Abstract
TLR3 is a key receptor for recognition of double-stranded RNA and initiation of immune responses against viral infections. However, hyperactive responses can have adverse effects, such as virus-induced asthma. Strategies to prevent TLR3-mediated pathology are therefore desired. We investigated the effect of single-stranded DNA oligonucleotides (ssDNA-ODNs) on TLR3 activation. Human monocyte-derived dendritic cells up-regulate maturation markers and secrete proinflammatory cytokines on treatment with the synthetic TLR3 ligand polyinosine-polycytidylic acid (poly I:C). These events were inhibited in cultures with ssDNA-ODNs. Poly I:C activation of nonhematopoietic cells was also inhibited by ssDNA-ODNs. The uptake of poly I:C into cells was reduced in the presence of ssDNA-ODNs, preventing TLR3 engagement from occurring. To confirm this inhibition in vivo, we administered ssDNA-ODNs and poly I:C, alone or in combination, via the intranasal route in cynomolgus macaques. Proinflammatory cytokines were detected in nasal secretions in the poly I:C group, while the levels were reduced in the groups receiving ssDNA-ODNs or both substances. Our results demonstrate that TLR3-triggered immune activation can be modulated by ssDNA-ODNs and provide evidence of dampening proinflammatory cytokine release in the airways of cynomolgus macaques. These findings may open novel perspectives for clinical strategies to prevent or treat inflammatory conditions exacerbated by TLR3 signaling.
Introduction
The immune system distinguishes between self and non-self with a variety of pattern recognition receptors such as Toll-like receptors (TLRs), C-type lectins, RIG-I–like receptors, and Nod-like receptors.1–4 Ten genes for TLRs exist in humans and they are expressed on the cellular surface or in the intracellular endosomal compartment.2 For activation to take place, the receptors form homodimers or heterodimers and transmit a signal via the adaptor proteins MyD88 and/or TIR domain-containing adapter-inducing INF-β (TIRF). The extracellular TLRs recognize conserved glycoprotein and lipopeptide structures specific to microbial organisms, while the endosomal receptors recognize nucleic acids.2
TLR3 is the only receptor transmitting its signal merely via the adaptor protein TRIF and it does so in response to double-stranded RNA (dsRNA) present in, for example, viral genomes or their intermediates during the replication cycle.5,6 However, self-produced dsRNA can also engage TLR3 when released from damaged tissues.7–9 The synthetic ligand polyinosine-polycytidylic acid (poly I:C) is commonly used to target TLR3 in vaccine adjuvant constructs and in different experimental conditions.4,7,10 Immune activation mediated via TLR3 has been shown to be important in clearance of viral infections because of a robust induction of IL-12 and type I interferons.5 Furthermore, studies revealing its ability to mediate efficient cross-presentation of endocytosed Ags11,12 have made TLR3 an interesting target in vaccine adjuvant development.4,13,14 However, several reports have also implicated TLR3 in having detrimental effects during certain infections or conditions.5,6 Cells infected with Epstein-Barr virus have been shown to secrete virally encoded RNA detected by TLR3 that results in systemic pathologic immune activation.15 The role of TLR3 in viral CNS infections is debated, where rabies virus, herpes simplex virus 1, or West Nile virus all have been implied to benefit from strong TLR3 responses.6 Recently, an epidemiologic study suggested that a functional TLR3 gene is a risk factor for severe tick-borne encephalitis virus infection.16 TLR3 activation has also been reported to have negative effects in the respiratory tract.17,18 Influenza infection mediates pathogenic effects in the lungs of wild-type mice, while less tissue damage was detected in TLR3−/− animals.19 However, damage of the lung does not necessarily have to be because of infection, but can also be enhanced by sterile cell death, for example, during oxygen treatment of patients with acute respiratory distress syndrome.20 RNA release from necrotic cell death has also been shown to mediate pathologic effects in the joints of patients with rheumatoid arthritis.21 Sterile cell death can have severe consequences during organ transplantations, when strong immune responses against the donor organ increase the progression toward rejection.22 A recent study has, for example, identified that a loss-of-function mutation in the TLR3 gene is beneficial for patients receiving a liver transplant because of hepatitis C–related cirrhosis, as it seems to protect from acute graft rejection.23
Thus, strategies to prevent TLR3 hyperresponsiveness may constitute a therapeutic path. Indeed, aptamers, Abs, and small-molecule inhibitors to block the receptor interaction with dsRNA have been tested in mice.6,24–26 However, because TLR3 is a highly conserved protein expressed mainly in the endosomal compartment, the task has proven difficult and no clinical treatment has to our knowledge hitherto reached affected patients. Other TLRs have successfully been inhibited by different strategies. Oligodeoxynucleotides (ODNs) containing TTAGGG motifs to mimic telomeric DNA have previously been found to have a general immunosuppressive effect because of inhibition of STAT signaling,27 while single-stranded DNA-ODNs (ssDNA-ODNs) with poly G motifs inhibit TLR9 activation (and in some cases also TLR7 and TLR8), with the suggested mechanism of competitive antagonism.28–30 We have here investigated an alternative mechanism for inhibition of TLR3 signaling. Although not yet fully characterized, dsRNA has been shown to share an uptake mechanism with ssDNA-ODNs containing phosphorothioate backbones.31,32 These findings are used in present study.
We here report that TLR3-mediated activation of human monocyte-derived dendritic cells (DCs) and nonhematopoietic cells is inhibited in the presence of ssDNA-ODN, where a preferential uptake of ssDNA-ODNs over dsRNA is shown to be the underlying mechanism. We provide in vivo proof-of-principle in cynomolgus macaques, where poly I:C–induced cytokine production in the airways was significantly blocked by ssDNA-ODN. Single-stranded DNA-ODN with a non-CpG sequence is a nonimmunogenic molecule,28,29 and our results encourage the development of a novel strategy in treatment of TLR3-mediated pathogenesis.
Methods
Reagents and TLR ligands
The following ligands against TLR3, 4, 8, and 9 were used; poly I:C (25 μg/mL unless otherwise stated; Amersham Biosciences), lipopolysaccharide (LPS; 100 ng/mL; Sigma-Aldrich), monophosphoryl lipid A (MPLA; 1 μg/mL; InvivoGen), imidazoquinoline (R848, 5 μg/mL; InvivoGen), ODN 2006, ODN 2395, and ODN 2216 (5 μg/mL; all from InvivoGen), and synthetic, endotoxin-free, completely phosphorothioate-modified oligonucleotides (ssDNA-ODN, 5 μg/mL; DNA Technology A/S). The sequence of the ssCpG-ODN used was 5′-GTCGTTTTGTCGTTTTGTCGTTGTTGGTGGTGGTG-3′ and non-CpG ssDNA-ODN 5′-GAAGTTTTGAGGTTTTGAAGTTGTTGGTGGTGGTG-3′. Poly I:C was fluorescently marked using a Label IT Cy5 labeling kit (Mirus Bio) according to the manufacturer's instruction.
DC endosome acidification was inhibited with chloroquine (20μM; InvivoGen) 1 hour before stimulus addition. Endocytosis was measured by flow cytometry using pHrodo dextran beads (50 μg/mL; Molecular Probes).
DC differentiation
Monocytes were negatively selected from buffy coats using the RosetteSep Monocyte Enrichment Kit (1 mL/10 mL buffy coat; StemCell Technologies) and differentiated into DCs, as shown previously,33,34 at a density of 5 × 105 cells/mL in RPMI 1640 completed with 10% FCS, 1mM sodium pyruvate, 10mM HEPES, 2mM l-glutamine, and 1% streptomycin and penicillin (all from Invitrogen Life Technologies), with GM-CSF (250 ng/mL; PeproTech) and IL-4 (6.5 ng/mL; R&D Systems) for 6 or 7 days. The cells were phenotyped with Abs against CD14, CD1a (both from DakoCytomation), CD3, and CD19 (both from BD Biosciences). Maturation was assessed 48 hours later using Abs targeting CD1a (DakoCytomation), CD80, and CD86 (both from BD Biosciences). Sample data were acquired on a FACSCalibur (BD Biosciences); the analysis was performed with FlowJo software (TreeStar) on viable cells, excluding dead cells with a 7-AAD stain (eBioscience).
Stromal cell cultures
N/TERT-transfected keratinocytes were obtained from Dr James Rheinwald (Brigham and Women's Hospital, Harvard Institutes of Medicine), and grown in EpiLife serum-free keratinocyte medium (Invitrogen Life Technologies) supplemented with 50 μg/mL Gentamicin. The cells were used when the confluence was approximately 80%. 16HBE epithelial cells were cultured in MEM medium (Sigma-Aldrich) complemented with 10% FCS in flasks precoated with a fibronectin-coating solution (LHC basal medium [Invitrogen] with 50 μg/mL BSA, Vitrogen 100 [30 μg/mL; BD Biosciences], and human fibronectin [10 μg/mL; BD Biosciences]). When 70%-100% confluence was reached, the cells were harvested and further cultured in precoated 24-well plates. Ligands were added when 90% confluence was reached, MRC5 lung fibroblasts were cultured in DMEM medium (Invitrogen Life Technologies) complemented with 10% FCS. Ligands were added when full confluence was reached. Supernatants were collected after 24 hours and cells were lysed for RNA collection.
Real-time PCR
RNA was isolated using an RNeasy kit (QIAGEN) according to the manufacturer's instructions. A High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used to produce cDNA.
The master mix and GAPDH, TLR3, and TLR9 primers used were from Applied Biosystems, and the real-time PCR was performed in a 7500 Real-Time PCR System (Applied Biosystems). The PCR products were mixed 4:1 in loading buffer (Fermentas) and run on a 2% EtBr PGE gel.
Cytokine detection
Secretion of cytokines and chemokines was measured with a Bio-Plex assay (Biosource), or with ELISA kits targeting TNF-α, IL-8 (both from BioLegend), or IFN-β (InterferonSource). Cytokine expression in macaque nose swaps was quantified with Bio-Plex (Millipore).
Western blot
Cells were lysed in radioimmunoprecipitation assay buffer (50mM Tris, pH 8.0, 150mM NaCl, 1.0% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 1mM vanadate, 100μM molybdate, 20mM sodium fluoride, and protease inhibitor cocktail [1 tablet per 10 mL of RIPA lysis buffer]) or directly in Laemmli buffer (Bio-Rad) at a concentration of 1 × 107 cells/mL. Total cell extract was resolved on 4%-12% NuPAGE or Tris-Glycine gradient gels (Invitrogen). The proteins were blotted onto nitrocellulose or polyvinylidene fluoride membranes using an iBlot Dry Blotting System (Invitrogen). Membranes were incubated with blocking buffer (5% dried milk in PBS and 0.05% Tween 20, or 3% BSA in PBS and 0.05% Tween 20, or Roti-Block solution; Carl Roth GmbH Co KG) and probed with anti–phospho-IRF-3, anti–IRF-3, anti–phospho-NFκB, anti-NFκB (all from Cell Signaling Technology), or anti–β-actin (Sigma-Aldrich). Horseradish peroxidase–conjugated Abs were used as secondary Abs (DakoCytomation). Immune complexes were detected using the Supersignal West Femto Chemiluminescence Western blotting detection system (Pierce) and visualized in an AGFA Curix 60 Film Processor or in a LAS-4000 mini (Fuji Film).
B-cell proliferation
B cells were negatively selected from buffy coats using RosetteSep Human B Cell Enrichment Cocktail (StemCell Technologies) and cultured in complete RPMI supplemented with IL-2 (1000 IU/mL; Chiron). Cultures were performed in triplicates (5 × 105 cells/mL). In some cultures, 3H-Thymidine (1 μCi/well; Amersham Biosciences) was added at day 3 for 16 hours before harvest. The number of cells in additional cultures was determined every 24 hours in a BD LSR Fortessa (BD Biosciences). This was performed by adding 50 μL of cell suspension from each well to 150 μL of a propidium iodide solution (10 μg/mL; Sigma-Aldrich) and then counting the number of negative cells in exactly 50 μL of the suspensions.
Confocal microscopy
Cells were cultured with 5 μg/mL of each ligand for 15 minutes, transferred to glass slides (Bio-Rad), and mounted in GOLD prolonged mounting media with 4′,6-diamidino-2-phenylindole (Invitrogen). The slides were analyzed in a Leica SP102 laser scanning confocal microscope with a 63× glycerol objective, using Leica confocal software. Images for figures were generated by reconstruction of multiple optical sections.
In vivo challenge
Adult cynomolgus macaques (Macaca fascicularis) were imported from Mauritius and housed in the facilities of the Commissariat à l'Energie Atomique et aux Energies Alternatives (CEA; Fontenay-aux-Roses, France). Macaques were treated intranasally with 500 μg of poly I:C, 500 μg of non-CpG ssDNA-ODN, or the combination of both in a volume of 550 μL, that is, 275 μL in each nostril. Nasal samples were collected after 18 or 24 hours using a Weck-cel sponge (Windsor Medical). Briefly, a complete sample collection tube containing the Weck-cel spear head plus the spin-X tube containing 300 μL of extraction buffer (protease inhibitor cocktail [Sigma-Aldrich] diluted in PBS) was housed within a sterile 30 mL of polypropylene tube. Weck-cel sponge was placed in the nostril for 5 minutes. Each sample collection tube was weighed before and after nasal fluid collection. A first centrifugation at approximately 13 000g for 15 minutes at 4°C and followed by another for 5 minutes were sufficient to extract approximately 450 μL from the top chamber. Sample collection tubes containing buffer and Weck-cel spear heads were stored at −80°C until sample use.
Ethical approval
Local ethical review boards approved all experiments in the present study. Buffy coats were obtained from the blood bank at Karolinska University Hospital Huddinge. Mouse experiments were performed under approval of the Stockholm North Ethical Committee on Animal Experiments. Cynomolgus macaques were used at CEA in accordance with French national regulation and under national veterinary inspectors (CEA permit no. A 92-032-02). CEA is in compliance with Standards for Human Care and Use of Laboratory of the Office for Laboratory Animal Welfare (OLAW; OLAW assurance no. A5826-01). The use of nonhuman primates at CEA is in accordance with recommendation with the European Directive (2010/63; recommendation no. 9). No suffering was specifically associated with the treatment and sample procedure to obtain nasal washes. The animals were used under the supervision of the veterinarians in charge of the animal facility and the Ethical Animal Committee of the CEA reviewed the protocols used.
Statistical analyzes
Nonparametric tests were used to compare the different culture conditions, animal groups, or repeated measures as specified in the figure legends.
Results
Single-stranded CpG-ODN inhibits poly I:C–induced DC activation
We investigated whether combinations of TLR3, 4, 7/8, and 9 ligands influenced maturation of monocyte-derived DCs by studying the expression of maturation markers CD80 and CD86. The TLR3 ligand poly I:C, TLR4 ligand LPS, and TLR7/8 ligand R848 induced significant up-regulation of DC maturation markers compared with untreated cells, while type B ssCpG-ODN, a TLR9 ligand, did not mature DCs (Figure 1A). None of the ligands had toxic effects (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), and analysis was only performed on viable cells. LPS alone induced such a strong response that none of the TLR3, 8, or 9 ligands augmented expression of DC maturation markers further when combined with the endotoxin (data not shown), while the combination of poly I:C and R848 had an additive effect on the up-regulation of CD80 and CD86 (data not shown). ssCpG-ODN did not influence the DC maturation induced by LPS or R848. However, in combination with poly I:C, ssCpG-ODN significantly inhibited up-regulation of CD80 and CD86 in a dose response–dependent manner (Figure 1B-D). In addition, poly I:C–transfected apoptotic cells, able to induce maturation in DCs, lost their activating effect in presence of ssCpG-ODN (supplemental Figure 2). The optimal concentration of ssCpG-ODN to block the stimulatory effect of poly I:C was in the order of 5 μg/mL (Figure 1D), which was subsequently used in vitro throughout the study. We furthermore assessed whether proinflammatory cytokine production by poly I:C–treated DCs was influenced by ssCpG-ODN. Poly I:C induced the release of IL-12p40, IL-12p70, and TNF-α, while secretion of these cytokines was significantly inhibited in DC cultures with ssCpG-ODN present (Figure 1E).
Poly I:C–induced DC activation is inhibited by ssCpG-ODN. (A) Immature DCs were treated for 48 hours with the TLR ligands ssCpG-ODN, LPS, R848, or poly I:C. Flow cytometry was used to measure CD80 and CD86 expression. The results are depicted as mean fluorescent intensity (MFI; mean ± SEM, n = 6). Significant differences compared with medium control were assessed by repeated measures ANOVA with Bonferroni multiple comparison; **P < .01 and ***P < .001. NS indicates nonsignificant. (B) DCs were treated for 48 hours with TLR ligands LPS, R848, and poly I:C as single treatment or in combination with ssCpG-ODN. DC maturation was determined by assessing CD80 and CD86. Flow cytometry results are depicted as the MFI (mean ± SEM values, n = 7 for LPS and R848, and n = 13 for medium and poly I:C treatments). Significant differences comparing with or without ssCpG-ODN were assessed by noparametric Mann-Whitney 2-tailed test; ***P < .001. (C) The expression of CD1a and up-regulation of CD80 and CD86 was measured with flow cytometry on DCs 48 hours after ligand addition and displayed as a representative figure. (D) Dose-response measurement (MFI mean ± SEM values, n = 7) of the ssCpG-ODN inhibitory effect on poly I:C–induced DC maturation. (E) Secretion of proinflammatory cytokines IL-12p40, IL-12p70, and TNF-α in the supernatants at 24 hours was measured using Bio-Plex technology. Significant differences comparing with or without ssCpG-ODN (n = 8) were assessed by nonparametric Mann-Whitney 2-tailed test; *P < .05 and **P < .01.
Poly I:C–induced DC activation is inhibited by ssCpG-ODN. (A) Immature DCs were treated for 48 hours with the TLR ligands ssCpG-ODN, LPS, R848, or poly I:C. Flow cytometry was used to measure CD80 and CD86 expression. The results are depicted as mean fluorescent intensity (MFI; mean ± SEM, n = 6). Significant differences compared with medium control were assessed by repeated measures ANOVA with Bonferroni multiple comparison; **P < .01 and ***P < .001. NS indicates nonsignificant. (B) DCs were treated for 48 hours with TLR ligands LPS, R848, and poly I:C as single treatment or in combination with ssCpG-ODN. DC maturation was determined by assessing CD80 and CD86. Flow cytometry results are depicted as the MFI (mean ± SEM values, n = 7 for LPS and R848, and n = 13 for medium and poly I:C treatments). Significant differences comparing with or without ssCpG-ODN were assessed by noparametric Mann-Whitney 2-tailed test; ***P < .001. (C) The expression of CD1a and up-regulation of CD80 and CD86 was measured with flow cytometry on DCs 48 hours after ligand addition and displayed as a representative figure. (D) Dose-response measurement (MFI mean ± SEM values, n = 7) of the ssCpG-ODN inhibitory effect on poly I:C–induced DC maturation. (E) Secretion of proinflammatory cytokines IL-12p40, IL-12p70, and TNF-α in the supernatants at 24 hours was measured using Bio-Plex technology. Significant differences comparing with or without ssCpG-ODN (n = 8) were assessed by nonparametric Mann-Whitney 2-tailed test; *P < .05 and **P < .01.
Poly I:C mediates activation via TLR3 and its intracellular signaling pathways in DC
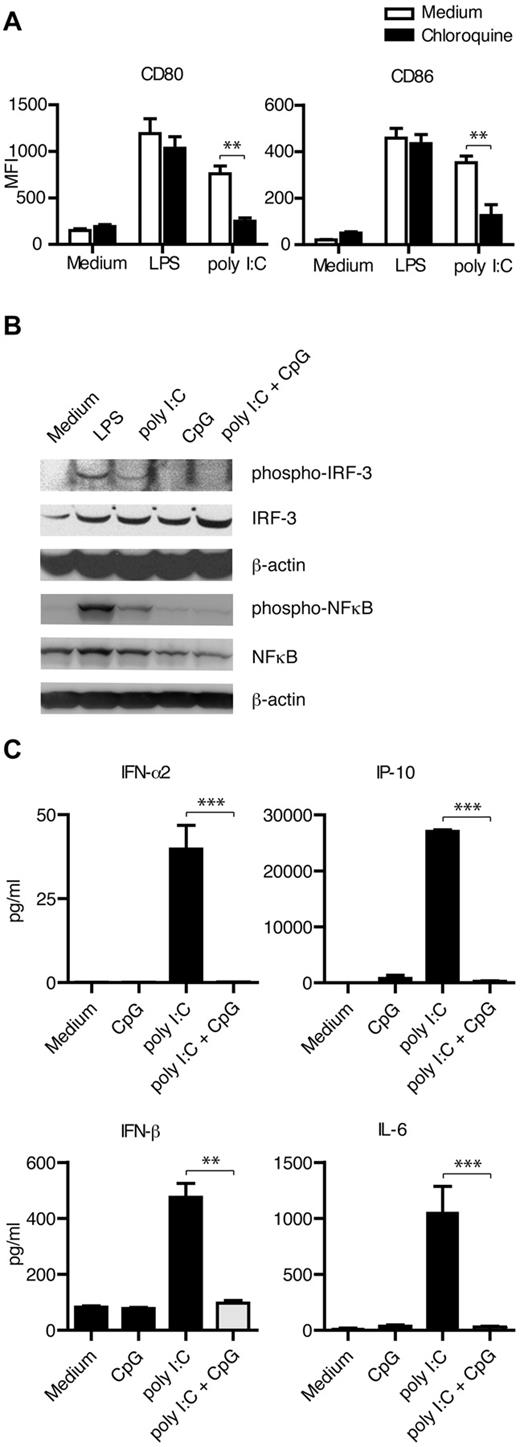
To investigate whether TLR3 was mediating the poly I:C–induced DC maturation, we pretreated cells with chloroquine 1 hour before addition of stimulus. TLR3 is located to the endosomes in DCs and acidification of the compartment is required for signaling to occur. Chloroquine inhibits this event, and thereby TLR signaling.35 DC maturation in response to poly I:C was significantly reduced in the presence of chloroquine, while the effect of LPS was unchanged (Figure 2A), indicating that TLR3 was inducing the signals for DC maturation on stimulation with poly I:C. TLR3 signaling mediates phosphorylation of both IRF-3 and NFκB, which we confirmed using Western blot. However, in cultures with ssCpG-ODN, poly I:C failed to activate these transcription factors (Figure 2B). We also measured the secretion of IRF-3- and NFκB-driven cytokine production by DCs. Poly I:C–induced secretion of IFN-α2, IFN-β, IP-10, and IL-6 was significantly inhibited by ssCpG-ODN (Figure 2C).
Single-stranded CpG-ODNs inhibit the intracellular TLR3 signaling pathways engaged by poly I:C in DCs. (A) DCs were treated for 1 hour with chloroquine before addition of TLR stimulus, and maturation was measured 48 hours later in 6 different donors. (B) Phosphorylation of the transcription factors IRF-3 and NFκB was detected by Western blot after TLR stimulation for 90 or 15 minutes, respectively. Representative data from 3 independent experiments is shown. (C) Poly I:C–induced secretion of the IRF-3- or NFκB-driven cytokines IFN-α2 (n = 8), IFN-β (n = 5), IP-10 (n = 8), and IL-6 (n = 8), as measured 24 hours after addition of stimuli. Quantity in culture supernatants is depicted in picograms per milliliter as mean ± SEM values. Significant differences comparing with or without addition of ssCpG-ODN were assessed by nonparametric Mann-Whitney 2-tailed test and are indicated by *P < .05, **P < .01, and ***P < .001.
Single-stranded CpG-ODNs inhibit the intracellular TLR3 signaling pathways engaged by poly I:C in DCs. (A) DCs were treated for 1 hour with chloroquine before addition of TLR stimulus, and maturation was measured 48 hours later in 6 different donors. (B) Phosphorylation of the transcription factors IRF-3 and NFκB was detected by Western blot after TLR stimulation for 90 or 15 minutes, respectively. Representative data from 3 independent experiments is shown. (C) Poly I:C–induced secretion of the IRF-3- or NFκB-driven cytokines IFN-α2 (n = 8), IFN-β (n = 5), IP-10 (n = 8), and IL-6 (n = 8), as measured 24 hours after addition of stimuli. Quantity in culture supernatants is depicted in picograms per milliliter as mean ± SEM values. Significant differences comparing with or without addition of ssCpG-ODN were assessed by nonparametric Mann-Whitney 2-tailed test and are indicated by *P < .05, **P < .01, and ***P < .001.
Altered sequence of ssDNA-ODN does not rescue poly I:C–driven DC maturation
To examine whether the CpG sequence in the ssDNA-ODN was important for the inhibition to occur, we stimulated DCs with poly I:C and non-CpG ssDNA-ODN. The poly I:C–induced DC maturation was effectively inhibited using either CpG or non-CpG ssDNA-ODNs (Figure 3A). We furthermore investigated whether the synthetically stabilized phosphorothioate backbone of the ssDNA-ODN used was required for the inhibitory effect. We found that an ssCpG-ODN with natural DNA phosphorodiester backbone had the ability to reduce poly I:C activation of DCs (Figure 3B), but the effect was less pronounced compared with ssDNA-ODNs built on phosphorothioate backbones. To gain insight into whether the class of ssCpG-ODN used was of significance, we tested the type A CpG ODN 2216 and type C CpG ODN 2395 (Figure 3C), as well as an additional type B CpG ODN 2006 (data not shown). Similar to both type B CpG-ODNs, the type C CpG-ODN had the ability to inhibit poly I:C–driven DC maturation, while the type A CpG-ODN, containing a mixed phosphorothioate-phosphorodiester backbone,36 failed to reduce the expression of maturation markers in DCs.
Non-CpG ssDNA-ODNs inhibit poly I:C–induced DC maturation. (A-C) DC maturation was determined in cultures with poly I:C and ssDNA-ODN with (A) a non-CpG sequence (data are shown as MFI mean ± SEM for 11 donors), (B) a phosphorodiester (PD) backbone (MFI mean ± SEM are shown for 12 donors), or (C) different classes of ODNs (MFI mean ± SEM are shown for 10 donors). Significant differences after poly I:C stimulation compared with and without ssDNA-ODNs were assessed by repeated measures ANOVA with Bonferroni Multiple Comparison and are indicated by **P < .01 and ***P < .001, respectively. NS indicates nonsignificant.
Non-CpG ssDNA-ODNs inhibit poly I:C–induced DC maturation. (A-C) DC maturation was determined in cultures with poly I:C and ssDNA-ODN with (A) a non-CpG sequence (data are shown as MFI mean ± SEM for 11 donors), (B) a phosphorodiester (PD) backbone (MFI mean ± SEM are shown for 12 donors), or (C) different classes of ODNs (MFI mean ± SEM are shown for 10 donors). Significant differences after poly I:C stimulation compared with and without ssDNA-ODNs were assessed by repeated measures ANOVA with Bonferroni Multiple Comparison and are indicated by **P < .01 and ***P < .001, respectively. NS indicates nonsignificant.
TLR3-mediated proinflammatory cytokine production is inhibited by ssDNA-ODN in nonhematopoietic cells
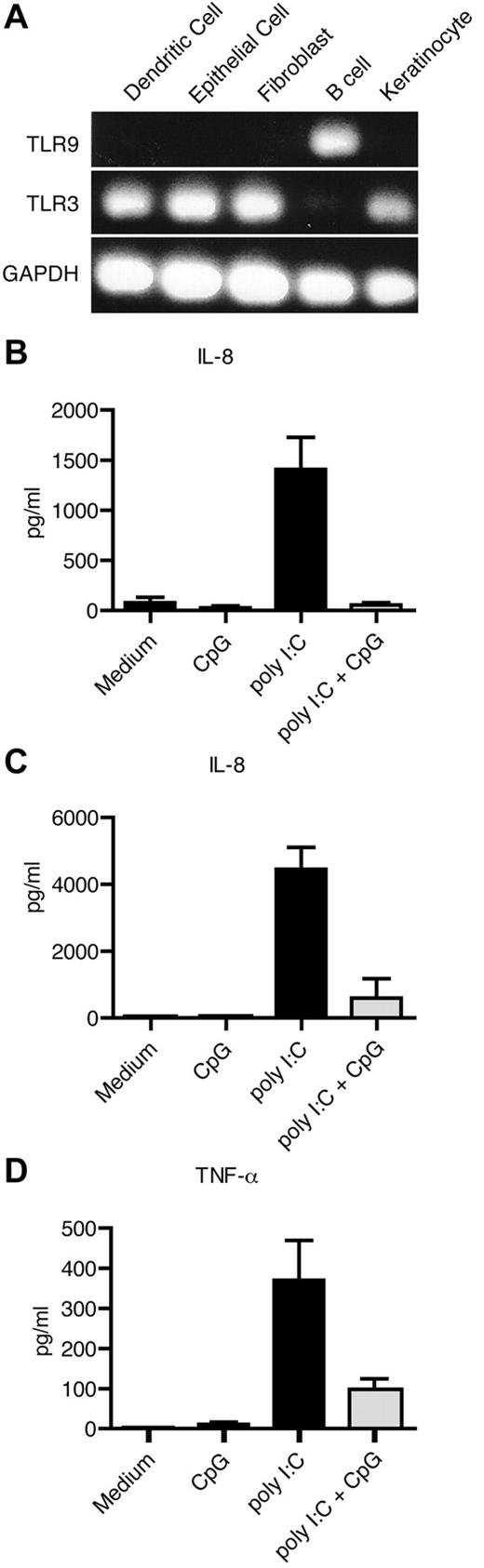
To confirm our observation in additional cell types, we investigated the expression of TLR3 and TLR9 in human primary keratinocytes, human secondary lung fibroblasts (MRC5), and a human bronchial epithelial cell line (16HBE; Figure 4A). As a positive control for TLR3 expression, monocyte-derived DCs were used. Primary CD19+ B cells were used as a negative control for TLR3 and positive control for TLR9 expression. Similar to DCs, keratinocytes, fibroblasts, and epithelial cells expressed TLR3, but not TLR9 (Figure 4A). A proinflammatory factor shown to be produced by nonhematopoietic cells is the chemokine IL-8.37 We therefore measured the secretion of IL-8 in the cultures after TLR stimulation. Poly I:C induced secretion of IL-8 by keratinocytes and fibroblasts, which was inhibited, or partly inhibited, in the cultures with ssCpG-ODN (Figure 4B-C). The epithelial cells, however, produced IL-8 even without addition of stimuli (data not shown). Therefore, we instead investigated the production of TNF-α, which was induced on poly I:C treatment and nearly returned to basal levels in epithelial cells cultured with ssCpG-ODN (Figure 4D).
Single-stranded DNA-ODNs inhibit the effect of poly I:C in human nonhematopoietic cells. (A) Expression of TLR3 and TLR9 mRNA was determined by PCR in DCs, 16HBE epithelial cells, MRC5 fibroblasts, primary B cells, and N/TERT human keratinocytes. (B-D) Secretion of IL-8 from keratinocytes (B) and fibroblasts (C), and TNF-α from epithelial cells (D) after 24 hours of TLR stimulation was measured with ELISA. Data from 3 independent experiments run in triplicates are shown.
Single-stranded DNA-ODNs inhibit the effect of poly I:C in human nonhematopoietic cells. (A) Expression of TLR3 and TLR9 mRNA was determined by PCR in DCs, 16HBE epithelial cells, MRC5 fibroblasts, primary B cells, and N/TERT human keratinocytes. (B-D) Secretion of IL-8 from keratinocytes (B) and fibroblasts (C), and TNF-α from epithelial cells (D) after 24 hours of TLR stimulation was measured with ELISA. Data from 3 independent experiments run in triplicates are shown.
Single-stranded CpG-ODN is endocytosed by DCs and inhibits the uptake of poly I:C
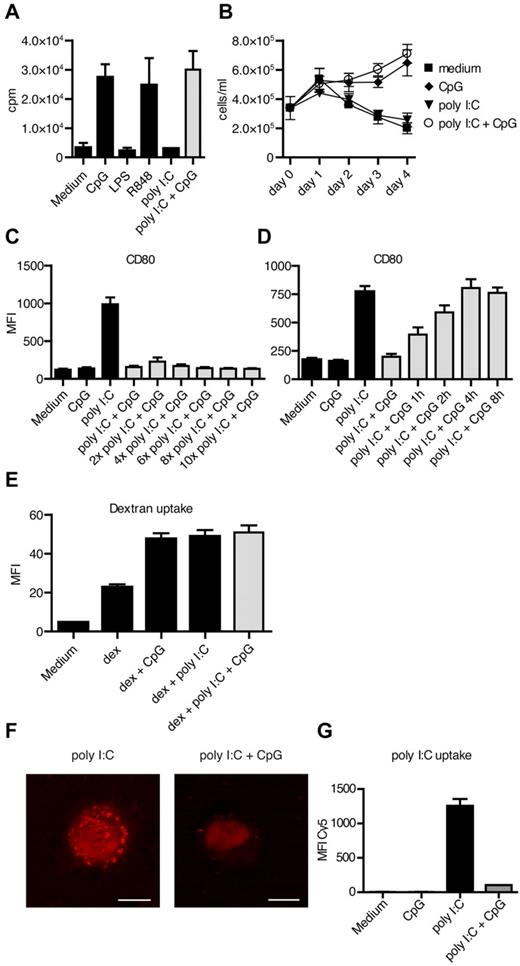
Next, we set out to study whether the inhibitory effect of ssCpG-ODN was occurring at the subcellular level or whether it was a phenomenon of ligand-ligand interaction. To study complex formation between the 2 ligands, we premixed poly I:C and ssCpG-ODN for 1 hour at 37°C and samples were subjected to 4%-20% polyacrylamide gradient gel electrophoresis, which distinguished bands representative for either poly I:C or ssCpG-ODN. However, we could not reveal any interaction (data not shown). We also treated primary human blood CD19+ B cells with poly I:C and ssCpG-ODN. CD19+ B cells express TLR9, but not TLR3 (Figure 4A), and proliferate and increase their survival on in vitro activation. In line with this, proliferation of B cells was induced with ssCpG-ODN, but not with poly I:C. When culturing B cells with the combination of poly I:C and ssCpG-ODN, the cells retained the ability to proliferate as measured by 3H-thymidine incorporation or counting by flow cytometry (Figure 5A-B), thus, again arguing against complex formation. In addition, increasing the poly I:C concentration in the DC cultures up to 10 times did not overcome the blocking effect of ssCpG-ODN (Figure 5C).
The uptake of poly I:C is prevented by ssDNA-ODN. (A-B) B-cell proliferation after TLR stimulation was measured by (A) 3H-thymidine incorporation and (B) flow cytometry-based cell counting. Data from 3 independent experiments run in triplicates are shown. (C) DC maturation was assessed after 48 hours of culture with ssCpG-ODN and increasing concentrations of poly I:C. The results are depicted as the MFI and are the mean ± SEM values from 6 donors. (D) Kinetic analyses of the inhibitory effect of ssCpG-ODN in poly I:C–stimulated DCs performed in 6 donors. (E) Endocytosis of pHrodo-labeled dextran beads by DCs was measured after 2 hours of culture with ssCpG-ODN, poly I:C, or combined treatment with ssCpG-ODN and poly I:C. (F) Single-stranded CpG-ODN and Cy5-labeled poly I:C (red) was cultured with DC for 15 minutes either alone or in combination and the subcellular location visualized with confocal microscopy. Scale bars are 5 μm. Representative data from 3 independent experiments are shown. (G) Quantitative measurement of Cy5-labeled poly I:C uptake into DC was performed with flow cytometry.
The uptake of poly I:C is prevented by ssDNA-ODN. (A-B) B-cell proliferation after TLR stimulation was measured by (A) 3H-thymidine incorporation and (B) flow cytometry-based cell counting. Data from 3 independent experiments run in triplicates are shown. (C) DC maturation was assessed after 48 hours of culture with ssCpG-ODN and increasing concentrations of poly I:C. The results are depicted as the MFI and are the mean ± SEM values from 6 donors. (D) Kinetic analyses of the inhibitory effect of ssCpG-ODN in poly I:C–stimulated DCs performed in 6 donors. (E) Endocytosis of pHrodo-labeled dextran beads by DCs was measured after 2 hours of culture with ssCpG-ODN, poly I:C, or combined treatment with ssCpG-ODN and poly I:C. (F) Single-stranded CpG-ODN and Cy5-labeled poly I:C (red) was cultured with DC for 15 minutes either alone or in combination and the subcellular location visualized with confocal microscopy. Scale bars are 5 μm. Representative data from 3 independent experiments are shown. (G) Quantitative measurement of Cy5-labeled poly I:C uptake into DC was performed with flow cytometry.
To further determine whether simultaneous addition of the ligands was necessary for the inhibitory effect of ssCpG-ODN in poly I:C–treated cells, we pretreated DCs with one of the ligands at varying time points before addition of the second ligand. The DC maturation was efficiently blocked when adding ssCpG-ODN up to 6 hours before poly I:C (data not shown). However, when DCs were first pretreated with poly I:C, there was a kinetic-dependent relative inhibition by ssCpG-ODN. Hence, DCs up-regulated maturation markers progressively the longer the cells were cultured with poly I:C alone (Figure 5D). This indicates that ssCpG-ODNs lack the ability to inhibit already initiated signaling from TLR3. Consequently, for single exposures to poly I:C, ssCpG-ODNs need to be present either before, or simultaneously with, addition of poly I:C to efficiently block the DC-maturing effect of the ligand, while under chronic stimulation, inhibition may be feasible at later time points. To examine the impact of ssCpG-ODNs on TRIF signaling, we stimulated DCs with MPLA, a TLR4 ligand known to mediate its effect largely via TRIF (supplemental Figure 3). The MPLA-induced DC maturation was not affected by ssCpG-ODN, indicating that it does not interfere with intracellular TRIF signaling.
Thereafter, we investigated whether the endocytic process of poly I:C–stimulated DCs was affected by ssCpG-ODN. However, ssCpG-ODN did not block the endocytic process enhanced by poly I:C, as measured by uptake of endocytosis-specific pHrodo-marked dextran beads (Figure 5E). On the contrary, ssCpG-ODN increased the uptake of dextran beads into DC per se. We further asked whether the uptake mechanism for ssCpG-ODN and poly I:C might be shared, as well as whether ssCpG-ODN was inhibiting uptake of poly I:C. Confocal microscopy revealed uptake of Cy5-labeled poly I:C in DCs 15 minutes after addition, while a lower uptake was detected in cultures with ssCpG-ODN (Figure 5F). This was confirmed in FACS, where the presence of ssCpG-ODN decreased the uptake of poly I:C into DCs (Figure 5G), while the uptake of FITC-tagged ODN 2006 was not altered in the presence of poly I:C (data not shown).
Single-stranded DNA-ODN inhibits proinflammatory cytokine release on intranasal administration of poly I:C in cynomolgus macaques
To investigate whether ssDNA-ODN can inhibit TLR3 signaling in vivo, we first treated C57Bl6 mice with a combination of non-CpG ssDNA-ODN, previously shown to have an inhibitory effect on human DC maturation (Figure 3A), and poly I:C (supplemental Figure 4). Mice injected intraperitoneally with poly I:C showed induction of IL-12p40 and IP-10 in serum. However, mice treated with the combination of poly I:C and ssDNA-ODN did not display lower IP-10 or IL-12p40 levels in serum than those treated with poly I:C alone (supplemental Figure 4A-B). To instead target the ligands to another tissue, we administered the poly I:C and non-CpG ssDNA-ODN intranasally and measured IL-12p40 secretion in bronchoalveolar lavage (data not shown). We could, however, not reveal any inhibition by ssDNA-ODN in this setting either.
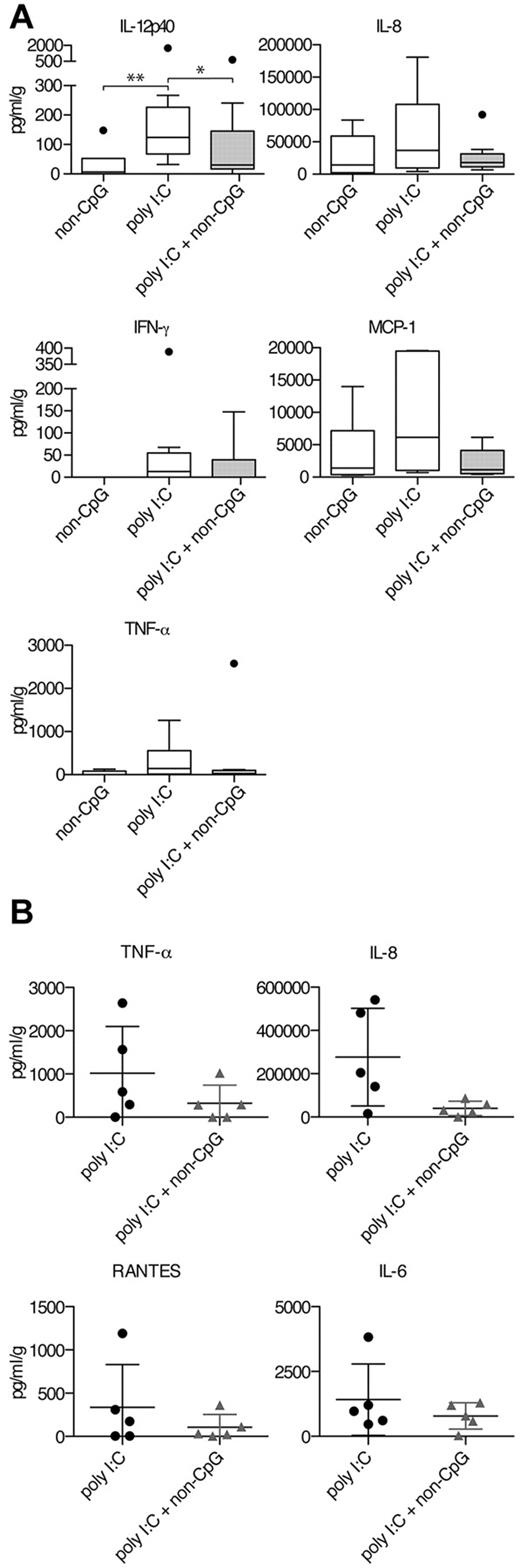
To overcome the differences between mice and primates, cynomolgus macaques were treated with poly I:C, non-CpG ssDNA-ODN, or a combination of the two via the intranasal route. Cytokine levels in nose swaps were measured by Bio-Plex technology 24 or 18 hours after administration. Cell infiltration and phenotype were evaluated in some animals by hematoxylin-eosin staining, but no significant differences between the groups were detected (data not shown). There was a large variation between the animals and to prevent skewing from outliers, these animals were excluded in the statistical analyses but depicted as individual filled circles in box plots (Figure 6A). After 24 hours, we detected induction of IL-12p40 after poly I:C treatment, which was significantly blocked in the presence of non-CpG ssDNA-ODNs (Figure 6A). IL-12 is an important regulator of IFN-γ, and we therefore assessed the production of IFN-γ in the nose of treated animals. Six of 9 animals showed induction of IFN-γ after intranasal administration of poly I:C, while 3 of the animals in the poly I:C and non-CpG ssDNA-ODN group displayed IFN-γ production (supplemental Figure 5). Lung fibroblasts and epithelial cells produced IL-8 and TNF-α on poly I:C treatment in vitro (Figure 4C-D). Similarly, there was a trend toward elevated IL-8- and TNF-α levels in the group receiving poly I:C compared with the poly I:C and non-CpG ssDNA-ODN–treated group (Figure 6A). To investigate the chemokine response, we measured levels of MCP-1 and found that poly I:C–mediated elevation was reduced by non-CpG ssDNA-ODNs. To investigate earlier kinetics, we administered poly I:C or poly I:C and non-CpG ssDNA-ODN in 10 additional macaques and measured the cytokine and chemokine release after 18 hours (Figure 6B). Again, the levels of IL-8 and TNF-α were reduced in the group receiving both ligands compared with poly I:C treatment only and a similar trend was detected for RANTES and IL-6. Altogether, these data show in vivo proof-of-principle of the capacity of ssDNA-ODN to block intranasal TLR3-induced responses in nonhuman primates, but not in mice, showing species differences in the regulation of TLR3-mediated responses.
Poly I:C–induced secretion of proinflammatory cytokines and chemokines is inhibited by non-CpG ssDNA-ODN in cynomolgus macaques. (A-B) Animals were treated intranasal with indicated TLR ligands. Nose swaps were collected at 24 or 18 hours. Proinflammatory cytokine and chemokine levels were measured using Bio-Plex technology. (A) Levels of IL-12p40, IL-8, TNF-α, IFN-γ, and MCP-1 were measured 24 hours after treatment. Data are depicted as box plots (range and median) with outliers marked as filled circles. Data in the poly I:C group and poly I:C + non-CpG group are from 3 independent experiments (n = 9 per group), while data in the non-CpG group and all data for MCP-1 are from 2 independent experiments (n = 6). (B) Levels of IL-8, TNF-α, RANTES, and IL-6 after 18 hours of treatment were measured from 10 additional animals. Dot plots display individual data, and mean ± SD is depicted. Significant differences between poly I:C–treated animals and animals treated either with ssDNA-ODN alone or in combination with poly I:C were assessed by nonparametric Mann-Whitney 2-tailed test and are indicated by *P < .05 and **P < .01.
Poly I:C–induced secretion of proinflammatory cytokines and chemokines is inhibited by non-CpG ssDNA-ODN in cynomolgus macaques. (A-B) Animals were treated intranasal with indicated TLR ligands. Nose swaps were collected at 24 or 18 hours. Proinflammatory cytokine and chemokine levels were measured using Bio-Plex technology. (A) Levels of IL-12p40, IL-8, TNF-α, IFN-γ, and MCP-1 were measured 24 hours after treatment. Data are depicted as box plots (range and median) with outliers marked as filled circles. Data in the poly I:C group and poly I:C + non-CpG group are from 3 independent experiments (n = 9 per group), while data in the non-CpG group and all data for MCP-1 are from 2 independent experiments (n = 6). (B) Levels of IL-8, TNF-α, RANTES, and IL-6 after 18 hours of treatment were measured from 10 additional animals. Dot plots display individual data, and mean ± SD is depicted. Significant differences between poly I:C–treated animals and animals treated either with ssDNA-ODN alone or in combination with poly I:C were assessed by nonparametric Mann-Whitney 2-tailed test and are indicated by *P < .05 and **P < .01.
Discussion
There has been a paradigm shift in the understanding of innate immune responses since the discovery of TLRs and other pattern recognition receptors.1–3 This knowledge has not only enabled new vaccine adjuvants to be developed, but the mechanisms behind many inflammatory conditions have also been characterized in greater detail.6 TLR3 has been shown to have a detrimental role in several infection models,15–19,38 in autoimmunity,21,39 and during sterile tissue damage.20,23 In the present study, we have characterized ssDNA-ODN–mediated inhibition of TLR3 activation in human DCs and in vivo in cynomolgus macaques.
We here provide evidence that poly I:C–induced DC maturation was inhibited by ssDNA-ODNs on all levels investigated: up-regulation of maturation markers, production of type I interferons and proinflammatory cytokines, and phosphorylation of vital transcription factors. Single-stranded DNA-ODNs have previously been shown to have an inhibitory effect on spontaneous IL-8 production from keratinocytes, irrespective of CpG sequence, but the mechanism for this phenomenon was not revealed.37
Necrotic and noncleared apoptotic cells induce immune activation because of exposed danger-associated molecular patterns,40 and the release of endogenous dsRNA activates TLR3.8,9,41 Virally infected or poly I:C–loaded apoptotic cells have also been shown to activate DCs via TLR3 leading to cross-presentation of Ags to CTLs.12,42 It is conceivable that cell death and the immunologic consequences induced by TLR3 signaling (in, for example, liver transplantation after hepatitis C–related cirrhosis) play a major role in acute graft rejection.23
Pathogenic responses to apoptosis and necrosis can have severe effects also in the lungs, where dying and infected cells have been shown to induce harmful responses via the TLR3 pathway.19,20 TLR3 responses to viral RNA can also have detrimental effects. Allergic asthma can, for example, be exacerbated by respiratory viral infections stimulating TLR3.17,18 We could indeed reduce the production of proinflammatory cytokines with ssCpG-ODN in both epithelial cells and fibroblasts originally derived from the lung. However, the cytokine production was not completely blocked. This might be because of engagement of TLR3 expressed on the cell surface or from MDA-5 located in the cytosol of the cells.5,7 In addition, apoptotic poly I:C–transfected HeLa cells had the ability to induce DC maturation, which in the presence of ssCpG-ODN was significantly reduced.
The CpG sequence in ssDNA-ODN ligands containing a phosphorothioate backbone has been shown to be indispensable for activation of TLR9, and the stimulatory effect of the ligand is lost if the CpG repeats are removed.43 Although non-CpG–containing ssDNA-ODNs retained the ability to inhibit poly I:C–mediated DC maturation in our experiments, and from this, as well as from the notion that DCs used in present study did not express TLR9 mRNA, we excluded inhibitory signaling via TLR9 as a mechanism for the observation. However, ssCpG-ODN with a phosphorodiester backbone only partly reduced the DC maturation after poly I:C treatment. In line with this, we only detected inhibition of poly I:C–induced DC maturation with class B and C ssCpG-ODNs, but not with the phosphorodiester containing class A ssCpG-ODN.44 In DCs, poly I:C is taken up and activates TLR3 via the endosomal route.45 This we could confirm because the response to poly I:C was absent when endosomal acidification was blocked. Uptake has been shown to be mediated via clathrin and dependent on the cytoplasmic lipid raft protein Raftlin, which is also essential for uptake of class B and C, but not class A, ssCpG-ODNs.31,32,35,46 Taking this into consideration, combined with the lack of complete inhibition of proinflammatory cytokine production from fibroblasts and epithelial cells also expressing TLR3 on their cell surface,7 the mechanism of ssDNA-ODN–mediated block of TLR3 signaling is likely to take place within the endosomal compartment.
When we investigated the endocytic uptake by DCs treated with poly I:C, ssCpG-ODN, or the combination of the two, we found that ssCpG-ODN induced similar uptake of dextran beads as poly I:C–stimulated cells and that the combination of the 2 ligands did not alter the dextran uptake. From this, we concluded that ssDNA-ODNs did not inhibit the endocytic process. Rather the opposite was detected, where endocytosis, but not DC maturation, was stimulated by ssCpG-ODNs. Confocal imaging and FACS quantification of the uptake of poly I:C revealed that ssCpG-ODN inhibited the uptake when added simultaneously. It can be speculated that the system was saturated by ssCpG-ODN, preventing poly I:C to be taken up. This was supported by the titration experiments, where increased concentration of poly I:C did not overcome the ssCpG-ODN–mediated block of DC maturation, while the inhibition indeed was lost in a concentration-dependent manner when reducing the concentration of ssCpG-ODN.
Single-stranded CpG-ODN is a vigorous activator of TLR943 and we have in the present study shown potent proliferation of human TLR9-expressing B cells after ssCpG-ODN treatment, which was not inhibited by the presence of poly I:C. In cells lacking expression of TLR9 mRNA, such as monocyte-derived DCs, no activation was observed. However, synergistic effects have previously been shown when coadministrating the ligands in animal models, both in vitro47,48 and in vivo.49,50 This is consistent with our in vivo data from mice, where the effect of poly I:C was not inhibited by type B ssDNA-ODNs. An explanation for this finding can be that the uptake mechanism for poly I:C has been shown to be redundantly dependent on Raftlin in mice.46 Hence, additional pathways of poly I:C uptake that are not shared with ssDNA-ODNs might exist in the murine system.
We therefore repeated the experiments in macaques, and administered the ligands via the intranasal route to imitate a viral infection, or the presence of damaged or dying cells. Poly I:C induced the production of proinflammatory cytokines in the majority of the animals and, in line with our human data, decreased in animals also receiving ssDNA-ODN. Significance was reached for ssDNA-ODN–mediated inhibition of IL-12p40 secretion after poly I:C stimulation. There was also a trend for the TNF-α, IL-8, MCP-1, and IFN-γ responses, where the highest responses were detected in the poly I:C group while the poly I:C and ssDNA-ODN group displayed levels similar to the ssDNA-ODN–treated group.
The findings in this study reveal a novel therapeutic approach for TLR3 inhibition. There are several inflammatory diseases associated with excessive TLR3 activation. Detrimental responses have been implicated both in response to sterile cell death8,17,21,39 and in infectious conditions.15,16,19,38 One option might be to inhibit TLR3-mediated responses triggered by dying cells during liver transplantation.23 A similar approach could be to temporarily treat patients with acute respiratory distress syndrome, where increased expression of TLR3 has been shown to mediate pathologic effects on oxygen treatment.20 We have here shown that the immune-activating effects of the TLR3 ligand poly I:C after intranasal administration in cynomolgus macaques can be effectively blocked by an intranasal dose of ssDNA-ODN. These findings have implications for novel treatment of TLR3-mediated pathology in both infections and inflammatory conditions.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Cecilia Kemi, Ulrika Johansson, and Lilian Walther Jallow for help with experimental procedures.
This work was supported by EUROPRISE, the Swedish Research Council, the Magnus Bergvalls Foundation, the Olle Engkvist Foundation, and the Åke Wibergs Foundation.
Authorship
Contribution: A.E.S. designed and performed research, analyzed data, and wrote the manuscript; M.H. contributed to experimental design, performed research, and analyzed data; L.V. contributed to experimental design and performed research; H.S. performed the macaque experiments; N.B. and R.L. contributed to experimental design for the macaque study; and C.I.E.S. and A-L.S. supervised the study and contributed to experimental design.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.H. is Department of Internal Medicine, The University of Texas Southwestern Medical Center, Dallas, TX.
Correspondence: Annette E. Sköld or Anna-Lena Spetz, Center for Infectious Medicine, Department of Medicine Huddinge, Karolinska University Hospital Huddinge F59, 14186 Stockholm, Sweden; e-mail: annette.skold@ki.se or anna-lena.spetz@ki.se.
References
Author notes
A.E.S. and M.H. contributed equally.