Abstract
Regulatory dendritic cells (DCs) play important roles in the induction of peripheral tolerance and control of adaptive immune response. Our previous studies demonstrate that splenic stroma can drive mature DCs to proliferate and further differentiate into a unique subset of CD11bhiIalow regulatory DCs, which could inhibit T-cell response, program generation of immunosuppressive memory CD4 T cells. However, the effect of regulatory DCs on B-cell function remains unclear. Here, we report that regulatory DCs can induce splenic B cells to differentiate into a distinct subtype of IL-10–producing regulatory B cells with unique phenotype CD19hiFcγIIbhi. CD19hiFcγIIbhi B cells inhibit CD4 T-cell response via IL-10. CD19hiFcγIIbhi B cells have enhanced phagocytic capacity compared with conventional CD19+ B cells, and FcγRIIb mediates the uptake of immune complex by CD19hiFcγIIbhi B cells. We found that regulatory DC-derived IFN-β and CD40 ligand are responsible for the differentiation of CD19hiFcγIIbhi B cells. Furthermore, an in vivo counterpart of CD19hiFcγIIbhi B cells in the spleen and lymph nodes with similar phenotype and regulatory function has been identified. Our results demonstrate a new manner for regulatory DCs to down-regulate immune response by, at least partially, programming B cells into regulatory B cells.
Introduction
B cells are generally considered to induce immune responses through antibody production and optimal CD4 T-cell activation.1 Now, more subsets of B cells with nonclassic functions have been identified.2-4 For example, innate effector B cells have been shown to be important in the initiation and effector phase of innate response against infections.5-7 B cells with regulatory functions exert the immune regulatory functions either through the secreted antibodies and/or cytokines with a suppressive effect or directly by cellular interactions.2,4,8 These B cells with regulatory functions, independently of secreted antibodies, are designated as regulatory B cells. Regulatory B cells have been identified to be involved in the regulation of several immune-mediated pathologic processes in both mice and humans, including autoimmune diseases and infections.9-12 The regulatory function of regulatory B cells may be mediated directly by the production of regulatory cytokines IL-10 and TGF-β and/or by the ability of B cells to interact with pathogenic T cells to inhibit harmful immune responses.2,13 Our previous study suggested that B cell–activating factor can induce marginal zone (MZ) B-cell differentiation into regulatory B cells, which could suppress autoimmunity in an IL-10–dependent fashion.14 So, more and more evidence shows the importance of regulatory B cells in the maintenance of immune homeostasis and the pathogenesis of immune disorders. However, the characteristics of regulatory B cells and the factors programming regulatory B-cell generation remain to be further identified.
Dendritic cells (DCs) are at the crossroads of innate and adaptive immunity and essential mediators of immunity and tolerance.15 The functional versatility of DCs is enabled in part by the various DC subsets with heterogeneous cell surface markers, distinct cytokine profiles, and different developmental stages.16 DCs with regulatory function, designated as regulatory DCs, have attracted much attention because they play important roles in maintaining immune homeostasis.17,18 Regulatory DCs negatively regulate immune response by inducing regulatory T cells, inhibiting T-cell proliferation, and inducing T-cell anergy.19-21 Distinct subsets of DCs have been found to regulate the function of B cells. For example, conventional DC-derived IL-12 induces naive follicular (FO) B cells to differentiate into IFN-γ and IL-12–producing effector B cells.22 In addition, plasmacytoid DCs regulate B-cell growth, differentiation, and immunoglobulin (Ig) secretion.23 However, there is no report about the effect of regulatory DCs on B-cell function to date.
Our previous studies show that splenic stromal cells, mimicking the secondary lymph organ microenvironment, can drive mature DCs (maDCs) to proliferate and differentiate into a novel subset of regulatory DCs (diffDCs, DCs differentiated from maDCs), with unique phenotype (CD11bhiIalow) and cytokine profile (higher IL-10 but minimal IL-12p70 production), which can inhibit maDC-initiated T-cell proliferation.24 There is considerable interaction between regulatory DCs and other immune cells in physiology and pathology conditions. For example, we showed that regulatory DCs can selectively recruit Th1 cells and inhibit Th1 proliferation, and program generation of IL-4–producing alternative memory CD4 T cells with suppressive activity.25,26 We have identified an in vivo counterpart of regulatory DCs in the spleen with similar phenotype and functions. Considering that B-cell development encompasses a continuum of stages that begin in primary lymphoid tissue, with subsequent functional maturation in secondary lymphoid tissue, such as spleen,1 these findings suggest that secondary lymphoid tissue may be potential locations for regulatory DC–B-cell interaction. However, whether regulatory DC and B cells interact in these locations and what the consequence of these interactions have not been elucidated.
In this study, we show that regulatory DCs can induce splenic T1, T2, MZ, and B1 B cells to differentiate into a distinct regulatory B subset, CD19hiFcγIIbhi B cells, which preferentially secret IL-10 and exert regulatory functions both in vitro and in vivo. We further investigated how the regulatory DCs program the generation of regulatory CD19hiFcγIIbhi B cells and how CD19hiFcγIIbhi B cells negatively regulate T-cell response. Our results provide a new manner of regulatory DCs for negative feedback control of T-cell immune response and maintenance of immune homeostasis by, at least partially, inducing differentiation of B cells into regulatory B cells. In addition, one new subset of regulatory B cells with CD19hiFcγIIbhi phenotype has been discovered, providing clues for investigating the roles of B-cell populations in the health and diseases.
Methods
Mice
C57BL/6J mice were obtained from Joint Ventures Sipper BK Experimental Animal Co. OVA323-339-specific TCR-transgenic DO11.10 mice or OT-2 mice, B6.SJL-Ptprca Pep3b/BoyJ mice (CD45.1 mice), FcγIIb−/− mice, and CD40−/− mice were obtained from The Jackson Laboratory. CD40L−/− mice were generated on C57BL/6J × 129SV/J (H-2b) mice as previously reported.27 The mice were bred in specific pathogen-free conditions. Animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of the Second Military Medical University, Shanghai, China.
Reagents
Recombinant mouse GM-CSF and IL-4 were from PeproTech. Neutralizing antimouse VEGF, IL-6, IP-10 antibodies, isotype control mAbs, and soluble CD40L (sCD40L) were from R&D Systems. The rabbit polyclonal antimouse IFN-β and recombinant mouse IFN-β were from PBL Biomedical Laboratories. Fluorescence-conjugated mAbs to CD1d, CD4, CD5, CD11c, B220, IgM, CD21, CD23, CD93, TIM-1, CD19, CD62L, CD16/CD32, and IL-10 were from BD PharMingen or BioLegend. GolgiStop and Cytofix/Cytoperm kit were from BD PharMingen. Phorbol myristate acetate, ionomycin, polyriboinosinic acid/polyribocytidylic acid (poly I:C), lipopolysaccharide (LPS), 7-amino-actinomycin D (7-AAD), OVA stock, and anti-OVA mAb were from Sigma-Aldrich.
Preparation of mouse imDCs, maDCs, and regulatory DCs
Purification of mouse splenic B cells
Splenic B cells were enriched using CD19-conjugated microbeads as previously described.14 Splenic transitional 1 (T1), T2, T3, FO, MZ, or B1 B cells were sorted respectively with a MOFLO High Speed Flow Cytometer (Beckman Coulter).
Coculture of regulatory DC and B cells
Splenic B cells were cocultured with DCs from wild-type mice. In some experiments, FcγIIb−/− B cells or CD40−/− B cells were cocultured with regulatory DCs or CD40L−/− regulatory DCs. First, purified splenic B cells were plated in 96-well U-bottom plates at a density of 2.0 × 105 per well. Then, purified imDCs, maDCs, or regulatory DCs were added at the indicated ratios (DC/B). In some cases, DCs were incubated with anti–IL-6, anti-VEGF, anti–IFN-β, or anti–IP-10 neutralizing Abs for 1 hour before coculture with B cells. After coculture, the supernatants were collected for ELISA or/and cells were harvested for flow cytometry analysis.
Cytokine assays
Mouse IL-6, IL-10, IL-12, IP-10, TGF-β, PGE2 and IFN-γ levels were assayed by ELISA kits. For intracellular cytokine staining, splenic B cells or specific B-cell subsets were cocultured with regulatory DCs for 48 hours and then stimulated with cytosine-phosphate-guanosine oligodeoxynucleotide (CpG ODN; 2 μg/mL), phorbol myristate acetate (50 ng/mL), ionomycin (500 ng/mL), and GolgiStop for additional 6 hours. Then cells were stained for surface markers and then fixed and permeabilized with the Cytofix/Cytoperm kit. Permeabilized cells were incubated with anti–IL-10 mAb. The cells were acquired with the BD LSRII flow cytometer (BD Biosciences) and analyzed by FlowJo Version 5.7.2 software (TreeStar).
RNA quantification
Total RNA was extracted with TRIzol (Invitrogen). LightCycler (Roche) and SYBR RT-PCR kits (Takara) were used for quantitative RT-PCR analysis. Date were normalized to β-actin expression.
Sorting of CD19hiFcγIIbhi B cells or CD19hiFcγIIb−/− B cells
Freshly purified splenic B cells or FcγIIb−/− B cells were cocultured with regulatory DCs at a ratio of 1:10 (DC/B) for 48 hours, and then the cells were labeled with anti-CD19 and anti-CD16/CD32 mAbs. The CD19hiFcγIIbhi and CD19hiFcγIIb−/− B cells were sorted, respectively.
Transwell experiments
Regulatory DCs were seeded at a density of 6 × 104 per 600 μL/well in 24-well plates. In addition, 6 × 105 B cells were either directly added or placed in transwell chambers (Millicell, 1.0 μm; Millipore) in the same well. IL-10 production was detected at 48 hours by ELISA.
Analysis of phagocytic ability of CD19hiFcγIIbhi B cells
FITC-OVA–containing immune complex (FITC-OVA-IC) were prepared as previously described.28 Purified B cells, sorted CD19hiFcγIIbhi B cells, or CD19hiFcγIIb−/− B cells were incubated with FITC-OVA-IC at 37°C for 2 hours. The cells were then washed and resuspended in chilled PBS for immediate flow cytometry. Cells incubated with FITC-OVA-IC at 4°C were used as a negative control. For confocal microscopy, cells were fixed with 4% formaldehyde and then washed with PBS. Nuclei were stained with Hoechst. Coverslips were mounted on slides using mounting media and read using Leica TCS SP2 confocal laser microscope (Wetzlar).
Assays for CD4 T-cell proliferation in vitro and in vivo
The in vitro and in vivo CD4 T-cell proliferation was measured as described previously.19,20 For in vitro assay, purified CD4 T cells from DO11.10 OVA323-339–specific TCR transgenic × C57BL/6J F1 hybrid mice cocultured with maDCs in the presence of OVA323-339 peptide for 24 hours; then the cells were washed. The collected activated CD4 T cells were cocultured with 1 × 105 CD19hiFcγIIbhi B cells or CD19+ B cells (1 × 105 activated CD4 T cells/200 μL/well). After 5 days of culture, cells stained for CD4 and 7-AAD were resuspended in exactly 200 μL PBS and cellular data acquired for 56 seconds by flow cytometry. The number of CD4+7-AAD− live cells was calculated to represent the altitude of CD4 T-cell proliferation. For in vivo assay, CFSE labeled-CD4 T cells from OT-2 × CD45.1 F1 hybrid mice were injected intravenously into C57BL/6J mice (5 × 105 cells/mouse) on day −1. On day 0, 1 × 106 OVA323-339 peptide-loaded maDCs were injected subcutaneously into the left footpad of C57BL/6J mice adoptively transferred with CD4 T cells. On day 1, CD19hiFcγIIbhi B cells or CD19loFcγIIblo B cells were also injected subcutaneously into the immunized mice's left footpad (5 × 105 cells/mouse). On day 5, single-cell suspensions prepared from the left popliteal lymph nodes were stained with anti-CD45.1 and anti-CD4 mAbs and were analyzed for CFSE dilution.
Identification of a natural counterpart of CD19hiFcγIIbhi B cells in vivo
DO11.10 OVA323-339–specific TCR transgenic × C57BL/6J F1 hybrid mice injected intraperitoneally with or without 40 μg OVA plus 10 μg CpG ODN were killed on days 3, 6, and 9. CD19+ cells enriched by CD19-conjugated microbeads from spleen and lymph nodes were double-stained with anti-CD19 and anti-CD16/CD32, and then CD19hiFcγIIbhi B cells and CD19loFcγIIblo B cells were sorted. In addition, the cytokine profile, phagocyotic capacity, and inhibitory function of the sorted CD19hiFcγIIbhi B cells and CD19loFcγIIblo B cells were analyzed.
Statistical analysis
Data are shown as mean ± SD for separate experiments. Statistical significance was determined by Student t test with a value of P less than .05 considered as statistically significant.
Results
Regulatory DCs induce splenic B cells to differentiate into IL-10–producing B cells
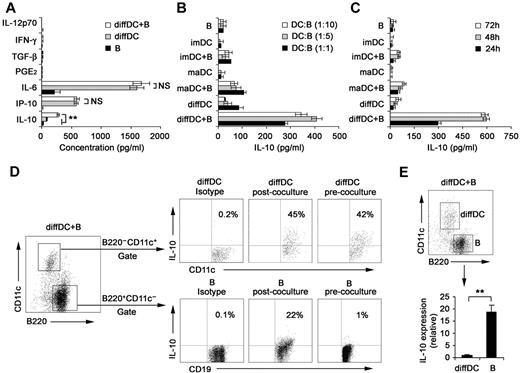
Our previous studies showed that regulatory DCs secrete higher levels of IL-10 but lower levels of IL-12p70 than imDCs and maDCs.21,24 DCs have been shown to be able to regulate B-cell functions.22,29,30 Therefore, we analyzed the proliferation, apoptosis, antibody secretion, and cytokine production by splenic CD19+ B cells after their interaction with various DCs. As shown in supplemental Figure 1A-B (available on the Blood Web site; see the Supplemental Materials link at the top of the online article), the proliferation and apoptosis of splenic B cells were not significantly affected after interaction with imDCs, maDCs, or regulatory DCs. Moreover, no difference in IgM and IgG2b production was detected among B cells cocultured with imDCs, maDCs, or regulatory DCs (supplemental Figure 1C). There was no significant difference in production of IL-12, IFN-γ, IL-6, IP-10, PGE2, and TGF-β by B cells cocultured with or without regulatory DCs (Figure 1A). Interestingly, B cells cocultured with regulatory DCs could produce higher levels of IL-10 than B cells cocultured with imDCs or maDCs (Figure 1B). IL-10 production by B cells cocultured with regulatory DCs increased gradually and reached a maximum after 48 hours of coculture (Figure 1C). To verify the cell source of IL-10, the IL-10 production by B220−CD11c+ regulatory DCs and CD19+B220+CD11c− B cells in coculture system was first confirmed to be positive by intracellular staining (Figure 1D). However, quantitative RT-PCR results showed that regulatory B cells expressed more IL-10 than diffDCs per cell (Figure 1E). These results demonstrated that more IL-10–producing B cells were generated in the B-cell/regulatory DC coculture system.
Regulatory DCs induce the differentiation of splenic B cells into IL-10–producing B cells. (A) Freshly purified splenic naive B cells were cocultured with regulatory DCs at a ratio of 1:1 for 24 hours. Culture supernatants were collected and assayed for cytokines (IL-10, IP-10, IL-6, IL-12p70, PGE2, TGF-β, and IFN-γ) by ELISA. (B) Splenic naive B cells were cocultured with various DCs at the indicated ratios (DC/B). Supernatants were collected at 24 hours and assayed for IL-10. (C) Splenic naive B cells were cocultured with various DCs at a ratio of 1:10 (DC/B), and supernatants were collected at different times for assay of IL-10. (D) Intracellular staining for IL-10 in splenic CD19+B220+CD11c− B cells and B220−CD11c+ regulatory DCs in coculture system. The percentages of each population are indicated in the plots. (E) B220+CD11c− B cells and B220−CD11c+ regulatory DCs were sorted after coculture, and IL-10 expression in the purified regulatory DCs and B cells was checked by quantitative RT-PCR. IL-10 expression in diffDCs was designated as 1 for the comparison of IL-10 expression between regulatory B cells and diffDCs. Results are representative of 3 independent experiments. Data are mean ± SD. **P < .01. NS indicates not significant.
Regulatory DCs induce the differentiation of splenic B cells into IL-10–producing B cells. (A) Freshly purified splenic naive B cells were cocultured with regulatory DCs at a ratio of 1:1 for 24 hours. Culture supernatants were collected and assayed for cytokines (IL-10, IP-10, IL-6, IL-12p70, PGE2, TGF-β, and IFN-γ) by ELISA. (B) Splenic naive B cells were cocultured with various DCs at the indicated ratios (DC/B). Supernatants were collected at 24 hours and assayed for IL-10. (C) Splenic naive B cells were cocultured with various DCs at a ratio of 1:10 (DC/B), and supernatants were collected at different times for assay of IL-10. (D) Intracellular staining for IL-10 in splenic CD19+B220+CD11c− B cells and B220−CD11c+ regulatory DCs in coculture system. The percentages of each population are indicated in the plots. (E) B220+CD11c− B cells and B220−CD11c+ regulatory DCs were sorted after coculture, and IL-10 expression in the purified regulatory DCs and B cells was checked by quantitative RT-PCR. IL-10 expression in diffDCs was designated as 1 for the comparison of IL-10 expression between regulatory B cells and diffDCs. Results are representative of 3 independent experiments. Data are mean ± SD. **P < .01. NS indicates not significant.
Considering that regulatory DCs are purified from coculture system of endothelial-like splenic stromal cells (ESSCs)/DCs, we analyzed IL-10 production by splenic B cells after interaction with ESSCs. As shown in supplemental Figure 2, ESSCs did not affect IL-10 secretion of B cells. In addition, regulatory DCs could promote preferential IL-10 production of mouse splenic B220+ B cells (supplemental Figure 3). Taken together, these results suggested that regulatory DCs can induce splenic B cells to differentiate into IL-10–producing B cells.
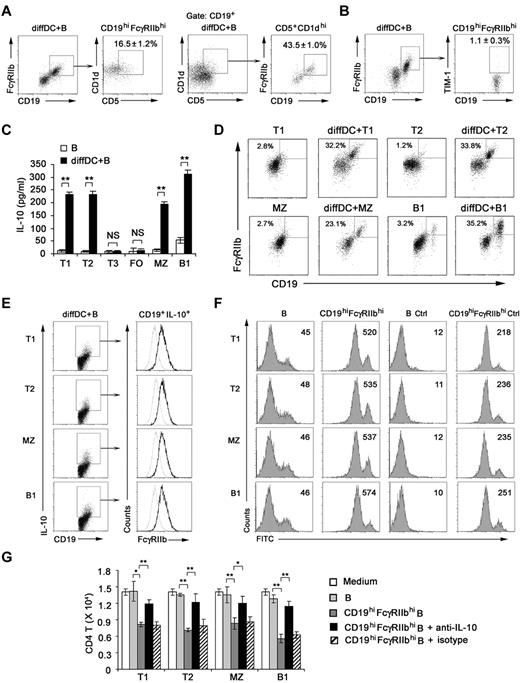
The regulatory DC-programmed, IL-10–secreting B cells exhibit unique phenotype (CD19hiFcγRIIbhi)
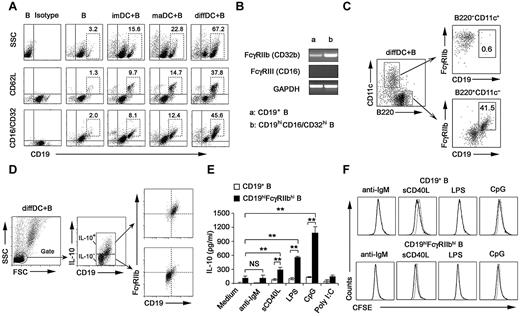
Next, we investigated the membrane molecule expression by IL-10–secreting B cells after being cocultured with regulatory DCs. After the coculture, B cells expressed higher levels of CD19, CD62L, and CD16/CD32 (Figure 2A; supplemental Figure 4), whereas the expression of CD40, CD80, CD86, B7H1, and Iab remained unchanged (data not shown). The available anti-CD16/CD32 mAb could recognize activating receptors FcγRIII (CD16) and inhibitory receptor FcγRIIb (CD32b). It is reported that FcγRIIb is the only Fc receptor expression on B cells.31 FcγRIIb expression in splenic CD19+ B cells and sorted CD19hiCD16/CD32hi B cells was analyzed at the mRNA level. The FcγRIIb but not FcγRIII (CD16) expression in sorted CD19hiCD16/CD32hi B cells was up-regulated more significantly than that in splenic CD19+ B cells (Figure 2B). These data suggested that it is FcγRIIb that is overexpressed on CD19hi B cells. A previous study showed that some regulatory DCs in mouse spleen are CD19+.32 We found that the CD19hiFcγRIIbhi population was a distinct population from cocultured B220−CD11c+ regulatory DCs (Figure 2C), indicating that the CD19hiFcγRIIbhi population belongs to B cells.
Regulatory DCs program generation of IL-10–producing B population with CD19hiFcγRIIbhi phenotype. (A) Purified splenic naive B cells cocultured with DCs at a ratio of 1:10 (DC/B) for 48 hours were stained with antibodies to CD19, CD62L, or CD16/CD32 and analyzed by flow cytometry. Numbers in plots indicate percentage of CD19hi cells or CD19hiCD62L+ cells or CD19hiCD16/CD32hi in the gated CD19+ B cells. (B) FcγIIb and FcγRIII mRNA expression by conventional splenic CD19+ B cells or sorted CD19hiCD16/CD32hi B cells was assessed by RT-PCR. The transcript of mouse GAPDH gene was used as the amplification control. (C) Purified splenic naive B cells were cocultured with regulatory DCs at a ratio of 1:10 (DC/B) for 48 hours. Cells were stained with antibodies to CD19, B220, CD11c, and CD16/CD32 and analyzed by flow cytometry. Numbers in plots indicate percentage of CD19hiFcγRIIbhi in the gated B220−CD11c+ regulatory DCs or B220+CD11c− B cells. (D) Purified splenic B cells were cocultured with regulatory DCs. CD19+IL-10+ B cells or CD19+IL-10− B cells were gated and assessed for FcγIIb expression. (E) Twenty-four hours after conventional splenic B cells or sorted CD19hiFcγRIIbhi B cells were stimulated with 20 μg/mL anti-IgM Ab, 2 μg/mL sCD40L, 200 ng/mL LPS, 2 μg/mL CpG ODN, or 2 μg/mL poly I:C, respectively, IL-10 production was measured by ELISA. (F) Conventional splenic B cells or sorted CD19hiFcγRIIbhi B cells were labeled with CFSE and cultured with anti-IgM Ab, sCD40L, LPS, or CpG ODN for 5 days. Histograms represent CFSE expression by the CD19+ or CD19hiFcγRIIbhi B cells. Gray lines represent CFSE staining of the unstimulated CD19+ or CD19hiFcγRIIbhi B cells. Results are representative of 3 independent experiments. Data are mean ± SD. **P < .01. NS indicates not significant.
Regulatory DCs program generation of IL-10–producing B population with CD19hiFcγRIIbhi phenotype. (A) Purified splenic naive B cells cocultured with DCs at a ratio of 1:10 (DC/B) for 48 hours were stained with antibodies to CD19, CD62L, or CD16/CD32 and analyzed by flow cytometry. Numbers in plots indicate percentage of CD19hi cells or CD19hiCD62L+ cells or CD19hiCD16/CD32hi in the gated CD19+ B cells. (B) FcγIIb and FcγRIII mRNA expression by conventional splenic CD19+ B cells or sorted CD19hiCD16/CD32hi B cells was assessed by RT-PCR. The transcript of mouse GAPDH gene was used as the amplification control. (C) Purified splenic naive B cells were cocultured with regulatory DCs at a ratio of 1:10 (DC/B) for 48 hours. Cells were stained with antibodies to CD19, B220, CD11c, and CD16/CD32 and analyzed by flow cytometry. Numbers in plots indicate percentage of CD19hiFcγRIIbhi in the gated B220−CD11c+ regulatory DCs or B220+CD11c− B cells. (D) Purified splenic B cells were cocultured with regulatory DCs. CD19+IL-10+ B cells or CD19+IL-10− B cells were gated and assessed for FcγIIb expression. (E) Twenty-four hours after conventional splenic B cells or sorted CD19hiFcγRIIbhi B cells were stimulated with 20 μg/mL anti-IgM Ab, 2 μg/mL sCD40L, 200 ng/mL LPS, 2 μg/mL CpG ODN, or 2 μg/mL poly I:C, respectively, IL-10 production was measured by ELISA. (F) Conventional splenic B cells or sorted CD19hiFcγRIIbhi B cells were labeled with CFSE and cultured with anti-IgM Ab, sCD40L, LPS, or CpG ODN for 5 days. Histograms represent CFSE expression by the CD19+ or CD19hiFcγRIIbhi B cells. Gray lines represent CFSE staining of the unstimulated CD19+ or CD19hiFcγRIIbhi B cells. Results are representative of 3 independent experiments. Data are mean ± SD. **P < .01. NS indicates not significant.
Then we wondered whether the preferential production of IL-10 we observed in Figure 1 was from CD19hiFcγRIIbhi B cells. The results showed that IL-10–producing B cells after coculture with regulatory DCs exhibited the CD19hiFcγRIIbhi phenotype (Figure 2D). Furthermore, we sorted the CD19hiFcγRIIbhi population and stimulated them with anti-IgM Ab, sCD40L, or a variety of TLR ligands, including poly I:C, LPS, and CpG ODN. As shown in Figure 2E, sCD40L, LPS, and CpG ODN, but not anti-IgM Ab and poly I:C, could stimulate the CD19hiFcγRIIbhi population to produce more IL-10, suggesting that CD40, TLR4, and TLR9 signaling may be critical for CD19hiFcγRIIbhi B-cell function. Moreover, there was no significant difference in expansion between CD19+ and CD19hiFcγRIIbhi B cells after sCD40L, LPS, or CpG ODN stimulation (Figure 2F). Together, these results demonstrated that regulatory DCs can program generation of a new population of IL-10–secreting CD19hiFcγRIIbhi B cells.
CD19hiFcγRIIbhi B cells have the increased phagocytic capacity
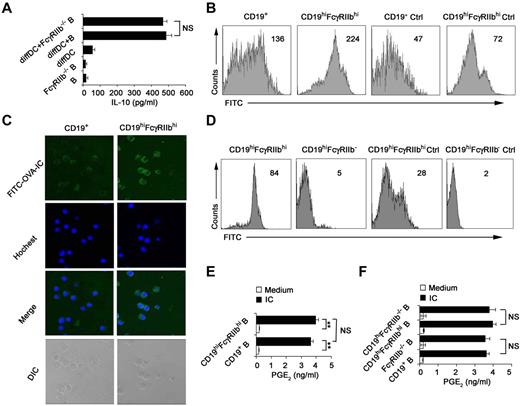
Because FcγRIIb was identified to be overexpressed on the IL-10–producing CD19hiFcγRIIbhi B cells in Figure 2A through D, we determined whether or not FcγRIIb is required for IL-10 production. We found that FcγRIIb−/− B cells were not compromised in their ability of IL-10 production once cocultured with regulatory DCs (Figure 3A), suggesting that FcγRIIb is not involved in preferential IL-10 production by CD19hiFcγRIIbhi B cells. It has been shown that the Fc receptor mediates effective internalization of Ag-IgG complexes.33 CD19hiFcγRIIbhi B cells were found to incorporate more FITC-OVA-IC than splenic B cells (Figure 3B-C), indicating that FcγRIIb is involved in the phagocytosis of IC by this new B-cell population. To further confirm that FcγRIIb mediates potent phagocytosis of FITC-OVA-IC by CD19hiFcγRIIbhi B cells, B cells from FcγRIIb−/− mice were cultured with regulatory DCs and then CD19hiFcγRIIb−/− B cells were sorted. As shown in Figure 3D, the ability of CD19hiFcγRIIb−/− B cells for the uptake of FITC-OVA-IC significantly decreased compared with that of CD19hiFcγRIIbhi B cells. So, CD19hiFcγRIIbhi B cells have the enhanced phagocytic capacity and FcγRIIb mediates the uptake of IC by CD19hiFcγIIbhi B cells.
CD19hiFcγRIIbhi B cells exhibit the increased phagocytic capacity. (A) Freshly purified splenic B cells or FcγRIIb−/− B cells were incubated with regulatory DCs for 48 hours at a ratio of 1:10 (DC:B). IL-10 in the supernatants was measured. (B) Phagocytic ability of CD19hiFcγRIIbhi B cells and conventional splenic CD19+ B cells was assessed by flow cytometry of FITC-OVA-IC phagocytosis. Ctrl indicates controls (cells incubated with FITC-OVA-IC at 4°C). (C) The confocal analysis of phagocytic ability of CD19hiFcγRIIbhi B cells and conventional splenic CD19+ B cells. Representative images of immunofluorescence were stained with Hoechst staining (nuclei). Images were acquired by Leica TCS SP2 confocal microscopy under a 20×/0.70 CS objective lens. (D) Phagocytic ability of CD19hiFcγRIIbhi B cells and CD19hiFcγRIIb−/− B cells was assessed by flow cytometry of FITC-OVA-IC phagocytosis. Ctrl indicates controls (cells incubated with FITC-OVA-IC at 4°C). (E-F) Splenic CD19+ B cells, sorted CD19hiFcγRIIbhi B cells, splenic CD19+FcγRIIb−/− B cells, and sorted CD19hiFcγRIIb−/− B cells were stimulated with or without IC for 24 hours, respectively, and then assayed for PGE2 production in the supernatants. Data represent 1 of at least 3 experiments with similar results. (B,D) Numbers indicate the mean fluorescence intensity of test samples. (A,E-F) Data are mean ± SD. **P < .01. NS indicates not significant.
CD19hiFcγRIIbhi B cells exhibit the increased phagocytic capacity. (A) Freshly purified splenic B cells or FcγRIIb−/− B cells were incubated with regulatory DCs for 48 hours at a ratio of 1:10 (DC:B). IL-10 in the supernatants was measured. (B) Phagocytic ability of CD19hiFcγRIIbhi B cells and conventional splenic CD19+ B cells was assessed by flow cytometry of FITC-OVA-IC phagocytosis. Ctrl indicates controls (cells incubated with FITC-OVA-IC at 4°C). (C) The confocal analysis of phagocytic ability of CD19hiFcγRIIbhi B cells and conventional splenic CD19+ B cells. Representative images of immunofluorescence were stained with Hoechst staining (nuclei). Images were acquired by Leica TCS SP2 confocal microscopy under a 20×/0.70 CS objective lens. (D) Phagocytic ability of CD19hiFcγRIIbhi B cells and CD19hiFcγRIIb−/− B cells was assessed by flow cytometry of FITC-OVA-IC phagocytosis. Ctrl indicates controls (cells incubated with FITC-OVA-IC at 4°C). (E-F) Splenic CD19+ B cells, sorted CD19hiFcγRIIbhi B cells, splenic CD19+FcγRIIb−/− B cells, and sorted CD19hiFcγRIIb−/− B cells were stimulated with or without IC for 24 hours, respectively, and then assayed for PGE2 production in the supernatants. Data represent 1 of at least 3 experiments with similar results. (B,D) Numbers indicate the mean fluorescence intensity of test samples. (A,E-F) Data are mean ± SD. **P < .01. NS indicates not significant.
IC can polarize macrophages to secrete high levels of IL-10 and PGE2.28,34 We wonder what roles FcγRIIb plays in the production of immunoregulatory cytokines by CD19hiFcγRIIbhi B cells once stimulated with IC. Interestingly, IL-10, TGF-β, PGE2, and IDO production by CD19hiFcγRIIbhi B cells remained unchanged as compared with splenic CD19+ B cells after stimulation with IC (Figure 3E; and data not shown). Furthermore, the production of PGE2 by CD19hiFcγRIIbhi B cells and splenic CD19+ B cells after stimulation with IC was independent of FcγRIIb (Figure 3F). Therefore, the biologic significance of the enhanced ability of CD19hiFcγRIIbhi B cells to uptake IC via FcγRIIb need to be further elucidated in the future.
IL-10 contributes to the regulatory function of CD19hiFcγRIIbhi B cells
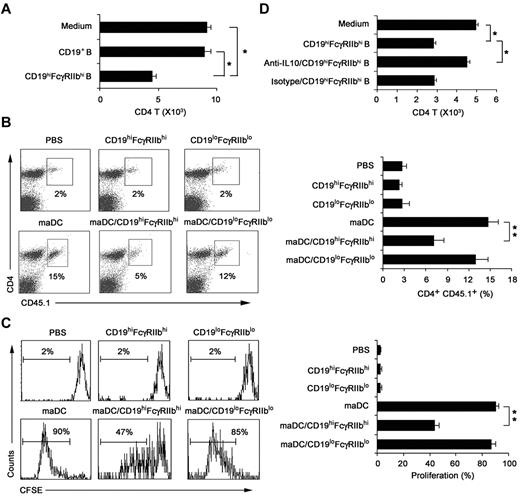
We investigated whether CD19hiFcγRIIbhi B cells have immune regulatory functions and what are the underlying mechanisms. The results showed that CD19hiFcγRIIbhi B cells inhibited proliferation of the activated CD4 T cells more significantly than CD19+ B cells (Figure 4A). To confirm their regulatory effect in vivo, mice transferred with CD45.1 × OT-2 naive CD4 T cells 1 day before being immunized by antigen-loaded maDCs to induce CD4 T-cell proliferation in vivo, and then CD19hiFcγRIIbhi B cells were adoptively transferred to the mice. As shown in Figure 4B and C, CD19hiFcγRIIbhi B cells could also inhibit maDC-induced CD4 T-cell proliferation in vivo. Taken together, CD19hiFcγRIIbhi B cells have a potent ability to suppress T-cell proliferation both in vitro and in vivo, demonstrating that CD19hiFcγRIIbhi B cells are a distinct subset of B cells with regulatory function.
CD19hiFcγRIIbhi B cells inhibit CD4 T-cell proliferation both in vitro and in vivo. (A) The effect of CD19hiFcγIIbhi B cells on in vitro maDC-primed proliferation of activated CD4 T cells was assayed. CD4 T cells were activated by maDCs for 24 hours in the presence of OVA323-339 peptide, and then CD19hiFcγIIbhi B cells were added. Five days later, the cells were collected and double-stained with anti-CD4-allophycocyanin and 7-AAD and counted by flow cytometry. (B-C) The effect of CD19hiFcγIIbhi B cells on in vivo maDC-initiated proliferation of CD4 T cells was assayed. CD4 T cells from OT-2 × CD45.1 F1 hybrid mice were labeled with 5μM CFSE and then transferred intravenously into C57BL/6J mice on day −1. On day 0, antigen-loaded maDCs were transferred subcutaneously into the left footpad. On day 1, CD19hiFcγIIbhi B cells or CD19loFcγIIblo B cells were also transferred into the left footpads. On day 5, mononuclear suspensions from the left popliteal lymph nodes were double-stained with anti–CD4-peridinin chlorophyll protein-Cy5.5 and anti–CD45.1-allophycocyanin for flow cytometry analysis. Representative dot plots showing the percentage of peptide-specific CD4 T cells (CD45.1+) among total CD4 T cells (B, left), and the percentage of peptide-specific CD4 T cells among total CD4 T cells was calculated (n = 3; B, right). Representative histograms showing CFSE expression by the CD4+CD45.1+ T cells (C, left), and the proliferation of CD4+CD45.1+ T cells was analyzed (n = 3; C, right). (D) As described in panel A, neutralizing anti–IL-10 antibody was added into the culture system. Data are mean ± SD. *P < .05. **P < .01.
CD19hiFcγRIIbhi B cells inhibit CD4 T-cell proliferation both in vitro and in vivo. (A) The effect of CD19hiFcγIIbhi B cells on in vitro maDC-primed proliferation of activated CD4 T cells was assayed. CD4 T cells were activated by maDCs for 24 hours in the presence of OVA323-339 peptide, and then CD19hiFcγIIbhi B cells were added. Five days later, the cells were collected and double-stained with anti-CD4-allophycocyanin and 7-AAD and counted by flow cytometry. (B-C) The effect of CD19hiFcγIIbhi B cells on in vivo maDC-initiated proliferation of CD4 T cells was assayed. CD4 T cells from OT-2 × CD45.1 F1 hybrid mice were labeled with 5μM CFSE and then transferred intravenously into C57BL/6J mice on day −1. On day 0, antigen-loaded maDCs were transferred subcutaneously into the left footpad. On day 1, CD19hiFcγIIbhi B cells or CD19loFcγIIblo B cells were also transferred into the left footpads. On day 5, mononuclear suspensions from the left popliteal lymph nodes were double-stained with anti–CD4-peridinin chlorophyll protein-Cy5.5 and anti–CD45.1-allophycocyanin for flow cytometry analysis. Representative dot plots showing the percentage of peptide-specific CD4 T cells (CD45.1+) among total CD4 T cells (B, left), and the percentage of peptide-specific CD4 T cells among total CD4 T cells was calculated (n = 3; B, right). Representative histograms showing CFSE expression by the CD4+CD45.1+ T cells (C, left), and the proliferation of CD4+CD45.1+ T cells was analyzed (n = 3; C, right). (D) As described in panel A, neutralizing anti–IL-10 antibody was added into the culture system. Data are mean ± SD. *P < .05. **P < .01.
To determine whether IL-10 could mediate the inhibition of CD4 T-cell proliferation by CD19hiFcγRIIbhi B cells, we added anti–IL-10 antibody into the activated-CD4 T/CD19hiFcγRIIbhi B coculture system and found that IL-10 blockade could significantly reverse the suppression of the activated CD4 T-cell proliferation by CD19hiFcγRIIbhi B cells (Figure 4D). In addition, IL-10 was found to be involved in the inhibition of T-cell proliferation by CD19hiFcγRIIbhi B cells in vivo (supplemental Figure 5). These results demonstrated that it is IL-10 that mediates the immunosuppressive effects of CD19hiFcγRIIbhi B cells.
Regulatory DC-derived IFN-β is involved in the differentiation of CD19hiFcγRIIbhi B cells
To determine the mechanism of CD19hiFcγRIIbhi B-cell differentiation driven by regulatory DCs, we first tested whether this differentiation process depends on soluble factors released by regulatory DCs or direct cell-cell contact with regulatory DCs. Splenic B cells were separated from regulatory DCs by a transwell system. We found that IL-10 production was significantly decreased but not completely abrogated (Figure 5A), suggesting that CD19hiFcγRIIbhi B-cell differentiation depends on soluble factors from regulatory DCs and cell-cell contact.
IFN-β and CD40L/CD40 interaction is required to induce regulatory B-cell generation driven by regulatory DCs. (A) Splenic naive B cells were cocultured with regulatory DCs either directly or separated from regulatory DCs by transwell system. (B) Splenic naive B cells were cocultured with regulatory DCs in the presence of the indicated neutralizing antibodies (5 μg/mL). (C) Neutralizing anti–IFN-β antibody was added to the B-cell culture system in the presence of regulatory DC supernatants (regulatory DC-SN, collected after 24 hours of culture). And the final concentration of supernatants used was 50%. (D) CD40L expression on regulatory DCs and CD40L−/− regulatory DCs was tested by flow cytometry. (E) Wild-type B cells or CD40−/− B cells were incubated with regulatory DCs or CD40L−/− regulatory DCs. Culture supernatants were collected at 48 hours and assayed for IL-10. (F-G) imDCs or maDCs were cocultured with purified splenic naive B cells in the presence of IFN-β (2 μg/mL) and sCD40L (2 μg/mL) for 48 hours. Culture supernatants were collected and assayed for IL-10 (F). Cells were stained with antibodies to CD19 and CD16/CD32 and analyzed by flow cytometry (G). The DC/B ratio in all these experiments was 1:10. Data are mean ± SD of triplicate wells. *P < .05. **P < .01.
IFN-β and CD40L/CD40 interaction is required to induce regulatory B-cell generation driven by regulatory DCs. (A) Splenic naive B cells were cocultured with regulatory DCs either directly or separated from regulatory DCs by transwell system. (B) Splenic naive B cells were cocultured with regulatory DCs in the presence of the indicated neutralizing antibodies (5 μg/mL). (C) Neutralizing anti–IFN-β antibody was added to the B-cell culture system in the presence of regulatory DC supernatants (regulatory DC-SN, collected after 24 hours of culture). And the final concentration of supernatants used was 50%. (D) CD40L expression on regulatory DCs and CD40L−/− regulatory DCs was tested by flow cytometry. (E) Wild-type B cells or CD40−/− B cells were incubated with regulatory DCs or CD40L−/− regulatory DCs. Culture supernatants were collected at 48 hours and assayed for IL-10. (F-G) imDCs or maDCs were cocultured with purified splenic naive B cells in the presence of IFN-β (2 μg/mL) and sCD40L (2 μg/mL) for 48 hours. Culture supernatants were collected and assayed for IL-10 (F). Cells were stained with antibodies to CD19 and CD16/CD32 and analyzed by flow cytometry (G). The DC/B ratio in all these experiments was 1:10. Data are mean ± SD of triplicate wells. *P < .05. **P < .01.
To further identify which soluble factor(s) was involved in CD19hiFcγRIIbhi B cell differentiation driven by regulatory DCs, IL-6, IP-10, VEGF, and IFN-β, highly secreted by regulatory DCs we observed before, were blocked, respectively, in the coculture system. As shown in Figure 5B, blockade of IFN-β, but not blockade of IL-6, IP-10, and VEGF, led to partial decrease of IL-10 production. Furthermore, neutralizing anti–IFN-β antibody was added into the B-cell culture system in the presence of 50% regulatory DCs' supernatant harvested after culture for 24 hours (50% diffDC-SN), and the decreased production of IL-10 was observed (Figure 5C). The results showed that regulatory DC-derived IFN-β contributes to the differentiation of CD19hiFcγRIIbhi B cells.
CD40L/CD40 interaction is involved in the differentiation of CD19hiFcγRIIbhi B cells programmed by regulatory DCs
We also wondered what membrane molecules involved in the cell-to-cell contact required for the differentiation of CD19hiFcγRIIbhi B cells. The CD40L-CD40 pathway is reported to be crucial for the differentiation of regulatory B cells.35 Regulatory DCs expressed CD40L (Figure 5D). To further determine whether expression of CD40L on regulatory DCs is required for the differentiation of CD19hiFcγRIIbhi B cells, B cells from CD40−/− mice were prepared to coculture with regulatory DCs or CD40L−/− regulatory DCs, and then the IL-10 production in the coculture system was measured (Figure 5E). The absence of CD40L on regulatory DCs or/and CD40 on B cells impaired production of IL-10, indicating that the CD40L-CD40 pathway was involved in the differentiation of CD19hiFcγRIIbhi B cells. Moreover, we added IFN-β plus sCD40L to imDCs or maDCs plus splenic B cells, and then found that imDCs or maDCs plus IFN-β/sCD40L could also induce B cells to differentiate into IL-10–producing B cells with CD19hiFcγIIbhi phenotype (Figure 5F-G). Together, these data suggested that direct regulatory DC-B cell contact in a CD40L-CD40–dependent way, together with the regulatory DC-derived IFN-β, plays a critical role in regulatory DC-programmed differentiation of CD19hiFcγRIIbhi B cells.
Regulatory DCs can induce splenic T1, T2, MZ, and B1 B cells to differentiate into CD19hiFcγIIbhi B cells
Although not defining markers, some markers enriched in regulatory B cells have been reported, such as CD5+CD1dhi and TIM-1+.36,37 So we checked these markers on CD19hiFcγRIIbhi B cells. We found that 16.5% of B cells were CD5+CD1dhi within the CD19hiFcγRIIbhi population, 43.5% of B cells were CD19hiFcγRIIbhi within the CD5+CD1dhi population (Figure 6A), and only approximately 1.1% of CD19hiFcγRIIbhi B cells were TIM-1+ (Figure 6B). These results demonstrated that CD19hiFcγRIIbhi cells are distinguished from TIM-1+ regulatory B cells and not identical with CD5+CD1dhi B cells by their phenotype.
Regulatory DCs induce CD19hiFcγIIbhi regulatory B-cell generation from T1, T2, MZ, and B1 B cells. (A) Splenic B cells cocultured with regulatory DCs were stained for CD19, FcγRIIb, CD5, and CD1d. Cells within the CD19hiFcγRIIbhi B-cell gate were assessed for CD1d and CD5 expression. Cells within the CD5+CD1dhi B-cell gate were assessed for CD19 and FcγRIIb expression. (B) Splenic B cells cocultured with regulatory DCs were stained for CD19, FcγRIIb, and TIM-1. Cells within the CD19hiFcγRIIbhi B-cell gate were assessed for TIM-1 expression. (C) Splenic T1 (CD93+IgMhiCD23lo), T2 (CD93+IgMhiCD23hi), T3 (CD93+IgMloCD23hi), MZ (CD93−CD21hiCD23lo), FO (CD93−CD21loCD23hi), and B1 (B220+CD5+) B cells were sorting-purified, respectively, and cultured with or without regulatory DCs for 48 hours. Culture supernatants were collected for IL-10 assay. (D) Splenic T1, T2, MZ, or B1 B cells cultured with regulatory DCs were stained for CD19 and CD16/CD32 and analyzed by flow cytometry. (E) Splenic T1, T2, MZ, or B1 B cells were cultured with regulatory DCs. CD19+IL-10+ B cells were gated and assessed for FcγIIb expression. Gray lines represent the expression of FcγIIb on T1, T2, MZ, and B1 B cells before coculture. (F) Splenic T1, T2, MZ, or B1 B cells were cultured with regulatory DCs for 48 hours. CD19hiFcγRIIbhi B cells derived from T1, T2, MZ, and B1 B cells were sorted by flow cytometry, respectively. The phagocytic ability of freshly isolated T1, T2, MZ, and B1 B cells and sorted CD19hiFcγIIbhi populations derived from T1, T2, MZ, and B1 B cells was assessed by flow cytometry of FITC-OVA-IC phagocytosis. Numbers in the histogram indicate the mean fluorescence intensity of test samples. Ctrl indicates controls (cells incubated with FITC-OVA-IC at 4°C). (G) Freshly isolated T1, T2, MZ, and B1 B cells or sorted CD19hiFcγIIbhi populations derived from T1, T2, MZ, and B1 B cells were cocultured with activated CD4 T cells (naive peptide-specific CD4 T cells were stimulated by maDCs for 24 hours). In some groups, neutralizing anti–IL-10 antibody was added into the activated CD4 T/B coculture system. After 5 days, the relative number of viable CD4 T cells in each well was detected by flow cytometry. Results are representative of 3 independent experiments. (C,G) Data are mean ± SD of triplicate cells. *P < .05. **P < .01. NS indicates not significant.
Regulatory DCs induce CD19hiFcγIIbhi regulatory B-cell generation from T1, T2, MZ, and B1 B cells. (A) Splenic B cells cocultured with regulatory DCs were stained for CD19, FcγRIIb, CD5, and CD1d. Cells within the CD19hiFcγRIIbhi B-cell gate were assessed for CD1d and CD5 expression. Cells within the CD5+CD1dhi B-cell gate were assessed for CD19 and FcγRIIb expression. (B) Splenic B cells cocultured with regulatory DCs were stained for CD19, FcγRIIb, and TIM-1. Cells within the CD19hiFcγRIIbhi B-cell gate were assessed for TIM-1 expression. (C) Splenic T1 (CD93+IgMhiCD23lo), T2 (CD93+IgMhiCD23hi), T3 (CD93+IgMloCD23hi), MZ (CD93−CD21hiCD23lo), FO (CD93−CD21loCD23hi), and B1 (B220+CD5+) B cells were sorting-purified, respectively, and cultured with or without regulatory DCs for 48 hours. Culture supernatants were collected for IL-10 assay. (D) Splenic T1, T2, MZ, or B1 B cells cultured with regulatory DCs were stained for CD19 and CD16/CD32 and analyzed by flow cytometry. (E) Splenic T1, T2, MZ, or B1 B cells were cultured with regulatory DCs. CD19+IL-10+ B cells were gated and assessed for FcγIIb expression. Gray lines represent the expression of FcγIIb on T1, T2, MZ, and B1 B cells before coculture. (F) Splenic T1, T2, MZ, or B1 B cells were cultured with regulatory DCs for 48 hours. CD19hiFcγRIIbhi B cells derived from T1, T2, MZ, and B1 B cells were sorted by flow cytometry, respectively. The phagocytic ability of freshly isolated T1, T2, MZ, and B1 B cells and sorted CD19hiFcγIIbhi populations derived from T1, T2, MZ, and B1 B cells was assessed by flow cytometry of FITC-OVA-IC phagocytosis. Numbers in the histogram indicate the mean fluorescence intensity of test samples. Ctrl indicates controls (cells incubated with FITC-OVA-IC at 4°C). (G) Freshly isolated T1, T2, MZ, and B1 B cells or sorted CD19hiFcγIIbhi populations derived from T1, T2, MZ, and B1 B cells were cocultured with activated CD4 T cells (naive peptide-specific CD4 T cells were stimulated by maDCs for 24 hours). In some groups, neutralizing anti–IL-10 antibody was added into the activated CD4 T/B coculture system. After 5 days, the relative number of viable CD4 T cells in each well was detected by flow cytometry. Results are representative of 3 independent experiments. (C,G) Data are mean ± SD of triplicate cells. *P < .05. **P < .01. NS indicates not significant.
To identify the B-cell subset(s) from which IL-10–producing CD19hiFcγRIIbhi B cells were differentiated, sorting-purified splenic T1, T2, T3, FO, MZ, or B1 B cells38 (supplemental Figure 6) were cultured with regulatory DCs. We found that IL-10–producing CD19hiFcγRIIbhi B cells were derived from T1, T2, MZ, and B1 B cells, but not T3 and FO B cells (Figure 6C-D). And CD19+IL-10+ T1, T2, MZ, and B1 B cells expressed higher level of FcγRIIb compared with that of freshly sorted T1, T2, MZ, and B1 B cells, respectively (Figure 6E). Furthermore, these sorted CD19hiFcγRIIbhi T1, T2, MZ, and B1 B cells had enhanced phagocytic capacity (Figure 6F) and significantly suppressed the activated CD4 T-cell proliferation via IL-10 (Figure 6G) compared with those of freshly sorted T1, T2, MZ, and B1 B cells, respectively. Together, these data indicate that regulatory DCs preferentially induce specific B-cell subsets, including T1, T2, MZ, and B1 B cells, to differentiate into IL-10–producing CD19hiFcγIIbhi B cells.
Phenotypic and functional identification of a natural counterpart of CD19hiFcγIIbhi B cells in vivo
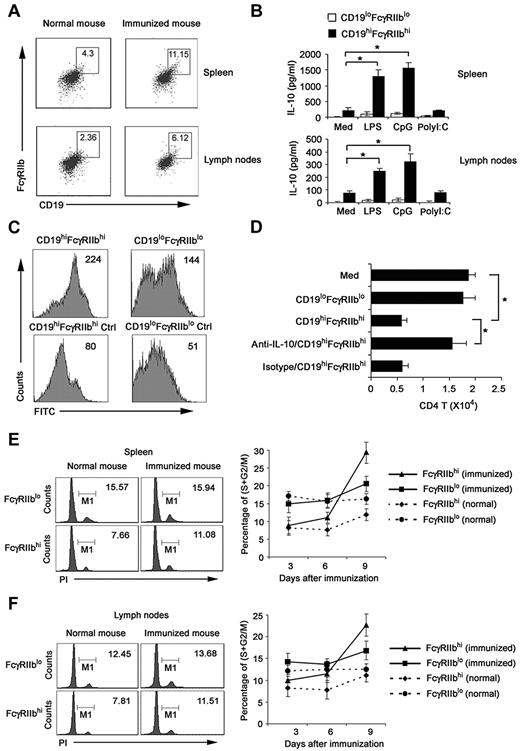
To improve the biologic relevance of the regulatory CD19hiFcγRIIbhi B cells, we investigated whether the natural counterpart of CD19hiFcγRIIbhi B cells exists in vivo. We analyzed CD19+ cells in spleen and lymph nodes of DO11.10 OVA323-339-specific TCR transgenic × C57BL/6J F1 hybrid mice after immunization with or without 40 μg OVA plus 10 μg CpG ODN on the basis of the phenotype of CD19hiFcγRIIbhi. As shown in Figure 7A and B, cells with a CD19hiFcγRIIbhi phenotype represented 4.30% of splenic CD19+ B cells and 2.36% of lymph nodes from normal mice; and they represented 11.15% of splenic CD19+ B cells and 6.12% of lymph nodes from immunized mice. CD19hiFcγRIIbhi B cells, directly sorted from spleen and lymph nodes of immunized mice, secreted high IL-10 with or without LPS and CpG ODN stimulation, which resembled the cytokine profile of regulatory CD19hiFcγRIIbhi B cells induced by regulatory DCs in vitro (Figure 7B). Furthermore, we found that these sorted CD19hiFcγRIIbhi B cells had enhanced phagocytic capacity compared with that of CD19loFcγIIblo B cells (Figure 7C). Moreover, these cells significantly suppressed the activated CD4 T-cell proliferation via IL-10 (Figure 7D), similar to the functional characteristics of CD19hiFcγRIIbhi B cells we identified in vitro.
Identification of the natural counterpart of CD19hiFcγIIbhi B cells in vivo. Splenocytes and lymph node cells enriched with anti-CD19 magnetic beads were double-stained. CD19hiFcγIIbhi B cells and CD19loFcγIIblo B cells were sorted by flow cytometry. (A) Numbers in plot regions indicate the percentage of CD19hiFcγIIbhi in the gated CD19+ B cells. (B) CD19hiFcγIIbhi and CD19loFcγIIblo were stimulated with 200 ng/mL LPS, 2 μg/mL CpG ODN, or 2 μg/mL poly I:C for 24 hours; then IL-10 production was measured by ELISA. (C) The phagocytic ability of sorted CD19hiFcγIIbhi and CD19loFcγIIblo B cells was assessed by flow cytometry of FITC-OVA-IC phagocytosis. Numbers in the histogram indicate the mean fluorescence intensity of test samples. Ctrl indicates controls (cells incubated with FITC-OVA-IC at 4°C). (D) CD19hiFcγIIbhi or CD19loFcγIIblo B cells were cocultured with activated CD4 T cells. In some groups, anti–IL-10 antibody was added into the activated CD4 T/B coculture system. After 5 days, the relative number of viable CD4 T cells in each well was detected by flow cytometry. (E-F) DO11.10 OVA323-339-specific TCR transgenic × C57BL/6J F1 hybrid mice were killed on days 3, 6, and 9 after immunization with 40 μg OVA plus 10 μg CpG ODN. CD19+ cells were sorted from spleen (E) and lymph nodes (F). Left: cell-cycle status of FcγIIbhi cells analyzed by propidium iodide (numbers indicate percentage of FcγIIbhi and FcγIIblo cells, respectively, in S/M/G2). Right: percentage of proliferating B cells in FcγIIbhi or FcγIIblo cell population of different phases of immune response by cycle analysis. (B,D) Data are mean ± SD of triplicate cells. *P < .05.
Identification of the natural counterpart of CD19hiFcγIIbhi B cells in vivo. Splenocytes and lymph node cells enriched with anti-CD19 magnetic beads were double-stained. CD19hiFcγIIbhi B cells and CD19loFcγIIblo B cells were sorted by flow cytometry. (A) Numbers in plot regions indicate the percentage of CD19hiFcγIIbhi in the gated CD19+ B cells. (B) CD19hiFcγIIbhi and CD19loFcγIIblo were stimulated with 200 ng/mL LPS, 2 μg/mL CpG ODN, or 2 μg/mL poly I:C for 24 hours; then IL-10 production was measured by ELISA. (C) The phagocytic ability of sorted CD19hiFcγIIbhi and CD19loFcγIIblo B cells was assessed by flow cytometry of FITC-OVA-IC phagocytosis. Numbers in the histogram indicate the mean fluorescence intensity of test samples. Ctrl indicates controls (cells incubated with FITC-OVA-IC at 4°C). (D) CD19hiFcγIIbhi or CD19loFcγIIblo B cells were cocultured with activated CD4 T cells. In some groups, anti–IL-10 antibody was added into the activated CD4 T/B coculture system. After 5 days, the relative number of viable CD4 T cells in each well was detected by flow cytometry. (E-F) DO11.10 OVA323-339-specific TCR transgenic × C57BL/6J F1 hybrid mice were killed on days 3, 6, and 9 after immunization with 40 μg OVA plus 10 μg CpG ODN. CD19+ cells were sorted from spleen (E) and lymph nodes (F). Left: cell-cycle status of FcγIIbhi cells analyzed by propidium iodide (numbers indicate percentage of FcγIIbhi and FcγIIblo cells, respectively, in S/M/G2). Right: percentage of proliferating B cells in FcγIIbhi or FcγIIblo cell population of different phases of immune response by cycle analysis. (B,D) Data are mean ± SD of triplicate cells. *P < .05.
We also enriched CD19+ cells from the spleen and lymph nodes at various times after immunization and analyzed the proliferation of FcγRIIbhi B and FcγRIIblo B cells by cell-cycle analysis. In both spleen and lymph nodes of normal mice, the percentage of FcγRIIbhi B cells in the S/M/G2 phages of cell cycle was less than that of FcγRIIblo B cells (Figure 7E-F). However, in mice that received OVA and CpG ODN, FcγRIIbhi B cells underwent stronger proliferation than FcγRIIblo B cells, as indicated by the greater increase in the proportion of cells undergoing S/M/G2 cycling. These results indicated that a subset of regulatory B cells with similar phenotype and functions to that of CD19hiFcγRIIbhi B cells identified in vitro do exist in mouse spleen and lymph nodes. Interestingly, the number of this subset increases after immunization, suggesting feedback negative regulation of immune response through induction of regulatory B cells may exist.
Discussion
Up to now, various regulatory B cells with different phenotypes have been reported, such as CD1dhiCD5+ B cells and TIM-1+ B cells.36,37,39 In this study, we identified a new subset of IL-10-producing regulatory B cells with high CD19 and FcγIIb expression. These CD19hiFcγRIIbhi B cells are distinguished from TIM-1+ B cells because only approximately 1.1% of CD19hiFcγRIIbhi B cells are positive for TIM-1. CD19hiFcγIIbhi B cells share surface markers with CD1dhiCD5+ B cells, such as high CD1d expression, but several differences exist between these 2 B-cell subsets. In particular, majority of CD19hiFcγIIbhi B cells are negative for CD5 and cellular origins of these 2 B-cell subsets were different. CD1dhiCD5+ B cells do not express CD93 or CD23, suggesting that they are unlikely to belong to T1 and T2-MZ precursor B-cell subsets,37 whereas CD19hiFcγRIIbhi B cells can emerge from splenic T1 (CD93+IgMhiCD23lo) and T2 (CD93+IgMhiCD23hi) B-cell subsets. All the differences showed that CD19hiFcγIIbhi B cells and CD1dhiCD5+ B cells are not the same subset of regulatory B cells. FcγIIb, the well-known unique inhibitory FcγR expressed on B cells, has never been reported as a surface marker of regulatory B cells. Th-2 cytokines, such as IL-10 or TGF-β, can up-regulate the expression of inhibitory Fc receptor on innate immune effector cells.31,40 However, blockade of IL-10, TGF-β, IFN-β, IP-10, or IL-6 in the regulatory DC/B coculture system could not reduce FcγRIIb expression on CD19hiFcγRIIbhi B cells (data not shown). Moreover, we demonstrate that CD19hiFcγIIbhi B cells have enhanced phagocytic capacity than CD19+ B cells, and FcγRIIb mediates the uptake of IC by CD19hiFcγIIbhi B cells, indicating that CD19hiFcγRIIbhi B cells may exert its negative regulatory function by attenuating the IC-mediated activation of immune response and provide potential new approach for treating IC-mediated autoimmune diseases. Therefore, which factor(s) is responsible for high expression of FcγRIIb on CD19hiFcγRIIbhi B cells and what biologic significance of high expression of FcγRIIb on regulatory B cells need to be further investigated.
Functionally, regulating T-cell function is the hallmark of regulatory B cells, but the molecular mechanisms of regulatory B cells are still poorly defined so far. There are at least 3 kinds of mechanisms that may account for regulatory properties of B cells, including the production of IL-10, the ability to suppress the activated CD4 T-cell proliferation, or inducing Treg cell generation.2,41,42 Although a high level of IL-10 was secreted by CD19hiFcγRIIbhi B cells in our experiments, IL-10 did not promote generation of CD4+CD25+Foxp3+ Treg cells (data not shown). In addition, CD19hiFcγRIIbhi B cells did not inhibit Th1 cytokine secretion of CD4 T cells activated by maDCs (data not shown). Interestingly, we found that CD19hiFcγRIIbhi B cells could potently inhibit the proliferation of activated CD4 T cells via IL-10. These results show that CD19hiFcγRIIbhi B cells display the inhibitory function by directly secreting IL-10, instead of indirectly inducing Treg cells, functionally confirming that CD19hiFcγRIIbhi B cells are a distinct subset of regulatory B cells.
Mechanisms for regulatory B-cell differentiation have not yet been fully identified. Our results demonstrate that CD40L/CD40 interaction between regulatory DC/B cells is required for optimal IL-10 production by CD19hiFcγRIIbhi B cells. Furthermore, regulatory DC-derived IFN-β is essential for the differentiation of CD19hiFcγRIIbhi B cells. Our results that B cells cocultured with imDCs or maDCs in presence of sCD40L and IFN-β also exhibited CD19hiFcγRIIbhi phenotype further confirm that CD40L and IFN-β are important for the differentiation of CD19hiFcγRIIbhi B cells. IFN-β is the major drug for treatment of multiple sclerosis patients and experimental autoimmune encephalomyelitis (EAE) mouse models. The molecular mechanisms responsible for the beneficial effect of IFN-β in multiple sclerosis/EAE are not fully elucidated. Putative mechanisms include alterations of the Th1/Th2 balance, induction of regulatory T cells, negative regulation of Th17 differentiation, suppression of maDC migration, and elimination of maDCs.43,44 Recent data indicate that IFN-β is effective in reducing EAE symptoms induced by Th1 cells, which is correlated with increased IL-10 production by splenocytes.45 More recently, induction of IL-10 secretion from human B cells by IFN-β has been reported.46 It was also reported that IFN-β helped to control neonatal acute inflammation by stimulating the secretion of IL-10 by neonatal B cells.47 Our work, together with others, demonstrated that IFN-β may play important roles in driving differentiation of IL-10–producing regulatory B cells; thus, IFN-β–driven differentiation of regulatory B cells might represent a possible molecular mechanism for the therapeutic effect of IFN-β in multiple sclerosis/EAE.
Regulatory IL-10-producing B cells have been shown to attenuate autoimmune diseases in several mouse models. For example, adoptive transfer of CD1dhiCD5+ IL-10–producing B cells into B cell–deficient mice suppressed T cell–mediated inflammatory response in a contact hypersensitivity mode 1.37 Transfer of regulatory T2-like-B cells is found to be effective in the suppression of lupus in MRL/lpr mice via an IL-10–dependent mechanism.35 In addition, a subset of human B cells with CD19+CD24hiCD38hi phenotype was recently found to possess regulatory activity in healthy persons but is functionally impaired in systemic lupus erythematosus patients.9 Thus, investigating the preventive or pathogenic roles of CD19hiFcγIIbhi B cells in different disease models and identification of CD19hiFcγIIbhi B cells in humans that similarly regulate inflammatory responses may add insights to the mechanisms of immune disorders and provide therapeutic approaches for treating autoimmune diseases.
In conclusion, we demonstrate that regulatory DCs could program splenic T1, T2, MZ, and B1 B cells to differentiation into CD19hiFcγIIbhi B cells, a novel regulatory B subset with high IL-10 production. Such B cells can regulate immune response by inhibiting CD4 T-cell proliferation. Regulatory DC-derived IFN-β and CD40L/CD40 interaction between regulatory DC/B are required for the differentiation of CD19hiFcγRIIbhi regulatory B cells programmed by regulatory DCs. So, regulatory DCs may have strategy to regulate immune responses by, at least partially, inducing generation of regulatory B cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Taoyong Chen for critical reading of the manuscript and Drs Xiongfei Xu, Xingguang Liu, and Yan Gu for excellent technical assistance.
This work was supported by the National High-Biotech Program (grants 2012AA020901 and 2012ZX10002014), the National Natural Science Foundation of China (grants 30972704, 81001308, and 30731160623), and a Foundation for the Author of National Excellent Doctoral Dissertation of China (grant 201180).
Authorship
Contribution: L.Q., C.Q., Y.C., Y. Bai, and Y. Bao performed the experiments; and X.C., L.Q., C.Q., and L.L. designed the experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xuetao Cao, National Key Laboratory of Medical Immunology & Institute of Immunology, Second Military Medical University, 800 Xiangyin Road, Shanghai 200433, China; e-mail: caoxt@immunol.org.
References
Author notes
L.Q. and C.Q. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal