hours compared with 100% at t = 0), whereas WEDE15 reactivity remained relatively constant (85%-90%), indicating cleavage of PAR1 by APC (Figure 2F). Interestingly, a significant fraction of PAR1 molecules on cells was cleaved by APC at Arg46, as is evident from the reduction of ATAP2 reactivity (70% at 60 minutes and ∼ 50% at 180 minutes). In contrast, incubation of EA.hy.926 endothelial cells with thrombin resulted in a rapid decease of SPAN12 reactivity to < 5% after 15 minutes, whereas ATAP2 and WEDE15 reactivity remained relatively constant (90% and 80% at 15 minutes and 75% and 65% at 180 minutes, respectively), consistent with cleavage of PAR1 by thrombin at Arg41 but not at Arg46 (Figure 2G). Thus, both data derived from SEAP-PAR1 mutant cleavage constructs on transfected cells as well as data from cleavage site sensitive antibodies on endothelial cells were consistent with noncanonical cleavage of PAR1 by APC at Arg46. These data provide strong evidence leading to a novel paradigm in which the opposite PAR1-dependent cellular effects of APC versus thrombin are because of differential activation of PAR1 caused by cleavage at Arg46 by APC versus cleavage at Arg41 by thrombin.
APC-like signaling by an agonist peptide derived from the novel tethered ligand sequence that arises from cleavage of PAR1 at Arg46
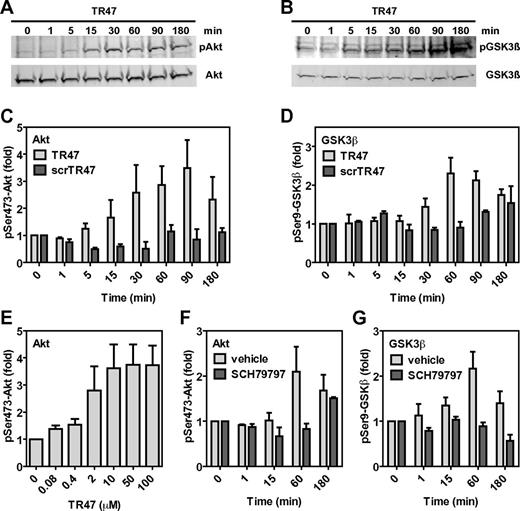
Cleavage of PAR1 by APC at Arg46 destroys the classic TRAP tethered ligand sequence (S42FLLRNPNDKY …) and creates a novel PAR1 N-terminus (N47PNDKY …). To test the hypothesis that this newly generated PAR1 N-terminus can act as a tethered ligand and mediate APC-specific signaling, a peptide (TR47, PAR1 residues 47-66; see supplemental Figure 2) was synthesized and tested on EA.hy.926 endothelial cells for activation of the cytoprotective Akt signaling cassette. The TR47 peptide induced robust and sustained activation of Akt as determined by phosphorylation of Ser473, starting as early as 15 minutes after addition of TR47 and lasting > 180 minutes (Figure 3A,C). To confirm activation of Akt by TR47, Akt-mediated inactivation of GSK3β via phosphorylation at Ser9 was determined, a well-known downstream substrate for Akt.32 TR47 induced significant Ser9-GSK3β phosphorylation starting at 30 minutes with a time course that fell well within the time course of TR47-mediated Akt activation (Figure 3B,D). No phosphorylation of Akt at Ser473 (Figure 3C) or GSK3β at Ser9 (Figure 3D) was observed for cells treated with a scrambled control peptide (scrTR47, supplemental Figure 2), indicating that the effects of TR47 were specific for the newly created N-tethered ligand sequence of PAR1 after cleavage at Arg46. Concentrations of TR47 (EC50 = 1.2μM) that were required to induce Akt phosphorylation were comparable with those reported for TRAP-mediated induction of signaling in endothelial cells with noticeable TR47-mediated Akt phosphorylation in the high nanomolar range and maximal effects achieved at 10μM TR47 (Figure 3E). Activation of Akt by TR47 was dependent on PAR1 because TR47 failed to induce Akt phosphorylation at Ser473 in the presence of the PAR1 inhibitor SCH79797 (Figure 3F). Similarly, induction of Ser9-GSK3β phosphorylation by TR47 required PAR1 because this effect was inhibited by the PAR1 antagonist SCH79797 (Figure 3G). Thus, a peptide with the sequence of the new N-terminus of PAR1 generated on cleavage at Arg46 induced PAR1-dependent activation of Akt and PAR1-dependent phosphorylation of GSK3β, strongly suggesting that activation of PAR1 at Arg46 creates a novel functional tethered ligand that is capable of PAR1-dependent activation of specific cell-signaling pathways.
The novel tethered ligand TR47 peptide induces PAR1-dependent signaling in endothelial cells. Phosphorylation of pSer473-Akt and pSer9-GSK3β by TR47 peptide was monitored in endothelial cells. (A) Time course of Akt phosphorylation at Ser473 by TR47 (50μM). (B) Time course of GSK3β phosphorylation at Ser9 by TR47 (50μM). (C) Phosphorylation of Akt at Ser473 by TR47 (50μM) or a control scrTR47 peptide (50μM) over time. (D) Phosphorylation of GSK3β at Ser9 by TR47 (50μM) and a control scrTR47 peptide (50μM) over time. (E) TR47 peptide dose dependence for phosphorylation of Akt at Ser473 at 60 minutes. (F) Time course for phosphorylation of Akt at Ser473 by TR47 (50μM) in the presence of the PAR1 inhibitor, SCH79797, or vehicle control (DMSO). (G) Time course for phosphorylation of GSK3β at Ser9 by TR47 (50μM) in the presence of the PAR1 inhibitor, SCH79797, or vehicle control (DMSO). (A-B) Representative experiments. (C-G) Data points represent the mean ± SEM (N ≥ 3).
The novel tethered ligand TR47 peptide induces PAR1-dependent signaling in endothelial cells. Phosphorylation of pSer473-Akt and pSer9-GSK3β by TR47 peptide was monitored in endothelial cells. (A) Time course of Akt phosphorylation at Ser473 by TR47 (50μM). (B) Time course of GSK3β phosphorylation at Ser9 by TR47 (50μM). (C) Phosphorylation of Akt at Ser473 by TR47 (50μM) or a control scrTR47 peptide (50μM) over time. (D) Phosphorylation of GSK3β at Ser9 by TR47 (50μM) and a control scrTR47 peptide (50μM) over time. (E) TR47 peptide dose dependence for phosphorylation of Akt at Ser473 at 60 minutes. (F) Time course for phosphorylation of Akt at Ser473 by TR47 (50μM) in the presence of the PAR1 inhibitor, SCH79797, or vehicle control (DMSO). (G) Time course for phosphorylation of GSK3β at Ser9 by TR47 (50μM) in the presence of the PAR1 inhibitor, SCH79797, or vehicle control (DMSO). (A-B) Representative experiments. (C-G) Data points represent the mean ± SEM (N ≥ 3).
The TR47 peptide mimics APC-induced signaling but does not mimic thrombin-induced or TRAP-induced cell signaling
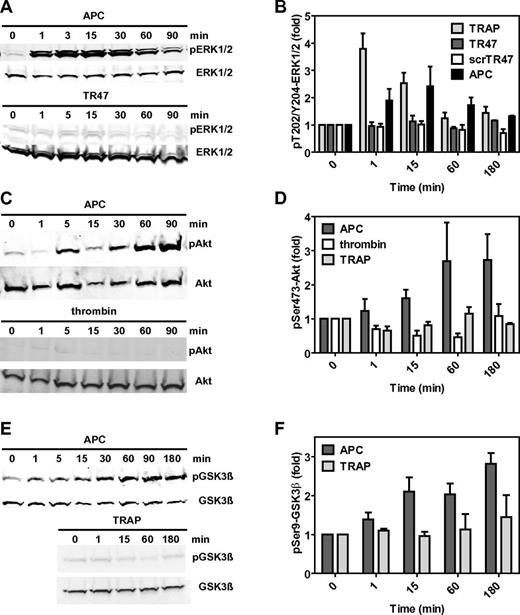
Classic activation of PAR1 by thrombin or TRAP results in the activation of the MAPK pathway as typically demonstrated by rapid phosphorylation of ERK1/2 at Thr202 and Tyr204 or Thr185 and Tyr187, respectively. Consistent with earlier reports, APC induced a transient but modest activation of ERK1/2 (Figure 4A-B).14 Interestingly, TR47 failed to induce any noticeable activation of ERK1/2 under conditions where TRAP and APC did so (Figure 4B). Thus, TR47 does not activate the classic PAR1 MAPK pathway that is activated by thrombin and TRAP. Remarkably, TR47 induced activation of Akt (Figure 3C) with a time course that mimicked APC's robust activation of Akt (Figure 4C-D). In contrast and reflecting the striking functional selectivity of TR47 and APC, neither thrombin nor TRAP induced Ser473-Akt phosphorylation under the experimental conditions used. Similar to activation of Akt, APC induced a sustained phosphorylation of GSK3β at Ser9 (Figure 4E-F), mimicking the effect of TR47 (Figure 3D). Unlike this effect of APC and TR47, incubation of endothelial cells with TRAP did not result in Ser9-GSK3β phosphorylation (Figure 4F). Together, these signaling data indicated that the N-terminus generated by cleavage of PAR1 at Arg41 causes activation of the MAPK pathway, whereas the N-terminus generated by cleavage of PAR1 at Arg46 causes activation of the Akt pathway. Thus, the different N-termini that arise from different cleavages of PAR1 by APC are biased agonists with remarkable functional selectivity.
The TR47 peptide and TRAP are PAR1 biased agonists. Differential phosphorylation was determined for pThr202/Tyr204-ERK1/2, pSer473-Akt, and pSer9-GSK3β that was induced by TRAP, TR47 peptide, control scrTR47 peptide, APC, and thrombin. (A) Time course of ERK1/2 phosphorylation at Thr202/Tyr204 by APC (70nM) and TR47 (50μM). (B) Phosphorylation of ERK1/2 at Thr202/Tyr204 by TRAP (50μM), TR47 (50μM), scrTR47 (50μM), and APC (70nM) over time. (C) Time course of Akt phosphorylation at Ser473 by APC (70nM) and thrombin (54nM). (D) Phosphorylation of Akt at Ser473 by APC (70nM), thrombin (54nM), and TRAP (50μM) over time. (E) Time course of GSK3β phosphorylation at Ser9 by APC (70nM) and TRAP (50μM). (F) Phosphorylation of GSK3β at Ser9 by APC (70nM) and TRAP (50μM) over time. (A,C,E) Representative experiments. (B,D,F) Data points represent the mean ± SEM (N ≥ 3).
The TR47 peptide and TRAP are PAR1 biased agonists. Differential phosphorylation was determined for pThr202/Tyr204-ERK1/2, pSer473-Akt, and pSer9-GSK3β that was induced by TRAP, TR47 peptide, control scrTR47 peptide, APC, and thrombin. (A) Time course of ERK1/2 phosphorylation at Thr202/Tyr204 by APC (70nM) and TR47 (50μM). (B) Phosphorylation of ERK1/2 at Thr202/Tyr204 by TRAP (50μM), TR47 (50μM), scrTR47 (50μM), and APC (70nM) over time. (C) Time course of Akt phosphorylation at Ser473 by APC (70nM) and thrombin (54nM). (D) Phosphorylation of Akt at Ser473 by APC (70nM), thrombin (54nM), and TRAP (50μM) over time. (E) Time course of GSK3β phosphorylation at Ser9 by APC (70nM) and TRAP (50μM). (F) Phosphorylation of GSK3β at Ser9 by APC (70nM) and TRAP (50μM) over time. (A,C,E) Representative experiments. (B,D,F) Data points represent the mean ± SEM (N ≥ 3).
TR47-induced vascular-endothelial protective effects in vitro and in vivo
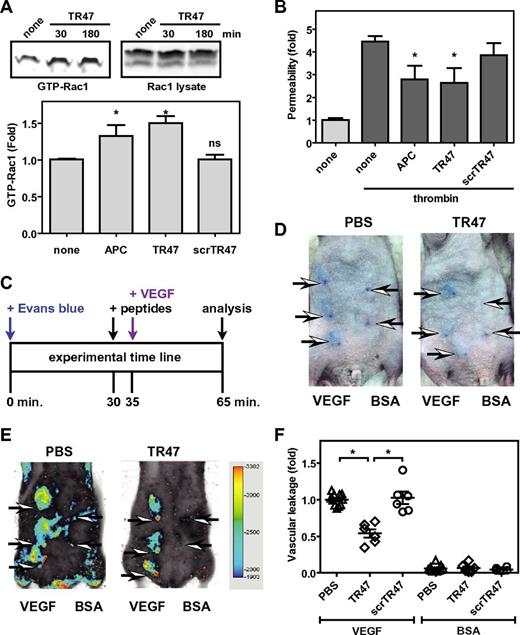
Using β-arrestin-mediated signaling versus G protein-dependent signaling is a hallmark of biased signaling by GPCRs. Recently, PAR1 was shown to exhibit biased signaling because activation of Rac1 by APC and APC-mediated endothelial barrier protection requires β-arrestin-2 and dishevelled-2 scaffolding, whereas thrombin-induced vascular leakage and RhoA activation requires G proteins but not β-arrestins.18 Accordingly, the TR47 peptide, but not the scrTR47 control peptide, induced APC-like activation of Rac1 (Figure 5A). In addition, mimicking the well-known vasculoprotective activity of APC, TR47 protected confluent endothelial barriers against thrombin-induced permeability, whereas the control scrTR47 peptide was without effect (Figure 5B).
The TR47 peptide inhibits endothelial permeability in vitro and in vivo vascular leakage in mice. (A) Activation of Rac1 by TR47 peptide, control peptide, and APC was determined and is shown as pull-down of active Rac1 (GTP-Rac1) with PAK1 (top left), total Rac1 (top right panel), and quantification of Rac1 activation at 180 minutes (bottom panel) using APC (70nM), TR47 (50μM), and scrTR47 (50μM). *P < .05. ns indicates not significant. (B) In vitro protection against thrombin-induced endothelial permeability by APC (25nM), TR47 (50μM), and scrTR47 (50μM). (C) Experimental time line for the in vivo VEGF-induced vascular leakage model. (D) Photographs are seen for Evans blue extravasation in the skin of TR47-treated mice (right) or of control PBS-treated mice (left) injected subcutaneously with VEGF (3 left arrows) or with BSA control (2 right arrows). (E) Heat map display of Evans blue extravasation quantified by the Odyssey near-infrared imager at 700 nm. TR47-treated mice (right) or control PBS-treated (left) mice were injected subcutaneously with VEGF (3 left arrows) or with BSA control (2 right arrows). (F) In vivo VEGF-induced vascular leakage in mice is shown for mice treated with PBS control, TR47 peptide (125 μg), or control scrTR47 peptide (125 μg). Data are also shown for BSA-treated control mice that received PBS, TR47, or scrTR47. (A top panels, D-E) Representative experiments. (A bottom panel, B,F) Data points represent the mean ± SEM (N ≥ 3). *P < .05.
The TR47 peptide inhibits endothelial permeability in vitro and in vivo vascular leakage in mice. (A) Activation of Rac1 by TR47 peptide, control peptide, and APC was determined and is shown as pull-down of active Rac1 (GTP-Rac1) with PAK1 (top left), total Rac1 (top right panel), and quantification of Rac1 activation at 180 minutes (bottom panel) using APC (70nM), TR47 (50μM), and scrTR47 (50μM). *P < .05. ns indicates not significant. (B) In vitro protection against thrombin-induced endothelial permeability by APC (25nM), TR47 (50μM), and scrTR47 (50μM). (C) Experimental time line for the in vivo VEGF-induced vascular leakage model. (D) Photographs are seen for Evans blue extravasation in the skin of TR47-treated mice (right) or of control PBS-treated mice (left) injected subcutaneously with VEGF (3 left arrows) or with BSA control (2 right arrows). (E) Heat map display of Evans blue extravasation quantified by the Odyssey near-infrared imager at 700 nm. TR47-treated mice (right) or control PBS-treated (left) mice were injected subcutaneously with VEGF (3 left arrows) or with BSA control (2 right arrows). (F) In vivo VEGF-induced vascular leakage in mice is shown for mice treated with PBS control, TR47 peptide (125 μg), or control scrTR47 peptide (125 μg). Data are also shown for BSA-treated control mice that received PBS, TR47, or scrTR47. (A top panels, D-E) Representative experiments. (A bottom panel, B,F) Data points represent the mean ± SEM (N ≥ 3). *P < .05.
To probe whether TR47 could induce vascular protective effects in vivo, a modification of the modified Miles assay was used in which vascular permeability is measured by the extravasation of Evans blue dye in the skin induced by local subcutaneous injection of VEGF165. Immunocompetent SKH1 hairless mice were used to avoid the need for hair removal that often can result in artifactual leakage resulting from inflammation of the skin. Evans blue extravasation in the skin was quantified using the Odyssey near-infrared fluorescent imager at 700 nm. TR47 or scrTR47 were injected intravenously in the retro-orbital sinus 5 minutes before local subcutaneous injection of recombinant murine VEGF or BSA control and 30 minutes after intravenous administration of Evans blue (Figure 5C). In the absence of TR47 (PBS control), VEGF induced clearly distinguishable areas of Evans blue extravasation, whereas after injection of BSA vehicle control only the needle points marking the injection site could be observed (Figure 5D). Quantification of Evans blue extravasation by near-infrared fluorescence at 700 nm eliminated the time-consuming need for Evans blue extraction from punch biopsy and provided reliable and reproducible results (Figure 5E). The TR47 peptide significantly decreased vascular leakage by 45% compared with PBS control, whereas vascular leakage in the presence of the scrTR47 control peptide was indiscriminately from PBS control (Figure 5F). Neither TR47 nor scrTR47 affected vascular leakage in the absence of VEGF. Thus, like APC, the TR47 peptide causes activation of Rac1, stabilizes endothelial barriers in vitro, and in vivo can markedly reduce vascular leakage.
Discussion
This study provides in vitro and in vivo experimentation to assess the hypothesis that PAR1-dependent signaling by APC involves a noncanonical cleavage of the receptor's N-terminal domain that reveals a novel cryptic intramolecular pharmacophore, which causes APC's cytoprotective, biased signaling. The results show that APC can cleave a synthetic PAR1 N-terminal polypeptide at both Arg41 and Arg46. Cleavage by APC of transfected R41Q-SEAP-PAR1 but not R41Q/R46Q-SEAP-PAR1 constructs supports the notion that APC can cleave PAR1 at Arg46 on cells, a concept that is consistent with epitope mapping of PAR1 on endothelial cells after APC cleavage of PAR1. Thus, data support the hypothesis that APC can cleave PAR1 on endothelial cells at Arg46.
After our initial report last year33,34 of the novel concept that APC cleaves PAR1 at Arg46 to generate a unique biased agonist N-terminal peptide, but before this present paper was submitted, a recent report provided data consistent with this concept.35 That recent report and our studies coincide in advancing the paradigm that APC's PAR1-dependent protective actions are based on Arg46 cleavage. This present paper provides a number of novel aspects for this new paradigm. Our studies uniquely provide direct evidence that APC can cleave the PAR1 polypeptide at Arg46, that the unique N-terminal peptide generated by this cleavage (TR47) triggers signaling via the Akt pathway and via Rac1 activation, that TR47 signaling requires PAR1, and an in vivo proof of concept showing that the TR47 peptide protects against vascular leakage in vivo.
What are the functional consequences of APC's cleavage at Arg46 in PAR1? After Arg46 cleavage, the known PAR1 agonist sequence containing SFLLR is no longer tethered to PAR1. Our studies showed that the TR47 peptide representing the sequence of the novel N-terminus that is generated by cleavage at Arg46 exerts remarkable biologic activities, including signaling reflected in phosphorylation of Akt and GSK3β and endothelial barrier stabilization. These cytoprotective activities caused by TR47 peptide reflect the general activity profile of APC but not that of thrombin or of the thrombin-generated N-terminal sequence represented in TRAP. Distinctions between the 2 peptides are plentiful, including the rapid phosphorylation of ERK1/2 caused by TRAP but not TR47 and the slower phosphorylation of Akt and GSK3β caused by TR47 but not TRAP. The PAR1 inhibitor SCH79797 ablates the phosphorylation of Akt and GSK3β by TR47, showing that these actions of TR47 require PAR1. Thus, data support the hypothesis that APC's cleavage of PAR1 at Arg46 generates a tethered ligand pharmacophore composing at least part of the amino acid sequence of PAR1 residues 47-66, which in vitro initiates a pattern of cytoprotective signaling that resembles the PAR1-dependent cytoprotective functional activities described for APC. However, whether another PAR or even a PAR-unrelated receptor can contribute to TR47-mediated signaling cannot be excluded.
What are the in vivo functional consequences of cleavage at Arg46 by APC? Our studies showed that, when the TR47 peptide is given to mice that are subjected to VEGF-induced vascular leakage, this APC-mimetic peptide significantly reduces vascular leakage. The in vivo data for TR47's barrier stabilization in a vascular leakage murine injury model resemble data for the ability of TR47 to stabilize endothelial cell barrier function in vitro. Thus, all data are consistent with the hypothesis that the novel PAR1 N-terminus generated by APC's cleavage at Arg46 functions as a biased tethered ligand, which causes PAR1 signaling that promotes cytoprotective functions.
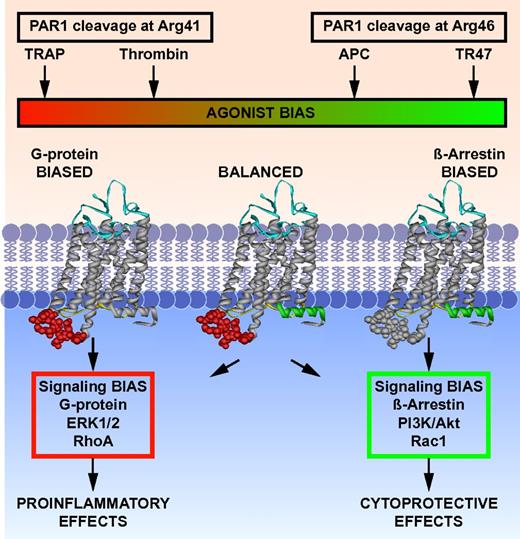
This study provokes a shift in the PAR1 paradigm. Recent insights into mechanisms for regulating signaling by GPCRs emphasize the complexities associated with biased agonism and allosteric modulation.36-39 Emerging structural and pharmacologic information supports the notion that GPCRs exist in dynamic conformational ensembles and that agonists, biased agonists, or allosteric ligands stabilize 1 or more conformational subsets at the expense of others with functional consequences and highly selective signaling outcomes.36,40-45 Proteolysis at Arg41 versus at Arg46 de facto alters the spectrum of conformations of PAR1 because of differences in sequence and N-terminus after cleavage at either Arg41 or Arg46. Consequently, cleavage by APC at Arg46 changes the readily accessible subsets of conformations of PAR1 with functional consequences (Figure 6). PAR1 is a biased receptor that can have multiple covalently linked cryptic tethered ligands that begin at Pro40, Ser42 or Asn47—the first 2 tethered ligands generate a very similar spectrum of signaling, whereas the N-terminal Asn47 tethered ligand stimulates different and very distinctive signal transduction pathways.6,18,24 Thus, PAR1 can understandably mediate the often opposite effects of thrombin and APC because each protease can generate very different tethered peptide agonists that mimic TRAP or TR47, which are intrinsically biased agonists.
Biased agonism of PAR1 resulting from canonical cleavage at Arg41 by thrombin or by noncanonical cleavage at Arg46 by APC. PAR1 is a biased receptor with biased agonists.18 Four different PAR1 agonists are shown along the top bar that can vary in their ability and efficacy to activate different signaling pathways in cells, thus depicting a signaling phenomenon that has been labeled “functional selectivity” or “biased agonism.” The receptor's bias is directly based on the spectrum of signaling that is mediated either via 1 or more G proteins or via β-arrestin-2, whereas the agonist bias is directly related to the sites of cleavage in the extracellular N-terminus (ie, resulting from cleavage either at Arg41 or at Arg46). The TRAP peptide(s) represent the N-terminal sequence of PAR1 that exists after cleavage at Arg41, whereas TR47 represents the N-terminal sequence of PAR1 that exists after cleavage at Arg46, as described in this report. Each agonist can have distinctly different properties because each one can stabilize a different subset of the dynamic conformational ensembles of PAR1. The conformer subsets that are stabilized by TRAP and thrombin promote signaling via different G proteins, whereas conformer subsets stabilized by APC's action, and presumably TR47, promote signaling via β-arrestins, especially β-arrestin-2, and dishevelled-2. However, one must note that TRAP is similar but not functionally equivalent to thrombin; and it is probable, but not yet shown, that the TR47 peptide does not have all the same activities and efficacy as APC. Not depicted are the effects of MMP1 cleavage at Asp39 in PAR1 or potential allosteric PAR1 modulatory factors, including binding of thrombin to the hirudin-like sequence of PAR1, localization of PAR1 in caveolae with caveolin-1, association with EPCR containing bound protein C or APC, dimerization with other PARs, or small molecular allosteric modulators.41,46
Biased agonism of PAR1 resulting from canonical cleavage at Arg41 by thrombin or by noncanonical cleavage at Arg46 by APC. PAR1 is a biased receptor with biased agonists.18 Four different PAR1 agonists are shown along the top bar that can vary in their ability and efficacy to activate different signaling pathways in cells, thus depicting a signaling phenomenon that has been labeled “functional selectivity” or “biased agonism.” The receptor's bias is directly based on the spectrum of signaling that is mediated either via 1 or more G proteins or via β-arrestin-2, whereas the agonist bias is directly related to the sites of cleavage in the extracellular N-terminus (ie, resulting from cleavage either at Arg41 or at Arg46). The TRAP peptide(s) represent the N-terminal sequence of PAR1 that exists after cleavage at Arg41, whereas TR47 represents the N-terminal sequence of PAR1 that exists after cleavage at Arg46, as described in this report. Each agonist can have distinctly different properties because each one can stabilize a different subset of the dynamic conformational ensembles of PAR1. The conformer subsets that are stabilized by TRAP and thrombin promote signaling via different G proteins, whereas conformer subsets stabilized by APC's action, and presumably TR47, promote signaling via β-arrestins, especially β-arrestin-2, and dishevelled-2. However, one must note that TRAP is similar but not functionally equivalent to thrombin; and it is probable, but not yet shown, that the TR47 peptide does not have all the same activities and efficacy as APC. Not depicted are the effects of MMP1 cleavage at Asp39 in PAR1 or potential allosteric PAR1 modulatory factors, including binding of thrombin to the hirudin-like sequence of PAR1, localization of PAR1 in caveolae with caveolin-1, association with EPCR containing bound protein C or APC, dimerization with other PARs, or small molecular allosteric modulators.41,46
Allosteric modulation of PAR1 by thrombin after cleavage at Arg41 can potentially arise because of the binding of thrombin's exosite I to the hirudin-like PAR1 residues 51-55. The binding of an EPCR-protein C complex before any PAR1 cleavage might allosterically alter PAR1 responses caused by thrombin's cleavage at Arg 41, thereby resulting in cytoprotective effects of thrombin.23,47 Allosteric modulation of PAR1 may also involve its association with 1 or more proteins that are located in caveolin 1-containing microdomains. Small molecular weight allosteric antagonists of PAR1 have recently been described.41 Much remains to be learned about PAR1's biased ligands and the allosteric regulation of PAR1 signaling.
This new mechanism for PAR1 cytoprotective actions involving APC's cleavage at Arg46 may be clinically relevant when considering potential explanations for the unfortunate serious side effect of hemorrhagic stroke caused by the PAR1 inhibitor, vorapaxar, in recent large phase 3 clinical trials.48,49 APC stabilizes endothelial barriers via β-arrestin–dependent signaling, whereas thrombin disrupts endothelial barriers via G proteins.10,18 Thus, it is possible that vorapaxar, as a PAR1 general antagonist, inhibits the beneficial effects of the endogenous protein C system on the blood-brain- barrier in addition to inhibiting thrombin's prothrombotic effects on platelets, thereby increasing risk for blood-brain-barrier leakage.5 Although studies are needed to clarify this potential mechanism for vorapaxar, it is clear that drug discovery efforts to identify compounds that antagonize PAR1's G protein-dependent signaling but not β-arrestin 2-dependent signaling may prove fruitful. Similarly, APC-mimetic compounds that promote cytoprotective PAR1-dependent signaling while avoiding thrombin-mimetic signaling would be very interesting biased PAR1 agonists.
In conclusion, an extensive combination of in vitro and in vivo data supports the novel paradigm for the biochemical mechanisms mediating APC's cytoprotective signaling via PAR1 in which APC's cleavage at Arg46 in PAR1 generates a new N-terminal tethered ligand beginning with Asn47, which is a biased agonist that initiates cytoprotective signaling pathways.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr L. Brass (University of Pennsylvania, Philadelphia, PA) for the gift of PAR1 antibodies; Dr C. J. S. Edgell (University of North Carolina, Chapel Hill, NC) for the EA.hy.926 endothelial cells; and M. Mathias and D. Rozenshteyn for excellent technical assistance.
This work was supported in part by the National Heart, Lung, and Blood Institute (grants HL087618 and HL104165, L.O.M.; and grants HL31950 and HL52246, J.H.G.), postdoctoral fellowships from the Swiss National Science Foundation, the Fondation Suisse pour les Bourses en Médecine et Biologie, and Novartis (PBGEP3-134242 and PASMP3_140065; L.B.), and the American Heart Association Western States Affiliate (postdoctoral fellowship; E.A.B.).
National Institutes of Health
Authorship
Contribution: L.O.M., R.K.S., L.B., and E.A.B. performed experiments; L.O.M., R.K.S., L.B., E.A.B., and J.H.G. designed the research and analyzed results; and L.O.M. and J.H.G. wrote the manuscript.
Conflict-of-interest disclosure: L.O.M. and J.H.G. are inventors on a pending patent application concerning PAR1 peptides that is the property of the Scripps Research Institute. The remaining authors declare no competing financial interests.
Correspondence: Laurent O. Mosnier, Department of Molecular and Experimental Medicine (MEM-180), Scripps Research Institute, 10550 North Torrey Pines Rd, La Jolla, CA 92037; e-mail: lmosnier@scripps.edu.