Abstract
Ly49-mediated recognition of MHC-I molecules on host cells is considered vital for natural killer (NK)–cell regulation and education; however, gene-deficient animal models are lacking because of the difficulty in deleting this large multigene family. Here, we describe NK gene complex knockdown (NKCKD) mice that lack expression of Ly49 and related MHC-I receptors on most NK cells. NKCKD NK cells exhibit defective killing of MHC-I–deficient, but otherwise normal, target cells, resulting in defective rejection by NKCKD mice of transplants from various types of MHC-I–deficient mice. Self–MHC-I immunosurveillance by NK cells in NKCKD mice can be rescued by self–MHC-I–specific Ly49 transgenes. Although NKCKD mice display defective recognition of MHC-I–deficient tumor cells, resulting in decreased in vivo tumor cell clearance, NKG2D- or antibody-dependent cell-mediated cytotoxicity–induced tumor cell cytotoxicity and cytokine production induced by activation receptors was efficient in Ly49-deficient NK cells, suggesting MHC-I education of NK cells is a single facet regulating their total potential. These results provide direct genetic evidence that Ly49 expression is necessary for NK-cell education to self–MHC-I molecules and that the absence of these receptors leads to loss of MHC-I–dependent “missing-self” immunosurveillance by NK cells.
Introduction
Natural killer (NK) cells are a unique and integral part of the innate immune system. Persons without NK cells or lacking normal NK-cell activity experience persistent and life-threatening infections of normally innocuous viruses.1,2 NK cells are able to distinguish normal cells from unhealthy cells by monitoring surface expression of a variety of molecules. The most well-characterized self-recognition system involves surveillance of host class I MHC (MHC-I) molecules, a process initially described by the missing-self hypothesis.3 This hypothesis states that target cells lacking normal expression of self–MHC-I molecules because of viral infection or transformation are specifically recognized and lysed by NK cells.
Several surface receptors are known to activate or inhibit the function of NK cells. Numerous NK-cell receptors such as the NKG2D, CD94/NKG2, NKR-P1, and Ly49 families of C-type lectin-like transmembrane proteins are encoded in a region on mouse chromosome 6 termed the NK gene complex (NKC). The most well-characterized MHC-I–specific receptors on mouse NK cells are the Ly49, which represent the mouse functional equivalents of the human killer-cell Ig-like receptor family. The Klra (Ly49) gene family is highly polymorphic, with significant variation in gene content between mouse strains.4 The Klra haplotype of 129-strain mice contains 19 genes that encode 3 activating and 9 inhibitory receptors; the remaining genes are pseudogenes.5
Ly49 receptors are divided into 2 main groups: activating and inhibitory receptors. Activating Ly49 receptors have been implicated in direct recognition of virally encoded MHC-I–like molecules on infected target cells.6 Most inhibitory Ly49 receptors recognize specific MHC-I molecules, resulting in some Ly49 that can bind “self” MHC-I and some that cannot. Rare self–MHC-I receptor-negative NK cells display hyporesponsive cytotoxic and cytokine potential in response to activation signals.7,8 Conversely, the greater the number of self–MHC-I receptors expressed by NK cells, the greater the response after activation.9 Therefore, in addition to target cell differentiation by mature NK cells, Ly49 molecules are hypothesized to also be required during NK-cell development, specifically for education to self-MHC expression. We have generated a mutant mouse strain in which the expression of Ly49 molecules is absent on most NK cells. In this study, we assess the development and the function of NK cells in Ly49-deficient mice and show that Ly49 receptors are directly responsible for NK-cell education and immunosurveillance to self–MHC-I in vivo.
Methods
Mice
C57BL/6 (B6), 129S1, and B2m−/− on the B6 background mice were purchased from The Jackson Laboratory. H2Kb−/−, H2Db−/−, and H2Kb−/−Db−/− mice on a B6 background were purchased from Taconic Farms. The generation of Ly49Atg (B6 background), Ly49Gtg (B6 background), Ly49Itg (FVB background), Rae-1ϵtg (B6 background), and B6.Ly49129 mice was previously described.10-14 Ly49Itg mice were kindly provided by Dr Michael Bennett (University of Texas, Dallas). NKCKD mice were generated by targeting Klra15 in Klra17lox/wt (Ly49qlox/wt) R1 embryonic stem (ES) cells. Neomycin-resistant ES cells were electroporated with CMV-Cre plasmid and were selected by PCR with the use of the following primers: 5′-GGCTTGAAGACTCAGGGTTTTGCTC and 5′-TCTTGACCCTTGATTGTCCTCAGGC. Homozygous Klra15-targeted mice were determined with an additional PCR for the absence of Klra17wt with the use of the following primers: 5′-CCTAAAAGTAATTGCTGTGACTATT and 3′-CTTTCTAACTAGCTAACAACAG. B6. NKCKD mice were produced by backcrossing NKCKD mice to the B6 background for 10 generations and selecting for the 129-specific Klra22 (Ly49v) gene as described,14 followed by single nucleotide polymorphism analysis with the use of an Illumina Beadstation 500G mouse medium density linkage panel (The Center for Applied Genomics–Sick Kids Hospital). The genome of B6.NKCKD mice is of B6 origin except for a region containing the NKC on chromosome 6 spanning nucleotides 127, 954, 449-138, 203, 431 deduced from single nucleotide polymorphism markers rs3681620 and rs13479071, respectively. Ly49 transgenes were introduced by breeding to B6.NKCKD mice. Ly49-transgene genotyping was performed as described.10-12 Ly49 transgene–positive, NKCKD heterozygous mice were then bred to homozygosity for the NKCKD chromosome. Third-generation mice homozygous for the Ly49 transgene were used for experiments. Ly49 expression was tested with Ly49 mAb to verify Ly49 transgenes and NKCKD backgrounds. All breeding and manipulations performed on animals were in accordance with university guidelines and approved by the University of Ottawa Animal Ethics Committee.
Cells
RMA-S cells were stably transfected with Rae-1β-pEF6 vector with the use of Lipofectamine 2000 (Invitrogen). The C1498 MHC-I–negative variant was produced by 3 rounds of ethylmethane-sulfonate3 (Sigma-Aldrich) treatment and sorting β2mlo cells. In parallel, C1498 cells were treated with EMS but not sorted to generate C1498 MHC-I–positive cells. Adherent lymphokine-activated killer (LAK) cells were generated as previously described.15
Semiquantitative RT-PCR and genomic PCR
RNA was isolated from LAK cells with the use of TRIzol reagent (Invitrogen). cDNA was synthesized with the Superscript First-Strand cDNA synthesis kit (Invitrogen). Semiquantitative RT-PCR was performed on serial 5-fold dilutions of cDNA with the use of gene-specific primers.5,16-18 Semiquantitative PCR to verify the presence of the concatemer was performed on genomic DNA with the use of the following primers as depicted in Figure 1D: 5′ forward out, 5′-CCAGTCCTAAATCTGATAGG; 5′ forward in, 5′-GCTGGCCGTTATCATGTTGA; 5′ reverse, 3′-ACAGTCTCCCTTGCCTGTAG; 3′ forward, 5′-GGTATGGTTTAACCCACTCA; 3′ reverse in, 3′-GGTGCCTCGAGTGGTTTCTA; and 3′ reverse out, 3′-TTCTGTAGCCAGCTTCTCTG.
Antibodies and flow cytometry
Anti-CD49b, anti-TCRβ, anti-CD16, anti–LFA-1, anti-CD44, anti-CD62L, anti-CD2, anti-CD11b, anti-CD122, anti-NKp46, anti-CD43, anti-CD27, anti-CD69, anti-Thy1.2, anti-NKG2D, anti-CD94, anti-Ly49A/D, anti-Ly49C/I/F/H, anti-CD107a, anti–granzyme B, anti-KLRG1, anti–IFN-γ, rIgG1, and rIgG2b were purchased from eBioscience. Anti-NKG2A/C/E, anti-Ly49D, anti-Ly49G, and anti-β2m were purchased from BD Biosciences. Anti-B220 was purchased from Miltenyi Biotec. Anti–granzyme A was purchased from Santa Cruz Biotechnology. Anti-CRACC was a gift from Dr André Veillette (Clinical Research Institute of Montreal). Fc receptors were blocked with rat serum (Sigma-Aldrich), and dead cells were excluded with propidium iodide (BD Biosciences). Flow cytometry was performed on a CyAN-ADP with the use of Summit Version 4.3 software (Beckman Coulter). Data were analyzed with Kaluza Version 1.2 software.
In vitro NK-cell assays
Cytotoxicity experiments were performed as previously described.15 ConA blast target cells were prepared by culturing 2.5 × 106 cells/mL of splenocytes for 2 days in cRPMI containing 5 μg/mL ConA (Sigma-Aldrich). To measure production of IFN-γ, 1 × 106 splenocytes isolated from poly(I:C)–treated mice (150 μg; 18 hours earlier) were incubated for 5 hours with target cells (1:1 ratio) or in plates coated with 1 μg/mL anti-NKp46 antibody. GolgiPlug (BD Biosciences) was added to the wells after 1 hour. Cells were stained for CD49b, TCRβ, and for Ly49I, Ly49O, Ly49V, and NKG2A; fixed; permeabilized with the Cytofix/Cytoperm kit (BD Biosciences); and then stained for IFN-γ or with an isotype-matched control mAb. For antibody-dependent cellular cytotoxicity (ADCC), after 51Cr-labeling, target cells were incubated in the presence of 10 μg/mL anti-Thy1.2 mAb for 30 minutes and then washed before use. For blocking NKG2D-ligand interactions, anti-NKG2D mAb was added directly to wells containing effector cells for a final concentration of 10 μg/mL and incubated 20 minutes after which time targets were added.
In vivo NK-cell assays
The splenocyte rejection assay was performed as previously described.14 NK cells were depleted by intraperitoneal injection of 40 μL of anti–asialo-GM1 (Wako Chemicals) 2 days before poly(I:C) injection. MHC-I–positive and MHC-I–negative tumor cells were differentially labeled with 0.5μM and 5μM of CFSE (Invitrogen). A mixture of 1 × 106 cells of each type was injected intraperitoneally into B6-background recipient mice. After 6 hours (C1498 and C1498 MHC-I) or 18 hours (RMA and RMA-S), peritoneal cells were harvested and analyzed for the presence of CFSE-labeled tumor cells by flow cytometry.
Southern blot analyses
Restriction fragment-length polymorphism analysis was performed on DNA isolated from the thymus and digested with EcoRI, KpnI, or BamHI. Blots were probed with a mixture of Klra7 (Ly49g) and Klra5 (Ly49e) cDNAs. The presence of the concatemer was discovered by probing with a fragment of the Klra15-targeting construct.
Genome analysis
Oligonucleotides for tiling microarrays were designed with a custom Perl script on the Repeatmasked (Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996-2010; http://www.repeatmasker.org) 129S6 genomic sequence spanning the Klra cluster and flanking genes (693 835 bp). In addition, a 5-Mbp Repeatmasked region, centered on the B6 Klra cluster (National Center for Biotechnology Information 37:chr6:127300000-132300000), was also used to design tiling array probes. All 60-bp probes were tiled at approximately every 15 bp, and the custom arrays were subsequently generated by NimbleGen. Hybridizations were performed by the functional genomic platform at the Institute for Research in Immunology and Cancer. Genomic DNA from wild-type (WT) and NKCKD mice were labeled with cyanine 5 (Cy5)– and Cy3-labeled 9mers (Trilink Technologies) with the use of 3′-5′ exo-Klenow fragment (New England Biolabs), and a dye swap was performed. Two-color hybridizations were performed with 6 μg of Cy5-labeled and 6 μg of Cy3-labeled gDNA with the use of the NimbleGen hybridization kit as recommended by the manufacturer. Arrays were scanned at 5-μm resolution with the use of a GenePix4000B scanner (Molecular Devices). The data discussed in this article have been deposited in Gene Expression Omnibus from the National Center for Biotechnology Information and are accessible through GEO Series accession no. GSE38372 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE38372). Data from scanned images were extracted and analyzed with NimbleScan 2.5 extraction software (NimbleGen Systems).
Statistical analysis
Statistical significance was determined by a 2-tailed unpaired Student t test with a cutoff P value of .05. Data are presented as ± SD.
Results
Generation of mice containing a concatemer upstream of the Klra gene cluster
In an effort to delete the Klra gene cluster, the Klra15 (Ly49o) promoter region was targeted with a floxed PGK-Neo selection cassette in ES cells carrying a Klra17lox allele previously used to make Ly49Q-deficient mice (Figure 1A).19 After generating mice from double-targeted, CMV-Cre electroporated ES cells, it was discovered that the long-range deletion was unsuccessful because of nonhomologous recombination of the Klra15-targeting construct, resulting in a total gain of DNA sequence. When a portion of the Klra15-targeting construct was used to probe digested genomic DNA from WT and mutant mice, a 10- to 12-kb DNA fragment hybridized very strongly only in mutant mouse DNA (Figure 1B). Array comparative genomic hybridization analyses confirmed that the gained sequence originated from the Klra15-targeting construct (Figure 1C). These data suggest that the targeting construct integrated as a concatemer. The location of the concatemer was verified to be at the intended recombination site of the Klra15 promoter region by semiquantitative PCR analysis of genomic DNA with the use of primers either within or outside the concatemerized targeting construct (Figure 1D).
The Klra gene cluster of Klra15-targeted (mutant) mice contains a concatemer insertion of the Klra15-targeting construct. (A) A schematic representation of the natural killer gene complex (NKC) and the targeting-construct produced for the Klra15 gene. (B) Southern blot analysis of wild-type (WT) and mutant thymus DNA digested with EcoRI, KpnI, or BamHI and probed with the Klra15-targeting construct depicted in panel A. (C) Array comparative genomic hybridization profile of the Klra cluster in mutant mice with a blow-up of the Klra15 gene (below). The x-axis represents the genomic region tiled on the microarrays, and the y-axis Shows differences in copy number between WT and mutant genomic DNA (log2 ratio [MT/WT]). Positive values indicate regions showing copy number increases in the NKC knockdown (NKCKD) genome. (D) PCR analysis of the Klra15-targeting construct copy number in mutant genomic DNA. Data are representative of ≥ 3 similar experiments, except for array comparative genomic hybridization analysis, which was performed twice. These experiments were performed with mice on the 129S1 background.
The Klra gene cluster of Klra15-targeted (mutant) mice contains a concatemer insertion of the Klra15-targeting construct. (A) A schematic representation of the natural killer gene complex (NKC) and the targeting-construct produced for the Klra15 gene. (B) Southern blot analysis of wild-type (WT) and mutant thymus DNA digested with EcoRI, KpnI, or BamHI and probed with the Klra15-targeting construct depicted in panel A. (C) Array comparative genomic hybridization profile of the Klra cluster in mutant mice with a blow-up of the Klra15 gene (below). The x-axis represents the genomic region tiled on the microarrays, and the y-axis Shows differences in copy number between WT and mutant genomic DNA (log2 ratio [MT/WT]). Positive values indicate regions showing copy number increases in the NKC knockdown (NKCKD) genome. (D) PCR analysis of the Klra15-targeting construct copy number in mutant genomic DNA. Data are representative of ≥ 3 similar experiments, except for array comparative genomic hybridization analysis, which was performed twice. These experiments were performed with mice on the 129S1 background.
Ly49, NKG2, CD94, and KLRI expression is silenced in NKCKD mice
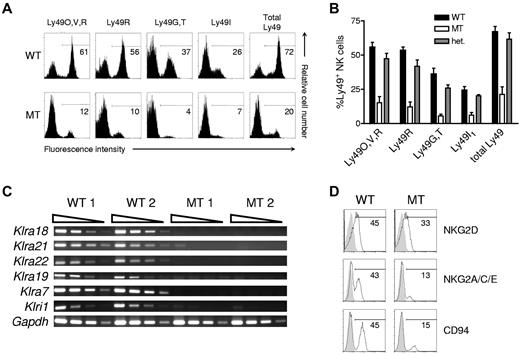
Initial analysis of Ly490 surface expression showed significant loss of positively staining NK cells from homozygous mutant mice relative to WT littermate controls (Figure 2A). Unexpectedly, additional staining of NK cells from Klra15-targeted mice with mAb specific for Ly49V, Ly49R, Ly49G, Ly49T, and Ly49I, individually or as a cocktail, showed that these subsets were also decreased by 70%-80% (Figure 2A-B). These results were surprising, because no other Klra genes were intentionally targeted during ES cell manipulation. Interestingly, heterozygous mice have near normal Ly49+ NK cells subsets, although a completely stochastic model of Ly49 acquisition would predict intermediate numbers (Figure 2B). Klra down-regulation was detectable at the level of mRNA (Figure 2C). Thus, the mechanism of Ly49 down-regulation may involve multilocus transcriptional silencing.
Klra15-targeted mice show a drastic reduction in telomeric NKC gene expression. (A) NK cells (DX5+TCRβ−) from the spleens of WT or mutant mice were analyzed for Ly49 expression with mAbs 4E5 (which binds Ly49O/V/R in mice with a 129-strain Klra gene cluster), 12A8 (Ly49R), 4D11 (Ly49G/T), and 14B11 (Ly49I) individually or as a cocktail (Total Ly49). The percentage of NK cells positively stained is indicated. (B) The mean ± SD percentage of different splenic NK-cell subsets present in WT, mutant, and heterozygous littermates (n = 5) is shown graphically. (C) Semiquantitative RT-PCR of cDNA for the indicated genes was performed on total RNA obtained from lymphokine-activated killer cells prepared from 2 individual WT or mutant mouse spleens. (D) The expression of NKG2/CD94 family of receptors on splenic NK cells from WT or mutant littermates. Data are representative of ≥ 3 similar experiments. These experiments were performed with mice on the 129S1 background.
Klra15-targeted mice show a drastic reduction in telomeric NKC gene expression. (A) NK cells (DX5+TCRβ−) from the spleens of WT or mutant mice were analyzed for Ly49 expression with mAbs 4E5 (which binds Ly49O/V/R in mice with a 129-strain Klra gene cluster), 12A8 (Ly49R), 4D11 (Ly49G/T), and 14B11 (Ly49I) individually or as a cocktail (Total Ly49). The percentage of NK cells positively stained is indicated. (B) The mean ± SD percentage of different splenic NK-cell subsets present in WT, mutant, and heterozygous littermates (n = 5) is shown graphically. (C) Semiquantitative RT-PCR of cDNA for the indicated genes was performed on total RNA obtained from lymphokine-activated killer cells prepared from 2 individual WT or mutant mouse spleens. (D) The expression of NKG2/CD94 family of receptors on splenic NK cells from WT or mutant littermates. Data are representative of ≥ 3 similar experiments. These experiments were performed with mice on the 129S1 background.
The residual Ly49+ NK cells from Klra15-targeted mice lost on average ∼ 50% of Ly49 receptor coexpression (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Therefore, NK cells from mutant mice are mostly Ly49− and the remaining Ly49+ cells have a greatly decreased probability of expressing multiple Ly49. Restriction fragment-length polymorphism analysis of WT and mutant mouse genomic DNA with Klra cDNAs as probes resulted in highly similar patterns (supplemental Figure 1B), suggesting that Ly49 down-regulation is not because of gene deletion. The observed gene down-regulation was not limited to Klra genes; neighboring genes in the NKC were also affected, albeit to a degree dependent on the proximity to the Klra locus (Figure 2C-D). Because of the overall NK-cell receptor expression phenotype of these mice, they are referred to as NKC-knockdown (NKCKD) mice hereafter.
In summary, the attempted gene targeting of Klra15 resulted in an insertion of the Klra15-targeting construct as a concatemer upstream of the Klra gene cluster. This insertion leads to regional transcriptional silencing and subsequent loss of most Ly49 receptor expression on NKCKD NK cells, including additional down-regulation of neighboring genes encoding the CD94/NKG2 and KLRI receptor families.
NKCKD NK cells display normal development and maturation markers
Next, we determined whether NKCKD NK-cell development, localization, or differentiation marker expression was affected. NK-cell numbers and percentages in the spleen, lungs, liver, and blood are similar in WT and NKCKD mice (supplemental Figure 2A). Twelve markers expressed by NK cells were found to be normally expressed (supplemental Figure 2B). In addition, the NKC-resident Klrb1b, Clec2d, and Clec2i genes were unaffected (supplemental Figure 3A). NKCKD NK cells cultured in IL-2 for 3 days contained normal levels of granzymes A and B (supplemental Figure 3B). Similarly, up-regulation of CD69 was normal, but the number of NKCKD NK cells expressing the KLRG1 activation marker was reduced (supplemental Figure 3B-C), similar to B6 strain Ly49C, Ly49I, NKG2A-negative NK cells, and NK cells from B2m−/− mice.7,20 In summary, the deficiency of NKCKD mice appears to be restricted to only NKC-encoded MHC-I–specific receptor expression on mature NK cells.
NKCKD NK cells exhibit defective natural killing of MHC-I–deficient hematopoietic cells
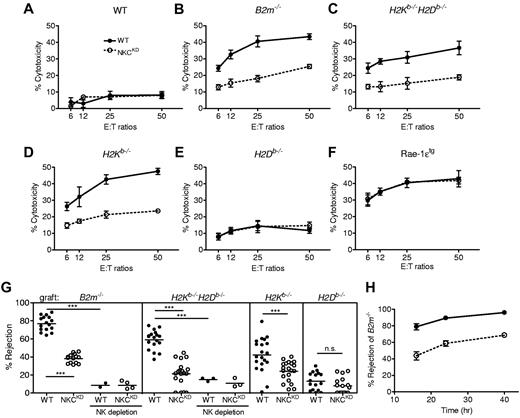
NKCKD mice provide a genetic model to directly test the “missing-self” hypothesis. First, in vitro cytotoxicity assays were used with ConA-stimulated lymphoblasts from MHC-I–sufficient or MHC-I–deficient mice as target cells. Effector LAK cells were produced from B6 background WT (B6.Ly49129) and NKCKD mice as initial experiments with 129-background LAK cells yielded low in vitro killing, as previously observed for this strain.14 Both WT and NKCKD LAK cells were unable to kill WT ConA blasts (Figure 3A). In contrast, B2m−/− ConA blasts were lysed efficiently by WT LAK cells (Figure 3B). However, NKCKD LAK killing of B2m−/− blasts was considerably lower (Figure 3B). Similarly, the killing of H2Kb−/−H2Db−/− ConA blasts was high for WT LAK cells and ∼ 50% lower for NKCKD LAK cells (Figure 3C). H2Kb−/− ConA blasts were also efficiently killed by WT LAK cells, but poorly lysed by NKCKD LAK cells (Figure 3D). In contrast, low killing of H2Db−/− ConA blasts was exhibited by either WT or NKCKD LAK cells (Figure 3E). To control for cytotoxic potential independent of MHC-I, ConA blasts were prepared from Rae-1ϵ–transgenic mice and used as target cells. These were equally and efficiently lysed by both WT and NKCKD LAK cells (Figure 3F) as well as freshly prepared NK cells (data not shown), showing that the decreased killing of MHC-I–deficient blasts by NKCKD LAK cells is not because of lower overall killing ability.
Natural killer gene complex knockdown (NKCKD) NK cells exhibit defective in vitro killing of MHC-I–deficient ConA blasts, and NKCKD mice exhibit reduced rejection of MHC-I–deficient splenocytes in vivo. The ability of lymphokine-activated killer (LAK) cells from WT and NKCKD to kill splenic ConA blast target cells from (A) B6 (WT), (B) B2m−/−, (C) H2Kb−/−H2Db−/−, (D) H2Kb−/−, (E) H2Db−/−, and (F) Rae-1ϵ transgenic (Rae-1ϵtg) mice was tested by 51Cr-release assay. Data are represented as the mean ± SD percentage of chromium release from triplicate wells. Data are representative of 3 similar experiments. Cytotoxicity experiments were performed with mice on the B6 background. (G) The ability of WT and NKCKD mice to reject CFSE-labeled splenocytes from B2m−/−, H2Kb−/−H2Db−/−, H2Kb−/−, and H2Db−/− mice was tested by flow cytometry of recipient splenocytes 16 hours after injection. Each symbol represents the data from an individual mouse, and the small horizontal lines indicate the mean. Some mice of each strain were pretreated with anti–asialo-GM1 Ab to deplete NK cells before injection of CFSE-labeled cells. Data are pooled from 3 to 5 independent experiments. Groups differed significantly as shown (***P < .001; NS, not significant). (H) Time-course rejection assay of B2m−/− splenocytes by WT and NKCKD mice. Data are displayed as the mean ± SEM rejection of 6 individual mice. Splenocyte rejection experiments were performed with 129S1 background mice.
Natural killer gene complex knockdown (NKCKD) NK cells exhibit defective in vitro killing of MHC-I–deficient ConA blasts, and NKCKD mice exhibit reduced rejection of MHC-I–deficient splenocytes in vivo. The ability of lymphokine-activated killer (LAK) cells from WT and NKCKD to kill splenic ConA blast target cells from (A) B6 (WT), (B) B2m−/−, (C) H2Kb−/−H2Db−/−, (D) H2Kb−/−, (E) H2Db−/−, and (F) Rae-1ϵ transgenic (Rae-1ϵtg) mice was tested by 51Cr-release assay. Data are represented as the mean ± SD percentage of chromium release from triplicate wells. Data are representative of 3 similar experiments. Cytotoxicity experiments were performed with mice on the B6 background. (G) The ability of WT and NKCKD mice to reject CFSE-labeled splenocytes from B2m−/−, H2Kb−/−H2Db−/−, H2Kb−/−, and H2Db−/− mice was tested by flow cytometry of recipient splenocytes 16 hours after injection. Each symbol represents the data from an individual mouse, and the small horizontal lines indicate the mean. Some mice of each strain were pretreated with anti–asialo-GM1 Ab to deplete NK cells before injection of CFSE-labeled cells. Data are pooled from 3 to 5 independent experiments. Groups differed significantly as shown (***P < .001; NS, not significant). (H) Time-course rejection assay of B2m−/− splenocytes by WT and NKCKD mice. Data are displayed as the mean ± SEM rejection of 6 individual mice. Splenocyte rejection experiments were performed with 129S1 background mice.
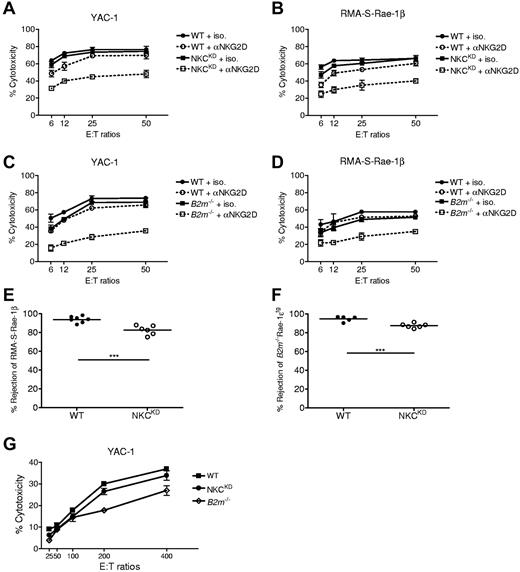
To determine whether NKCKD NK cells effectively monitor the expression of MHC-I in vivo, splenocyte rejection assays were conducted. Approximately 80% of B2m−/− splenocytes are rejected by WT mice after 16 hours; however, NKCKD mice displayed only half as much rejection (Figure 3G). The weak rejection displayed by NKCKD mice is probably because of residual Ly49 expression, resulting in low numbers of MHC-I–educated NK cells. The rejection by NKCKD mice of splenocytes specifically deficient in MHC-Ia molecules was also tested. Rejection of H2Kb−/−H2Db−/− splenocytes by WT mice was ∼ 20% lower than rejection of B2m−/− splenocytes but still 3 times as much as that mediated by NKCKD mice (Figure 3G). Rejection of both B2m−/− and H2Kb−/−H2Db−/− splenocytes was due to NK cells as shown by pretreatment of recipient mice with anti–asialo-GM1 antiserum (Figure 3G).
The ability of NKCKD mice to destroy cells missing single MHC-Ia molecules was determined. The rejection of H2Kb−/− splenocytes by WT mice was less than that observed for H2Kb−/−H2b−/− splenocytes, but NKCKD mice rejected H2Kb−/− splenocytes significantly less well than WT mice (Figure 3G). The rejection of H2Db−/− splenocytes by both WT and NKCKD mice was low (Figure 3G). To determine whether the rejection of MHC-I–deficient cells was simply delayed, a time-course experiment was performed. As late as 40 hours after injection MHC-I–deficient cells are still present in NKCKD mice even though there is close to 100% rejection in WT mice (Figure 3H). In summary, these data show that NKCKD NK cells display specific defective in vitro and in vivo killing of MHC-I–deficient cells.
A self-specific Ly49 transgene restores MHC-I immunosurveillance in NKCKD mice
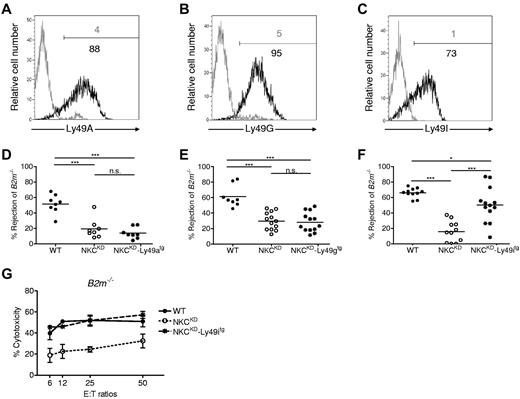
To determine the direct contribution of Ly49 receptors to the deficient missing-self responses of NKCKD mice, 3 different Ly49 transgenes were individually introduced onto the NKCKD background via backcrossing. Ly49a, Ly49g, and Ly49i transgenes were expressed on most NK cells from NKCKD mice (Figure 4A-C). Ly49a and Ly49g transgenes did not rescue the ability of NKCKD mice to reject MHC-I–deficient cells (Figure 4D-E), in agreement with the known lack of binding of Ly49A and Ly49G to either Kb or Db. However, transgenic expression of Ly49I, which binds to both Kb and Db,11,21 was able to restore most missing self-responses (Figure 4F). Furthermore, Ly49I expression on NK cells from NKCKD mice restored in vitro cytotoxicity toward B2m−/− ConA blast target cells to normal levels (Figure 4G). Thus, Ly49 silencing is responsible for most deficient missing self-responses in NKCKD mice. These data further support the hypothesis that Ly49 expression by NK cells for self-MHC-I during development results in the acquisition of cytotoxic potential to cells lacking self–MHC-I molecules.
Loss of MHC-I-immunosurveillance in NKCKD mice is because of silencing of Ly49 that bind to self–MHC-I. Three different Ly49-transgenes were introduced into NKCKD mice by breeding. (A-C) Flow cytometric analysis of Ly49 expression in Ly49tg-NKCKD NK cells. Ly49 staining of NK cells is shown for (A) NKCKD-Ly49Atg, (B) NKCKD-Ly49Gtg, and (C) NKCKD-Ly49Itg mice as a black histogram. The gray histogram shows control staining of splenic NK cells from NKCKDmice. (D-F) Specific rejection of B2m−/− splenocytes by (D) Ly49Atg-NKCKD mice, (E) Ly49Gtg-NKCKD mice, (F) Ly49Itg-NKCKD mice in comparison to WT or NKCKD mice. Each symbol represents the data from a single mouse. Data are pooled from 3 to 5 independent experiments. Groups differed significantly as shown (*P < .05; ***P < .001; NS indicates not significant). (G) LAK prepared from WT, NKCKD, and NKCKD-Ly49itg mice were used as effector cells in a 51Cr-release assay against B2m−/− ConA blasts. These experiments were performed with mice on the B6 background.
Loss of MHC-I-immunosurveillance in NKCKD mice is because of silencing of Ly49 that bind to self–MHC-I. Three different Ly49-transgenes were introduced into NKCKD mice by breeding. (A-C) Flow cytometric analysis of Ly49 expression in Ly49tg-NKCKD NK cells. Ly49 staining of NK cells is shown for (A) NKCKD-Ly49Atg, (B) NKCKD-Ly49Gtg, and (C) NKCKD-Ly49Itg mice as a black histogram. The gray histogram shows control staining of splenic NK cells from NKCKDmice. (D-F) Specific rejection of B2m−/− splenocytes by (D) Ly49Atg-NKCKD mice, (E) Ly49Gtg-NKCKD mice, (F) Ly49Itg-NKCKD mice in comparison to WT or NKCKD mice. Each symbol represents the data from a single mouse. Data are pooled from 3 to 5 independent experiments. Groups differed significantly as shown (*P < .05; ***P < .001; NS indicates not significant). (G) LAK prepared from WT, NKCKD, and NKCKD-Ly49itg mice were used as effector cells in a 51Cr-release assay against B2m−/− ConA blasts. These experiments were performed with mice on the B6 background.
Loss of tumor cell MHC-I surveillance in NKCKD mice
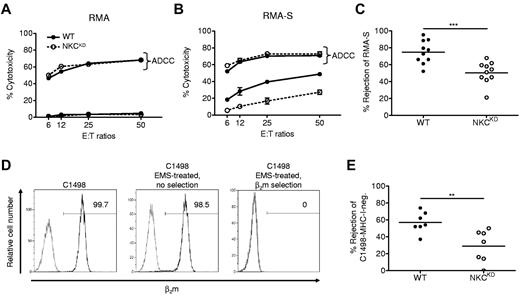
The ability of NKCKD NK cells to kill tumor cells was determined. It was observed that WT and NKCKD LAK killing of MHC-I–sufficient RMA tumor cells was low (Figure 5A), showing that the diminished expression of MHC-I–specific receptors by NKCKD NK cells does not result in a loss of inhibition. In contrast, the killing of the MHC-I–deficient variant RMA-S was higher; however, NKCKD LAK cells killed RMA-S only half as well as WT LAK cells (Figure 5B), suggesting that the ability to respond to MHC-I–deficient cells requires MHC-I receptor expression during NK-cell development. ADCC of both RMA and RMA-S was equal between WT and NKCKD LAK cells (Figure 5A-B), again showing that the intrinsic MHC-independent killing potential of NKCKD NK cells is intact.
NKCKD NK cells exhibit reduced cytotoxicity toward MHC-I–deficient tumor cells in vitro and in vivo. (A-B) The ability of WT versus NKCKD lymphokine-activated killer (LAK) cells to kill tumor target cells was tested by 51Cr-release assay. LAK cells generated from WT and NKCKD mice were used as effectors cells against untreated (A) RMA, (B) RMA-S target cells, or targets precoated with anti-Thy1.2 mAb to test antibody-dependent cellular cytotoxicity (ADCC) function. The data are displayed as the mean ± SD percentage of chromium release from triplicate wells. (C) In vivo rejection of RMA-S relative to RMA cells was assessed. The mean rejection is indicated by a horizontal line. Each symbol represents the data from a single mouse. (D) Production of a MHC-I–negative C1498 subline. Flow cytometry for β2m (dark histogram) is shown for the indicated C1498 lines. The gray histogram indicates isotype control mAb staining. The percentage of cells positively staining for β2m is indicated. (E) In vivo rejection of C1498-MHC-I–negative cells versus C1498-MHC-I–positive tumor cells. Note that both cell lines received EMS treatment. Data are representative of ≥ 3 similar experiments. Groups differed significantly as shown (**P < .01; ***P < .001). These experiments were performed with mice on the B6 background.
NKCKD NK cells exhibit reduced cytotoxicity toward MHC-I–deficient tumor cells in vitro and in vivo. (A-B) The ability of WT versus NKCKD lymphokine-activated killer (LAK) cells to kill tumor target cells was tested by 51Cr-release assay. LAK cells generated from WT and NKCKD mice were used as effectors cells against untreated (A) RMA, (B) RMA-S target cells, or targets precoated with anti-Thy1.2 mAb to test antibody-dependent cellular cytotoxicity (ADCC) function. The data are displayed as the mean ± SD percentage of chromium release from triplicate wells. (C) In vivo rejection of RMA-S relative to RMA cells was assessed. The mean rejection is indicated by a horizontal line. Each symbol represents the data from a single mouse. (D) Production of a MHC-I–negative C1498 subline. Flow cytometry for β2m (dark histogram) is shown for the indicated C1498 lines. The gray histogram indicates isotype control mAb staining. The percentage of cells positively staining for β2m is indicated. (E) In vivo rejection of C1498-MHC-I–negative cells versus C1498-MHC-I–positive tumor cells. Note that both cell lines received EMS treatment. Data are representative of ≥ 3 similar experiments. Groups differed significantly as shown (**P < .01; ***P < .001). These experiments were performed with mice on the B6 background.
Similar to in vitro cytotoxicity results, NKCKD mice displayed a significant reduction in the ability to reject RMA-S in vivo compared with WT mice (Figure 5C). Because RMA and RMA-S have been cultured separately for almost 30 years, it is possible that they have differentially acquired additional mutations, possibly contributing to NK-cell susceptibility. We produced an MHC-negative variant of the C1498 lymphoma by EMS treatment, followed by multiple rounds of cell sorting (Figure 5D). Similar to RMA-S, C1498–MHC-negative tumor cells were rejected only half as well by NKCKD mice compared with WT controls (Figure 5E), emphasizing that Ly49 expression by NK cells is necessary for MHC-I immunosurveillance against cancer.
Defective killing of MHC-I–deficient tumor cells by NKCKD NK cells is restored by activation through NKG2D
To determine the effect of activating ligands expressed by MHC-I–negative tumors we tested the ability of NKCKD NK cells to kill the prototypical mouse NK tumor target cell YAC-1, which are MHC-low and express high levels of NKG2D ligands.22,23 In contrast to RMA-S results, the killing of YAC-1 was found to be almost equally high by both WT and NKCKD LAK cells (Figure 6A), most probably because of the near normal expression of NKG2D (Figure 2D). We hypothesized that in the absence of overriding NKG2D activation signals YAC-1 should be differentially killed by WT versus NKCKD LAK cells. WT LAK killing decreased marginally in the presence of anti-NKG2D mAb, but NKCKD LAK killing was dramatically reduced to levels similar to NKCKD LAK killing of RMA-S (Figure 6A). To further test this hypothesis, RMA-S was stably transfected with Rae-1β. Similar to YAC-1, both WT and NKCKD NK cells exhibited high killing of RMA-S-Rae-1β target cells, with NKCKD killing being only slightly less than WT killing (Figure 6B). In addition, anti-NKG2D blocking of WT LAK decreased killing of RMA-S-Rae-1β only slightly, but killing by NKCKD LAK cells was considerably lower (Figure 6B). These results suggest that, although missing self-immunosurveillance against tumor cells is deficient in NKCKD mice, this deficiency can be masked by strong activating signals such as through NKG2D or CD16/CD32 (Figure 5B).
Activation via NKG2D overcomes deficient missing-self responses by natural killer gene complex knockdown (NKCKD) and B2m−/− NK cells to tumor cells. (A-B) The cytotoxic potential of WT versus NKCKD LAK cells against (A) YAC-1 and (B) RMA-S-Rae-1β target cells was assessed by 51Cr-release assay in the presence or absence of blocking anti-NKG2D. (C-D) Similarly, cytotoxicity of WT versus B2m−/− LAK cells was assessed against (C) YAC-1 and (D) RMA-S-Rae-1β target cells in the presence or absence of blocking anti-NKG2D. The data are displayed as the mean ± SD percentage of chromium release from triplicate wells. (E-F) In vivo rejection of (E) RMA-S-Rae-1β relative to RMA cells or of (F) B2m−/−Rae-1etg splenocytes relative to WT splenocytes was assessed. The mean rejection is indicated by a horizontal line. Each symbol represents the data from a single mouse. Data are representative of ≥ 3 similar experiments. Groups differed significantly as shown (***P < .001). (G) Splenocytes from the indicated mouse strains were used in cytotoxicity assays against 51Cr-labeled YAC-1 cells at the indicated effector-to-target ratios. The data are displayed as the mean ± SD of chromium release from triplicate wells. These experiments were performed with mice on the B6 background.
Activation via NKG2D overcomes deficient missing-self responses by natural killer gene complex knockdown (NKCKD) and B2m−/− NK cells to tumor cells. (A-B) The cytotoxic potential of WT versus NKCKD LAK cells against (A) YAC-1 and (B) RMA-S-Rae-1β target cells was assessed by 51Cr-release assay in the presence or absence of blocking anti-NKG2D. (C-D) Similarly, cytotoxicity of WT versus B2m−/− LAK cells was assessed against (C) YAC-1 and (D) RMA-S-Rae-1β target cells in the presence or absence of blocking anti-NKG2D. The data are displayed as the mean ± SD percentage of chromium release from triplicate wells. (E-F) In vivo rejection of (E) RMA-S-Rae-1β relative to RMA cells or of (F) B2m−/−Rae-1etg splenocytes relative to WT splenocytes was assessed. The mean rejection is indicated by a horizontal line. Each symbol represents the data from a single mouse. Data are representative of ≥ 3 similar experiments. Groups differed significantly as shown (***P < .001). (G) Splenocytes from the indicated mouse strains were used in cytotoxicity assays against 51Cr-labeled YAC-1 cells at the indicated effector-to-target ratios. The data are displayed as the mean ± SD of chromium release from triplicate wells. These experiments were performed with mice on the B6 background.
To determine whether these findings were unique to NK cells from NKCKD mice or to MHC-I–uneducated NK cells in general, YAC-1 killing by LAK from B2m−/− mice in the presence or absence of anti-NKG2D mAb was tested. Similar to NKCKD LAK cells, B2m−/− LAK killing of YAC-1 was slightly less than WT, but, in the presence of anti-NKG2D mAb, only B2m−/− LAK killing was considerably decreased, showing deficient missing self-responses (Figure 6C). A similar pattern was observed when using RMA-S-Rae-1β tumor cells as targets; high killing by both WT and B2m−/− LAK cells, but significantly decreased killing by blocking NKG2D only with B2m−/− LAK cells (Figure 6D).
It is possible that hyporesponsiveness is being masked by IL-2 activation of LAK cells. However, the in vivo rejection of RMA-S-Rae-1β cells by both WT and NKCKD mice was considerably increased (Figure 6E) compared with rejection of parental RMA-S cells (Figure 5C) and mirrored in vitro cytotoxicity assay results (Figure 6B). Similarly, the rejection of B2m−/−Rae-1tg splenocytes was efficient in both WT and NKCKD mice, but slightly less for NKCKD mice (Figure 6F), suggesting a small effect by the self-education defect in NKCKD mice. Finally, the killing of YAC-1 by fresh ex-vivo NK cells from WT, NKCKD, and B2m−/− mice (Figure 6G) was tested. YAC-1 cells were killed by fresh NKCKD NK cells as efficiently as WT NK cells, arguing against hyporesponsiveness being masked by IL-2 activation in the LAK cytotoxicity experiments. However, B2m−/− NK cells showed a moderate defect in YAC-1 killing compared with WT NK cells (Figure 6G). In summary, these data suggest that defective self–MHC-I education, whether through absence of receptors (NKCKD mice) or absence of ligands (B2m−/− mice), still allows NK cells to display cytotoxicity toward tumor cells bearing strong activating ligands.
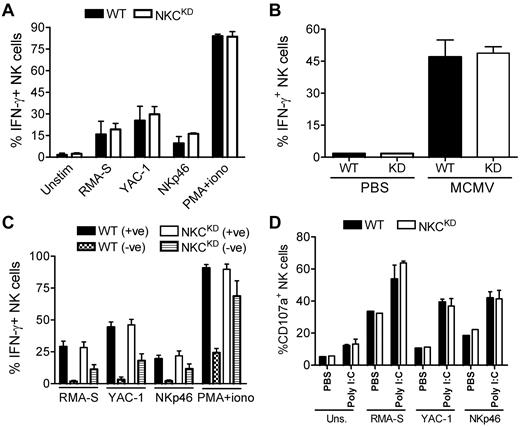
NKCKD NK cells display normal cytokine and degranulation responses
In contrast to killing of MHC-I–deficient target cells, de novo intracellular IFN-γ protein synthesis after stimulation with RMA-S or YAC-1 tumor cells, anti-NKp46 mAb, phorbol 12-myristate 13-acetate/ionomycin, or after murine CMV (MCMV) infection was normal in NKCKD NK cells compared with WT NK cells (Figure 7A-B). When the ability of specific NK subsets is analyzed, either positive or negative for inhibitory self-MHC-I–specific receptors, more IFN-γ is produced by the Ly49/NKG2+ subset than the Ly49/NKG2− subset in WT NK cells (Figure 7C), in accordance with the licensing/education hypothesis, regardless of the type of stimulus. The corresponding Ly49/NKG2+ subset had the same frequency of IFN-γ+ NK cells in NKCKD mice compared with WT mice (Figure 7C), although the Ly49/NKG2+ subset is 5 times smaller in NKCKD mice; thus, it cannot account for the normal levels of IFN-γ among bulk NK cells. Surprisingly, the Ly49/NKG2− subset of NKCKD NK cells produced significantly more IFN-γ than the corresponding subset in WT NK cells (Figure 7C).
NKCKD NK cells display normal IFN-γ production and degranulation responses. (A) Splenocytes from WT or NKCKD mice pretreated with poly(I:C) were incubated with the indicated tumor cells, on plates coated with anti-NKp46 mAb or with phorbol 12-myristate 13-acetate (PMA)/ionomycin. After 5 hours intracellular staining was performed to assess IFN-γ production by flow cytometry. (B) The frequency of IFN-γ+ NK cells was assessed in the spleens of the indicated mouse strains after 36 hours of infection with Smith strain murine CMV (MCMV). (C) The same assay as in panel A was performed, but splenocytes were additionally stained with anti-Ly49 and NKG2A mAb. Data are shown as the percentage of IFN-γ+ cells among Ly49/NKG2+ and Ly49/NKG2− subsets. (D) CD107a levels on the surface of NK cells were evaluated by flow cytometry after the indicated tumor or mAb stimulations. Mice were pretreated with poly(I:C) or with a PBS control. Data are representative of ≥ 3 similar experiments, except for degranulation assays which were performed twice. Three individual mice were used as a source of splenocytes for each experiment (n = 3). These experiments were performed with mice on the B6 background.
NKCKD NK cells display normal IFN-γ production and degranulation responses. (A) Splenocytes from WT or NKCKD mice pretreated with poly(I:C) were incubated with the indicated tumor cells, on plates coated with anti-NKp46 mAb or with phorbol 12-myristate 13-acetate (PMA)/ionomycin. After 5 hours intracellular staining was performed to assess IFN-γ production by flow cytometry. (B) The frequency of IFN-γ+ NK cells was assessed in the spleens of the indicated mouse strains after 36 hours of infection with Smith strain murine CMV (MCMV). (C) The same assay as in panel A was performed, but splenocytes were additionally stained with anti-Ly49 and NKG2A mAb. Data are shown as the percentage of IFN-γ+ cells among Ly49/NKG2+ and Ly49/NKG2− subsets. (D) CD107a levels on the surface of NK cells were evaluated by flow cytometry after the indicated tumor or mAb stimulations. Mice were pretreated with poly(I:C) or with a PBS control. Data are representative of ≥ 3 similar experiments, except for degranulation assays which were performed twice. Three individual mice were used as a source of splenocytes for each experiment (n = 3). These experiments were performed with mice on the B6 background.
Similar to IFN-γ induction, the degranulation potential of NKCKD NK cells was normal in response to tumor target cell coincubation or plate-bound anti-NKp46 mAb stimulation, as detected by CD107a surface expression (Figure 7D). The reason RMA-S did not elicit lower CD107a on NKCKD NK cells is not clear, although other studies have found discrepancies between CD107a levels and actual lysis of target cells.24 Thus, bulk NKCKD NK cells display normal IFN-γ and degranulation responses despite most of these cells being Ly49−, suggesting Ly49 expression is not strictly required for these functions.
Discussion
We show that NK-dependent in vitro killing and in vivo rejection of MHC-I–deficient cells is significantly lower in NKCKD mice and that this deficiency can be rescued by a Ly49 transgene. These results are consistent with the hypothesis that self–MHC-I–specific inhibitory Ly49 expression during NK-cell development is necessary for NK cells to achieve responsiveness toward MHC-I–deficient cells. If the only function of Ly49 is inhibition in mature NK cells, then the loss of Ly49 should result in the increased killing of MHC-I+ cells. However, the lack of RMA tumor cell and WT ConA blast killing by NKCKD NK cells argues against this model. One explanation is that NKCKD NK cells are simply “abnormal” and lack cytotoxic ability, but their high and normal killing of YAC-1 and Rae-1ϵtg ConA blasts, as well as ADCC-mediated killing, shows that NKCKD NK cells have a normal cytotoxic potential. The data best fit a model whereby self–MHC-I–specific inhibitory Ly49 receptor expression is required during NK-cell development and education for the subsequent acquisition of cytotoxicity that can be specifically triggered by the loss of self–MHC-I on target cells. This new animal model provides direct genetic evidence of Ly49-mediated NK-cell education in addition to complementing and extending previous genetic reconstitution experiments with adoptively transferred bone marrow cells.8
The down-regulation of the whole Ly49 gene cluster after targeting of the most upstream/telomeric gene is reminiscent of the first granzyme- and globin-targeted mice, which unintentionally resulted in silencing of the other gene family members.25 In these cases, the neomycin selection cassettes are thought to have contributed to the silencing effect. However, Southern blot analysis showed that NKCKD mice contain no neomycin genes because of Cre-mediated deletion at the ES cell stage, in agreement with the inability of NKCKD NK cells to survive in neomycin-containing culture medium (data not shown). The granzyme and globin gene families are hypothesized to be under the control of a locus control region (LCR) that governs expression temporally and with regard to tissue/cell type.25 The silencing of the whole gene cluster is possibly a result of the concatemer insertion interrupting an LCR-mediated opening of the cluster. There is currently no evidence that the Ly49 gene cluster is under the control of an LCR, and genomic Ly49A transgenes displaying variegated expression argue against an LCR for this locus.26 Instead, Ly49 expression appears to be a stochastic event governed by bidirectional promoters upstream of each gene.27 Another possible silencing mechanism is long-range methylation caused by the repetitive elements in the original targeting construct, now amplified by concatemerization, turning on repeat-dependent DNA methylases whose function is to repress such regions of the genome.28
Because 3 related, adjacent gene families (Ly49, CD94/NKG2, KLRI) are silenced in NKCKD, any of the 3, or combinations thereof, may be responsible for the loss of MHC-I immunosurveillance. When both MHC-Ia and β2m-dependent MHC-Ib molecules are absent, as in the case of β2m-deficient cells, the rejection by WT NK cells is higher than that seen when only MHC-Ia molecules are absent (H2Kb−/−H2Db−/−; Figure 3G). This suggests that β2m-dependent MHC-Ib molecules are also used by NK cells to monitor missing self-MHC expression. Expression of MHC-Ib molecules such as Qa-1b, H-2Blastocyst, CD1d, and Q9 have all been shown to inhibit NK-cell cytotoxicity.29-32 However, only one inhibitory receptor for Qa-1b has been identified (NKG2A).33 The ligand for KLRI is unknown, but the structural and primary homology to the NKG2 family suggests that KLRI ligands may also be MHC-Ib proteins. In the specific case of target cells from H2Kb−/−H2Db−/− and H2Kb−/− mice, the defective in vitro and in vivo killing by NKCKD NK cells can be mostly attributed to the loss of Ly49 expression. Ly49 molecules are known to be direct receptors for the Kb and Db MHC-Ia molecules on NK cells. The absence of Db by itself did not elicit significant cytotoxicity or rejection (Figures 3E and 4D), in agreement with prior studies showing that Kb is a stronger self-educating ligand than Db in mice with B6- or 129-derived Ly49 haplotypes.14,34 Transgenic expression of a Ly49 that can bind to both H-2b MHC-Ia molecules restored most of the missing self-responses, suggesting that the loss of MHC-I immunosurveillance in NKCKD mice can be attributed mostly to Ly49 down-regulation.
Cytokine production is not strictly dependent on Ly49 expression; high levels of IFN-γ production are clearly evident in Ly49/NKG2-negative cells in NKCKD NK cells after stimulation by tumor cells, activating receptor crosslinking, or pharmacologic stimulation (Figure 7C). However, Ly49/NKG2-negative NK cells in WT mice do not produce significant amounts of IFN-γ after the same stimulation, suggests a possible compensatory mechanism when most NK cells do not express self–MHC-I–specific receptors, such that NK-cell cytokine production is preserved. Most of the IFN-γ produced during MCMV infection originates from “unlicensed” Ly49C/I-negative NK cells.35 Our findings showing that NK-cell IFN-γ production is normal in NKCKD mice after MCMV infection (Figure 7B) are consistent with this report.
NKCKD NK cells are not entirely hyporesponsive because they kill NKG2D-ligand–positive cells as efficiently as WT NK cells, in agreement with the near normal levels of NKG2D and normal levels of granzyme A and B in NKCKD NK cells. The high killing of YAC-1 and RMA-S-Rae-1β by NKCKD NK cells (as well as B2m−/− NK cells) suggests that the MHC-I requirement for education is less important for the recognition of MHC-I–deficient cells that possess additional activating signals, such as NKG2D ligands, or ligands for Ly49H. Thus, with respect to cytotoxicity NK cells from NKCKD mice are most hyporesponsive toward MHC-I–deficient cells, whereas cytotoxicity triggered through activating receptors is normal. Previous publications have reported that uneducated NK cells have decreased responses when directly stimulated through activating receptors and in response to tumor cells.7,36,37 Our contrasting data showing that NKCKD NK cells exhibit normal lysis of these cells suggest that responses to MHC-deficient cells are more sensitive to educating effects than responses to cells bearing activating ligands. However, because MHC-deficient NK cells typically show weaker responses to cells bearing activating ligands, our data may suggest that NKCKD NK cells display a limiting but still minimally active amount of MHC-specific inhibitory receptors. Those receptors could be small amounts of residual Ly49/NKG2 receptors or alternatively could be distinct receptors that compensate for the absence of Ly49/NKG2 receptors.
It has been proposed that MHC-I–uneducated NK cells are less sensitive to stimulatory ligands, but also cannot receive inhibitory signals in response to self–MHC-I molecules.38 Such a balance of signals is consistent with the normal lysis of RMA cells in ADCC assays (Figure 5A) or of Rae-1ϵTg cells (Figure 3F) by NKCKD NK cells. The use of IL-2–activated NK cells in cytotoxicity assays may explain the discrepancy between our results and previously published studies; IL-2 can render hyporesponsive NK cells functional.8 By contrast, the almost equal rejection of RMA-S-Rae-1β and B2m−/− Rae-1ϵtg cells by NKCKD mice (Figure 6E-F) and the near normal killing of YAC-1 shown by fresh ex vivo NKCKD NK cells (Figure 6G) suggest that strong activation signals override defective missing self-responses in naive NK cells as well. Our findings provide new insight into the contribution of Ly49 molecules to the missing-self and NK-cell education hypotheses. NKCKD mice will be a useful tool for future research into the role of MHC-I recognition by NK cells in various immune settings, including infectious disease, transplant rejection, and neoplasia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr A. Veillette for helpful discussion and Drs S. Anderson, M. Brown, L. Lanier, and J. Carlyle for critically evaluating the manuscript.
This work was supported by the Canadian Institutes of Health Research (CIHR; operating grant MOP 62841). S.B. is supported by a Fonds de la recherche en santé Québec scholarship. L.-H.T. was supported by a CIHR Cancer Training Program scholarship. A.P.M. is a Canada Research Chair in Innate Pathogen Resistance.
Authorship
Contribution: S.B. designed and performed most experiments and co-wrote the manuscript; M.M.T., M.M.A.R., A.B.M., R.P., L.-H.T., and A.D.T. performed experiments; B.T.W., J.-R.L., and Q.Z. designed and performed experiments; K.S.T. and D.H.R. provided reagents and analyzed results; and A.P.M. designed and performed experiments, supervised research, and wrote the manuscript.
Conflict-of-interest disclosure: D.H.R. is on the Scientific Advisory Board of a biotechnology company, Innate Pharma, that seeks to mobilize NK cells for cancer therapy and receives research funding from a pharmaceutical company, Novo Nordisk, to determine the role of NK cells and NK receptors in inflammatory disease. The remaining authors declare no competing financial interests.
Correspondence: Andrew P. Makrigiannis, Department of Biochemistry, Microbiology, and Immunology, University of Ottawa, Guindon Hall, Room 4226, 451 Smyth Road, Ottawa, ON, K1H 8M5, Canada; e-mail: amakrigi@uottawa.ca.

![Figure 1. The Klra gene cluster of Klra15-targeted (mutant) mice contains a concatemer insertion of the Klra15-targeting construct. (A) A schematic representation of the natural killer gene complex (NKC) and the targeting-construct produced for the Klra15 gene. (B) Southern blot analysis of wild-type (WT) and mutant thymus DNA digested with EcoRI, KpnI, or BamHI and probed with the Klra15-targeting construct depicted in panel A. (C) Array comparative genomic hybridization profile of the Klra cluster in mutant mice with a blow-up of the Klra15 gene (below). The x-axis represents the genomic region tiled on the microarrays, and the y-axis Shows differences in copy number between WT and mutant genomic DNA (log2 ratio [MT/WT]). Positive values indicate regions showing copy number increases in the NKC knockdown (NKCKD) genome. (D) PCR analysis of the Klra15-targeting construct copy number in mutant genomic DNA. Data are representative of ≥ 3 similar experiments, except for array comparative genomic hybridization analysis, which was performed twice. These experiments were performed with mice on the 129S1 background.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/3/10.1182_blood-2012-02-408732/4/m_zh89991293980001.jpeg?Expires=1766028136&Signature=Nx~4ME9vlQz9lQeYVYBtzzUBrzPCz-wK-~J-DVuNe3NArOGxYmuPIuaYPxdaiC0bBM-oqu1y-BNIicXDwC4JgaSTIYACCuD0VXAU4LIjt~TbChVtvmoApUnkMlWWKUMuwEyWVo2X3ej9eEi9ypGe6pM4eW5vgIla7VtN8ITaI3BredYbuW014nAJgzW4yz5I~I0mpeFBBa0AtcFLxzLWravTn~WSr8CF-MSFQmld6RfJ9IQ6uv07I1Yg1OcZUy-liqkHEC309btE09o4mF6V8d89lYPaCbgF2oVhtZCHXFmajTd-Yrn70wDILaDD~bjcOXZ1tphQS1uB-Mt38GJcnQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal