To the editor:
A recent study of single nucleotide polymorphisms (SNPs) in the B-cell activating factor (BAFF) gene in patients revealed the potential relevance of genetic disparity in the BAFF gene to chronic graft-versus-host disease (cGVHD) development.1 Four recipient SNPs were reported to be predictors of cGVHD development after allogeneic hematopoietic stem cell transplantation. Two of these SNPs were in intron 2 (rs16972217; rs7993590), and 2 were in intron 3 (rs12428930; rs2893321). To better understand the nature of the association of these SNPs with cGVHD development, we conducted further analyses.
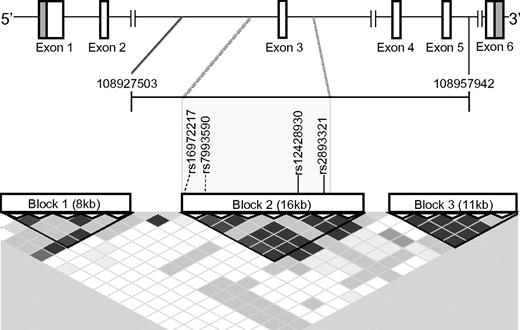
While these 4 SNPs predicted cGVHD, they did not do so independently of each other. Even though they were deemed “tagSNPs” by Clark et al, TagSNPs are typically chosen to represent SNPs in regions of high linkage disequilibrium (LD) that allow coverage of a region (or haplotype) using a minimum number of polymorphisms.1 Using the program Haploview2 or the Genome Variation Server (http://gvs.gs.washington.edu/GVS/) used in the report, we found that all 4 SNPs were in very strong LD in whites of northern European descent (CEU; Figure 1). Thus, presuming the allele frequencies in this European-American patient population are similar to those in the reference CEU population, these SNPs cannot be independently prognostic of the cGVHD phenotype, because each SNP is highly predictive of the others (r2 > 0.90). Therefore, rather than considering each SNP separately in a multivariate analysis, the haplotype(s) incorporating the 4 recipient SNPs should be viewed as a single predictor of cGVHD. Notably, this region encompasses exon 3 and flanking regions, prompting us to further study a potential functional role of these SNPs.
Linkage disequilibrium (LD) blocks within the BAFF gene. A schematic of the BAFF gene, TNFSF13B, is depicted at the top. The span of the 20 SNPs genotyped by Clark et al is demarcated by the genomic coordinates 109827503 and 108957942,1 shown at the ends of the horizontal bracket immediately below the gene. Coordinates were taken from the most recent human gene build (hg19) visualized by the NCBI genome browser (http://www.ncbi.nlm.nih.gov). The 3 LD blocks depicted were determined using the confidence interval method with Hapmap 3 data implemented by Haploview 4.2.2 The rs numbers of the 4 Clark et al cGVHD SNPs, shown in the gray box, are within the same LD block (Block 2). Solid vertical lines indicate a SNP that was plotted by Haploview; and dashed lines, a SNP that was not. The LD plot generated by Haploview was modified so that it aligns with the gene schematic above it. Degree of LD between 2 SNPs can be visualized on the LD plot shown in this figure as follows: (1) locate the SNP at the top of the LD plot and then follow diagonally along the diamond shaped boxes to the intersection with the diagonal from the other SNP of interest. The SNPs in high LD are represented by the darkest diamond shaped boxes (pairwise D' values shown; black indicates D' ≥ 0.95); D' is a measure of the frequency of association of alleles at 2 loci. D' ranges from 0 to 1, with 0 representing random association and 1 representing absolute association.
Linkage disequilibrium (LD) blocks within the BAFF gene. A schematic of the BAFF gene, TNFSF13B, is depicted at the top. The span of the 20 SNPs genotyped by Clark et al is demarcated by the genomic coordinates 109827503 and 108957942,1 shown at the ends of the horizontal bracket immediately below the gene. Coordinates were taken from the most recent human gene build (hg19) visualized by the NCBI genome browser (http://www.ncbi.nlm.nih.gov). The 3 LD blocks depicted were determined using the confidence interval method with Hapmap 3 data implemented by Haploview 4.2.2 The rs numbers of the 4 Clark et al cGVHD SNPs, shown in the gray box, are within the same LD block (Block 2). Solid vertical lines indicate a SNP that was plotted by Haploview; and dashed lines, a SNP that was not. The LD plot generated by Haploview was modified so that it aligns with the gene schematic above it. Degree of LD between 2 SNPs can be visualized on the LD plot shown in this figure as follows: (1) locate the SNP at the top of the LD plot and then follow diagonally along the diamond shaped boxes to the intersection with the diagonal from the other SNP of interest. The SNPs in high LD are represented by the darkest diamond shaped boxes (pairwise D' values shown; black indicates D' ≥ 0.95); D' is a measure of the frequency of association of alleles at 2 loci. D' ranges from 0 to 1, with 0 representing random association and 1 representing absolute association.
SNPs that result in increased BAFF production are potentially relevant for cGVHD pathophysiology, since excess BAFF has been significantly associated with disease status.3,4 BAFF SNPs associated with autoimmune diseases and lymphomas have been reported in the promoter region and demonstrated to be functional.5-8 We noted that all 20 SNPs tested by Clark et al1 were within introns 2 through 5, but none were in the promoter. LD between a known functional promoter SNP, rs9514828, and these 4 SNPs was very low (r2 ranged from 0.03-0.05). We therefore considered whether 1 of these SNPs might affect splicing as a potential mechanism leading to cGVHD, particularly because alternative splicing of exon 3 results in the only known functional BAFF isoform, ΔBAFF.9 Using the Human Splicing Finder (http://www.umd.be/HSF/),10 we located the splice sites flanking exon 3. Only 1 SNP, rs12428930, was < 1000 bp from an intron/exon boundary, but not within a predicted splice site. According to the dbSNP build 135 database (http://www.ncbi.nlm.nih.gov/SNP/), there are no known SNPs located within 200 bp of the exon/intron boundaries of exon 3; however, the SNPs identified by Clark et al flank exon 3, suggesting they may be markers of a functional SNP that affects alternative splicing, potentially leading to ΔBAFF production. Thus, further studies of promoter and alternative splicing SNPs and altered BAFF expression in cGVHD are warranted.
Authorship
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kristy L. Richards, 1062 Genetic Medicine Bldg, Department of Medicine, Division of Hematology and Oncology, University of North Carolina, CB no. 7361 UNC-CH, Chapel Hill, NC 27599; e-mail: kristy_richards@med.unc.edu.
References
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal