Abstract
Anaplastic large-cell lymphomas (ALCLs) are a group of clinically and biologically heterogeneous diseases including the ALK+ and ALK− systemic forms. Whereas ALK+ ALCLs are molecularly characterized and can be readily diagnosed, specific immunophenotypic or genetic features to define ALK− ALCL are missing, and their distinction from other T-cell non-Hodgkin lymphomas (T-NHLs) remains controversial. In the present study, we undertook a transcriptional profiling meta-analysis of 309 cases, including ALCL and other primary T-NHL samples. Pathway discovery and prediction analyses defined a minimum set of genes capable of recognizing ALK− ALCL. Application of quantitative RT-PCR in independent datasets from cryopreserved and formalin-fixed paraffin-embedded samples validated a 3-gene model (TNFRSF8, BATF3, and TMOD1) able to successfully separate ALK− ALCL from peripheral T-cell lymphoma not otherwise specified, with overall accuracy near 97%. In conclusion, our data justify the possibility of translating quantitative RT-PCR protocols to routine clinical settings as a new approach to objectively dissect T-NHL and to select more appropriate therapeutic protocols.
Introduction
Systemic anaplastic large-cell lymphomas (ALCLs) are a peripheral T cell–derived malignancy accounting for approximately 12% of all T-cell non-Hodgkin lymphomas (T-NHLs).1 Originally described by Stein et al in 1985 as ALCL-expressing CD30,2 its definition and relationship with other T-NHLs has undergone a series of revisions.1,3,4 Based on genetic and clinical features, 2 different entities are now recognized as systemic forms, the ALK-positive (ALK+) and ALK-negative (ALK−) ALCL.1,5 The first entity is characterized by recurrent chromosomal translocations involving the Anaplastic Lymphoma Kinase (ALK) gene, which leads to the expression and constitutive activation of anaplastic lymphoma kinase (ALK) fusion proteins.6,7
Whereas ALK+ ALCL are readily diagnosed by anti-ALK Abs, the recognition of ALK− ALCL is in some instances subjective. In fact, immunophenotypic or genetic features that define ALK− ALCL precisely are missing; accordingly, ALK− ALCL has been considered a provisional entity by the World Health Organization (WHO) classification.1
Although all ALCLs display strong and diffuse immunoreactivity for CD30, the expression of this marker is not specific for ALCL. Indeed, CD30 is found also in activated nonneoplastic lymphoid cells, in a subset of peripheral T-cell lymphoma not otherwise specified (PCTL-NOS), in Hodgkin lymphoma, and other neoplasms such as embryonal carcinoma.8 At present, the diagnosis of ALCL relies on the application of a panel of Abs for B- and T cell–restricted antigens, epithelial membrane antigen (EMA), granzyme B, perforin, and T cell–restricted intracellular antigen-1 (TIA1).5 ALCLs usually show an aberrant T-cell phenotype with frequent loss of the common T-cell markers such as the pan–T-cell antigens,9 although in approximately 85%-90% of ALCL clonal TCR gene rearrangement can be detected by PCR. Furthermore, PAX5 negativity is critical for the differentiation of ALCL from common Hodgkin lymphoma and CD30+ diffuse large B-cell lymphoma.10
Recently, chromosomal translocations affecting 6p25.3, which targets DUSP22 and/or IRF4 have been described in a subset of ALK− ALCLs, with predominant cutaneous involvement11,12 ; however, the effects on the pathogenesis of this lymphoma are still largely unknown.
ALK− ALCL distinction is supported by genetic criteria, epidemiologic data, and clinical features. The crude 5-year overall survival of this lymphoma is 49%, a value intermediate between 70% for ALK+ ALCL and 32% for PTCL-NOS.1 Nevertheless, when patients are stratified according to the clinical parameters (ie, age and/or stage), ALK+ and ALK− ALCL patients display a similar prognosis in terms of failure-free and overall survival.5 Accordingly, a retrospective study emphasized the primary impact of age and serum β2-microglobulin levels on the prognosis of ALCL patients.13 These data suggest that clinical factors other than biologic components might play a prominent role in outcome of ALCL patients and that clearcut distinction of ALK− ALCL from PTCL-NOS, in particular for those with strong and uniform CD30 expression, is essential for delivering the most appropriate therapies, thus avoiding unnecessary toxicities or suboptimal treatments.
Gene-expression profiling and comparative genomic hybridization studies have shown that ALK+ and ALK− ALCL share restricted genomic signatures and/or preferential genomic aberrations.14-19 Nevertheless, these studies have not identified exclusive/specific markers for ALK− ALCL. We uncovered several genes similarly expressed in ALK+ and ALK− ALCL samples that are capable of distinguishing ALCL tumors from PTCL-NOS and other NHL samples, suggesting the existence of a common ALCL signature.20
Because gene-expression profiling analysis on every patient is currently impractical and not standardized for routine clinical settings, alternative strategies should be considered. Ideally, the stratification of unique entities requires simple, reproducible, and low-cost tests. While this goal has been reached for ALK+ ALCL, effort is required for the unequivocal distinction of ALK− ALCL from CD30+ PTCL.
In the present study, we undertook a systematic approach to profile the expression signature of a large set of primary T-NHLs and defined a minimum set of genes useful for the stratification of ALK− ALCL. Quantitative RT-PCR (RT-qPCR) analysis performed in independent datasets of cryopreserved or formalin-fixed paraffin-embedded (FFPE) samples validated the gene-expression profiling predictions and suggested the possibility of translating RT-qPCR protocols to routine clinical settings as a new avenue to defining T-NHL and to selecting more appropriate therapeutic protocols.
Methods
Processing of microarray data
Gene-expression data were obtained from 3 publicly available (GSE6338, GSE14879, and GSE19069 at the National Center for Biotechnology Information Gene Expression Omnibus repository http://www.ncbi.nlm.nih.gov/geo/),16-18 proprietary,20 and unpublished datasets including T cells from healthy donors and those from ALCL, PTCL, angioimmunoblastic T-cell lymphoma (AITL), and adult T-cell leukemia/lymphoma patients.
Expression values were extracted from CEL files using Affymetrix chip definition files and manufacturer's annotations for HG-U133Plus arrays (release 31). Normalization was performed with the robust multiarray average procedure using the appropriate functions in the affy package for Bioconductor (http://bioconductor.org). To strengthen the robustness and the reproducibility of the whole analysis, the expression set has been analyzed using the arrayQualityMetrics function in the homonymous package in R software (http://www.r-project.org). Samples classified as outliers under standard analysis conditions were removed from computations.
Microarray data analysis
Differential analysis was carried out using the Comparative Marker Selection suite available as a GenePattern module (http://www.broad.mit.edu/genepattern). Genes were ranked based on the value of 2-sided t test statistics to assess differential expression. A total of 100 000 permutations were performed to compute the significance (nominal P value) of the rank assigned to each gene. The analysis was adjusted for multiple hypotheses testing using the q value as the statistical approach. The selected probe lists were visualized in a heat map format using Heat Map Viewer. The search of classifier genes was executed by Prediction Analysis of Microarrays, as described previously.21 The optimal value of Δ to obtain the minimum cross-validation error was chosen using a leave-one-out cross-validation process.
Patients and case selection for RT-qPCR analysis
Cryopreserved samples of 20 PCTL-NOS and 40 ALCL (20 ALK+ and 20 ALK−) patients were provided by the Universities of Leuven, Wuerzburg, Torino, Bologna, Verona, Brescia, and Napoli and by the San Raffaele Scientific Institute of Milan. T-NHLs were selected on the basis of stringent criteria: (1) lymph node biopsy site, (2) presence of at least 50% neoplastic cells, (3) RNA preservation, and (4) strong CD30 expression, presence of T cell–associated markers, granzyme B, and TIA-1 positivity and PAX-5 negativity (for ALCL cases).
FFPE tumor samples of 32 PCTL-NOS and 63 ALCL (29 ALK+ and 34 ALK−) patients were from our archive. All samples were obtained at the time of diagnosis before treatment. ALCL cases were submitted to central pathologic review by a panel of 2 expert hematopathologists (S.P. and G.I.). Final diagnoses were assigned according to the criteria of the WHO classification,1 as described previously.20 Two unclassifiable cases were excluded from the study. Because CD30+ PTCL/NOS is not yet characterized in the WHO classification, we defined this subgroup by the following features: (1) CD30 expression in more than 30% of neoplastic elements, (2) pleomorphic morphology with mainly medium-sized cells, and (3) lack of hallmark cells. Informed consent was obtained from all enrolled patients following the procedures approved by the local ethical committees of each participating institution.
Purification of total RNA and cDNA synthesis
Total RNA from cryopreserved and FFPE samples were extracted using TRIzol reagent (Invitrogen) or the miRNeasy FFPE Kit (QIAGEN), respectively, according to the manufacturer's protocols. cDNA was obtained from 0.5 μg of total RNA treated previously with RNase-free DNase (Roche Diagnostic) using reverse transcriptase SuperScript III and random hexamers (Invitrogen) or gene-specific reverse primers, as reported in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
RT-qPCR
RT-qPCR was performed with a Thermal iCycler (Bio-Rad) using the Bio-Rad iQ SYBR Green Supermix according to the manufacturer's instructions. The PCR cycling conditions were as follows: 95°C for 5 minutes, followed by 40 cycles at 94°C for 10 seconds and 60°C for 30 seconds.
The oligonucleotide primer pairs used for RT-qPCR were designed with the PrimerBLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) to obtain amplicons of 70-110 bp. Primer sequences are reported in supplemental Table 1.
To confirm the amplification specificity, the PCR products were subjected to the analysis of melting curve, linearity, and slope of standard curve. All PCR assays were performed in triplicate. Gene-expression results were normalized to GAPDH and HUPO expressions and calculated using the ΔCt method according to the manufacturer's instructions.
The quality of cryopreserved and FFPE samples was assessed according to the following qPCR parameters: PTCL-NOS with GAPDH/HUPO Ct value > 30, ALCL with either GAPDH/HUPO Ct value > 30 or TNFRSF8 ΔCt value > 5 were excluded from the analysis.
Statistical analysis
Wilcoxon rank-sum test was used to assess differences in the distribution of 2 sample populations. The predictive power of the investigated genes was tested using the Linear Discriminant Analysis for the classification of multivariate observations, with leave-one-out procedure. ROC analysis was performed using DiagnosisMed package. Beeswarm package was used for visualizing stripchart distributions. All calculations were performed in R software.
Calculation of metagene value
Metagene adjusted ΔCt value corresponds to the weighted mean of ΔCt values of TNFRSF8, BATF3 and TMOD1. Every single gene weight was established as the averaged scaling factors calculated on the training sets for each left-out sample, with the scaling factor defined as the value that transform each ΔCt to discriminant functions in the Linear Discriminant Analysis model. ROC analysis of the metagene value was performed to identify the thresholds leading to the maximum classification accuracy. The unclassifiable samples area (metagene values between 1.64 and 1.66) was interposed between the whiskers of PTCL-NOS and ALK− ALCL distributions (delimiting 1.5 times the corresponding interquartile ranges and encompassing > 95% samples). For convenience, online tool is available for smart calculation of the predicted phenotype at http://cerms.altervista.org.
Results
Generation of a large T-NHL gene-expression profiling dataset
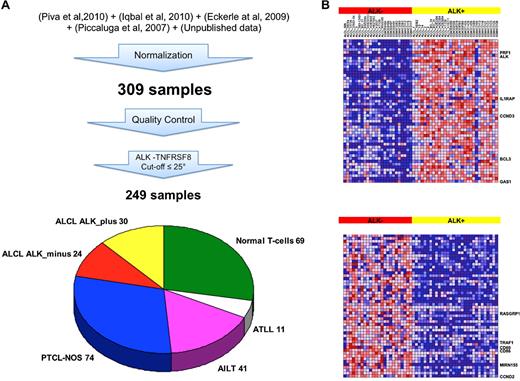
To unravel the regulatory network underlying the ALCL phenotype, and to discover new genomic lesions or biomarkers useful for the recognition of ALK− ALCL patients, we undertook a transcriptional meta-analysis assembling 4 available datasets of T-NHL patients,16-18,20 integrating them with some proprietary unpublished cases, for a total of 309 samples. Normal T-cell and T-NHL samples were included based on stringent quality controls of the array, followed by a molecular diagnostic verification as described in “Methods.” Substantial expression of CD30/TNFRSF8 and/or ALK over the 25th percentile was further required to select ALCL cases with higher neoplastic cell content, and therefore more likely to display a distinctive ALCL signature. As a matter of fact, exclusion of samples within the lower quartile led to the best false discovery rate values and to the largest number of differentially expressed genes between ALK+ and ALK− samples (supplemental Figure 1). This approach reduced batch effects because of cohort- or laboratory-specific biases, and steered to the selection of 249 samples, among which 69 normal T-cells, 11 T-ALL, 41 AITL, 74 PTCL-NOS, and 54 ALCL (30 ALK+, 24 ALK−; Figure 1A).
Generation of a large T-NHL gene-expression profiling dataset and transcriptional characterization of ALCL patients. (A) Gene-expression data were extracted from the indicated datasets and normalized with robust multiarray average procedure. Samples classified as outlier on standard quality control analyses were removed from following computations. Expression of TNFRSF8 and/or ALK over the 25th percentile was required to include ALCL cases with higher neoplastic cell content. (B) Heat map of the top 50 genes up-regulated in ALK+ and ALK− ALCL cases (30 ALK+ and 24 ALK−). Each colored cell in the heat map represents the gene-expression value for a probe in a sample. The highest expression values are displayed in red, the lowest values in blue.
Generation of a large T-NHL gene-expression profiling dataset and transcriptional characterization of ALCL patients. (A) Gene-expression data were extracted from the indicated datasets and normalized with robust multiarray average procedure. Samples classified as outlier on standard quality control analyses were removed from following computations. Expression of TNFRSF8 and/or ALK over the 25th percentile was required to include ALCL cases with higher neoplastic cell content. (B) Heat map of the top 50 genes up-regulated in ALK+ and ALK− ALCL cases (30 ALK+ and 24 ALK−). Each colored cell in the heat map represents the gene-expression value for a probe in a sample. The highest expression values are displayed in red, the lowest values in blue.
Identification of a minimum set of genes useful for the stratification of ALK− ALCL
In agreement with previous studies,20 the application of unsupervised analysis to this large dataset was not able to distinguish ALK− ALCL from other T-NHL categories (data not shown). First, we investigated the transcriptional pattern distinguishing ALK+ and ALK−ALCL in the large assembled dataset of 54 ALCL. Differential analysis identified 185 genes up-regulated in ALK+ and 95 in ALK− ALCL (Figure 1B). PRF1, IL1RAP, CCND3, BCL3, and GAS1 scored within the top up-regulated transcripts of ALK+ specimens (supplemental Table 2).20 In ALK− samples, CD80, DC86, CCND2 and MIR155HG were among the most significant over-expressed genes (Figure 1B). It has been recently proposed that mir-155 is part of a distinct miRNA expression pattern that characterizes ALK−ALCL.22 Because miR-155 is univocally processed from its noncoding MIR155HG host gene,23 we used the surrogate expression of MIR155HG to assess the transcriptional level of the mature miR-155 in the large dataset of T-NHL and normal T-cells. Surprisingly, we found that mir-155 was consistently over-expressed in the majority of samples with the exception of ALK+ ALCL, suggesting that mir-155 could not be used as a differential marker of ALK− ALCL (supplemental Figure 2).
To identify ALK− markers, we decided to perform a differential analysis comparing the expression profiles of either whole ALCL or ALK− ALCL subset with those of PTCL-NOS and AITL samples (supplemental Figures 3-5). TNFRSF8, BATF3, GGT1, and LGALS1 resulted among the top scorers for both ALK− and whole ALCL cases, indicating that a common ALCL signature could be dominant as result of the high heterogeneity of the ALK−subgroup.
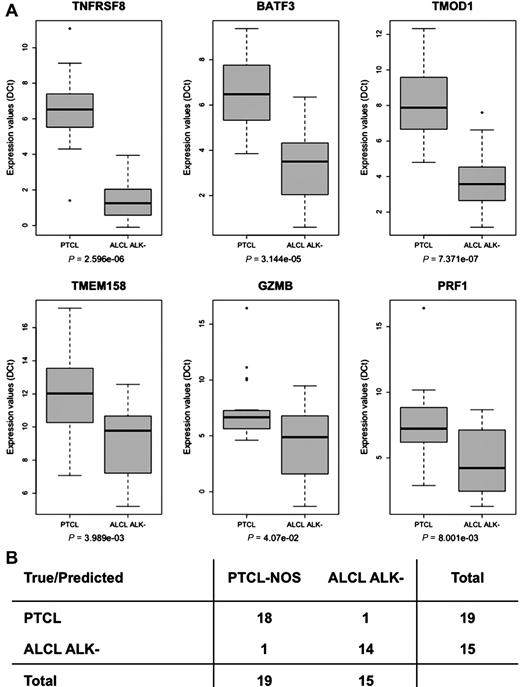
Provided that immunohistochemical detection of ALK protein unequivocally recognizes ALK+ ALCL, in the attempt to search for genes characterizing the ALK− ALCL fingerprint, we applied the Prediction Analysis of Microarrays software to the assembled dataset. The prediction led to the identification of a 6-gene classifier (TNFRSF8, BATF3, TMOD1, TMEM158, MSC, POPDC3), highly informative in discerning ALK− ALCL patients (overall error rate 4.1%). Importantly, the combined expression of the genes included in the classifier outperformed the evaluation of conventional ALCL markers such as granzyme B (GZMB) and perforin (PRF1), which are occasionally expressed also in normal T-cell and other T-NHL (Figure 2).
Class-prediction analysis identifies a genomic classifier for ALK− ALCL. (A) Heat map showing the expression of the 6 genes included in the Prediction Analysis of Microarrays classifier, together with ALK, GZMB, and PRF1 in ALCL, PTCL-NOS, and AITL used in the analysis. (B) Box-plot representation of the same genes in the full dataset of 249 samples, including normal donors and adult T-cell leukemia/lymphoma tumors. The Kruskal-Wallis P value is indicated for each transcript.
Class-prediction analysis identifies a genomic classifier for ALK− ALCL. (A) Heat map showing the expression of the 6 genes included in the Prediction Analysis of Microarrays classifier, together with ALK, GZMB, and PRF1 in ALCL, PTCL-NOS, and AITL used in the analysis. (B) Box-plot representation of the same genes in the full dataset of 249 samples, including normal donors and adult T-cell leukemia/lymphoma tumors. The Kruskal-Wallis P value is indicated for each transcript.
Validation of the classifier by RT-qPCR
The performance of the ALK− ALCL fingerprint was challenged using RT-qPCR in an independent set of cryopreserved samples. The expression of the 6 previously identified genes, together with GZMB and PRF1, was evaluated in 19 PTCL-NOS and 28 ALCL (13 ALK+ and 15 ALK−) cases. The Wilcoxon test of RT-qPCR data established that TNFRSF8, BATF3, and TMOD1 genes were expressed the most differentially in ALK− ALCL (Figure 3A). Therefore, we verified the predictive power of the 3-gene classifier model using leave-one-out and linear discriminant analysis as the cross-validation procedure and prediction method, respectively. This classifier separated robustly either ALK− ALCL (accuracy 94.1%) or ALCL overall (accuracy 93.6%) from PTCL-NOS, with higher power than larger sets of genes (Figure 3B and data not shown).
Validation of a 3-gene model classifier by RT-qPCR in cryopreserved samples. (A) Box plot showing RT-qPCR quantification of TNFRSF8, BATF3, TMOD1, TMEM158, GZMB, and PRF1 expression in PTCL-NOS and ALK− ALCL. Statistical significance calculated using the Wilcoxon rank-sum test are shown. Expression levels are given as ΔCt. (B) Classification accuracy measures. positive predictive value, 94.7%; negative predictive value, 93.3%.
Validation of a 3-gene model classifier by RT-qPCR in cryopreserved samples. (A) Box plot showing RT-qPCR quantification of TNFRSF8, BATF3, TMOD1, TMEM158, GZMB, and PRF1 expression in PTCL-NOS and ALK− ALCL. Statistical significance calculated using the Wilcoxon rank-sum test are shown. Expression levels are given as ΔCt. (B) Classification accuracy measures. positive predictive value, 94.7%; negative predictive value, 93.3%.
Translation of the classifier to routine FFPE samples
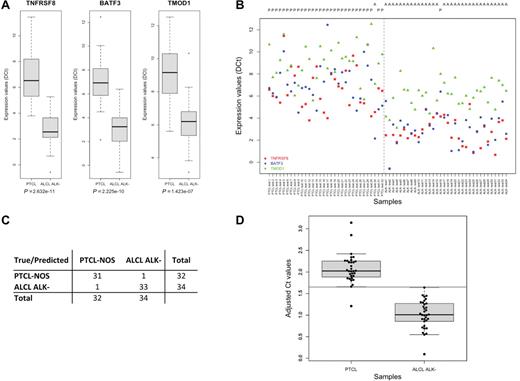
The translation of RT-qPCR protocols to the routine is impaired by the poor RNA quality of FFPE tissues, which represent the gold standard for diagnostic procedures. To achieve this goal, we developed a multiple gene-specific retrotranscription protocol using RNA from FFPE sections. This method increased the production of cDNA targets by 250 to 1000 fold, allowing the generation of biologic relevant RT-qPCR data from 80% of FFPE samples (supplemental Figure 6). We verified that up to 10 gene-specific primers could be used in the RT reaction with no impairment of the sensitivity or specificity of the assay (data not shown). The 3-gene classifier model was then evaluated in 32 PTCL-NOS and 63 ALCL (29 ALK+ and 34 ALK−) FFPE samples. Similarly to that data observed with frozen samples, TNFRSF8, BATF3, and TMOD1 genes exhibited significantly higher expression in ALK− ALCL compared with PTCL-NOS (Figure 4A-B). More importantly, either applying leave-one-out cross validation on FFPE samples or using cryopreserved samples and FFPE as training and test set, respectively, the 3-gene model separated either ALK− ALCL (accuracy 96.97%) or ALCL overall (accuracy 95.79%) from PTCL-NOS with a very high power (Figure 4C and data not shown). In addition, the molecular classifier was challenged for the efficiency to discriminate CD30+ PTCL/NOS cases because their distinction from ALK− ALCL often represents a major diagnostic challenge. In our panel, we found 7 CD30+ PTCL-NOS (supplemental Table 3). The classifier correctly identified 6 of 7 cases.
Validation of the 3-gene model classifier by RT-qPCR in FFPE tissues. (A) Box-plot and (B) point-wise distribution showing RT-qPCR quantification of TNFRSF8, BATF3, and TMOD1 expressions in FFPE samples of PTCL-NOS (n = 32) and ALK− ALCL (n = 34), together with the predicted output using LDA and leave one-out procedure. P indicates PTCL; and A, ALCL. (C) Classification accuracy measures. positive predictive value, 96.9%; negative predictive value, 97.1%; (D) Box-plot and strip-chart distributions of the metagene values. Gray band indicates the unclassifiable samples area.
Validation of the 3-gene model classifier by RT-qPCR in FFPE tissues. (A) Box-plot and (B) point-wise distribution showing RT-qPCR quantification of TNFRSF8, BATF3, and TMOD1 expressions in FFPE samples of PTCL-NOS (n = 32) and ALK− ALCL (n = 34), together with the predicted output using LDA and leave one-out procedure. P indicates PTCL; and A, ALCL. (C) Classification accuracy measures. positive predictive value, 96.9%; negative predictive value, 97.1%; (D) Box-plot and strip-chart distributions of the metagene values. Gray band indicates the unclassifiable samples area.
Finally, for convenience in diagnostic procedures, a “metagene” was calculated as the weighted mean of the ΔCt of the 3 genes and expressed by the following equation: 0.378 × ΔCtTNFRSF8, 0.303 × ΔCtBATF3, 0.177 × ΔCtTMOD1, as described in the “Methods” section. Therefore, the overall calculation generated a single value able to discriminate ALK− ALCL (< 1.64) from PTCL-NOS (> 1.66) with the maximum classification accuracy (Figure 4D).
Discussion
In the present study, we undertook a systematic approach to profiling the expression signatures of a large set of primary T-NHL cases, focusing our attention on systemic ALCL. Specifically, we wondered how related ALK+ and ALK− ALCL are and whether ALK− ALCL could be recognized as a distinct subset. To this aim, we identified a robust diagnostic classifier able to distinguish ALK− ALCL cases from other common PTCL. Such analysis was first performed using a comprehensive and centrally revised dataset of transcriptional profiles of T-NHL generated on high-density microarrays. The predictions based on gene-expression profiling were subsequently confirmed by RT-qPCR in an independent dataset of cryopreserved samples. These analyses led to the recognition of a 3-gene classifier that successfully separated ALK− ALCL from PTCL-NOS with high power. The development of an efficient retrotranscription protocol and the generation of an easy procedure to derive a single metagene value allowed the application of molecular analyses to FFPE tissues, suggesting the possibility of translating the 3-gene model to routine clinical settings as a new approach to precisely defining T-NHL and selecting more appropriate therapeutic strategies.
PTCLs are among the most aggressive NHLs, accounting for 10%-15% of lymphoid neoplasms. Clinically, their response to conventional chemotherapy is discouraging, with 5-year relapse-free and overall survival rates of 26% and 20%, respectively.1,24 Although the new WHO classification represents a step forward in the definition of these tumors, several issues remain open. First, PTCL-NOS is considered a basket category embracing an aggressive and heterogeneous group of nodal/systemic PTCL.25 Second, ALCL is a clinically and biologically heterogeneous disease including the ALK+ and ALK− systemic forms.5 Whereas ALK+ ALCLs are molecularly characterized and can be readily diagnosed, immunophenotypic or genetic features to precisely define ALK−ALCL are missing.1 Moreover, no causal molecular events leading to the transformation to ALK− ALCL have been demonstrated so far, and it is unclear whether they share common features with other PTCLs, including ALK+ ALCL,26 and/or with classic Hodgkin Lymphoma.27 These uncertainties hamper a successful and reliable diagnostic approach in daily clinical practice.25
Several studies focusing on the molecular profiling of PTCL postulated the existence of different subtypes characterized by distinct cellular derivations.15,16,18,20,28-32 However, most of these studies were largely underpowered to allow definitive diagnostic or prognostic statements. In addition, more convenient methods for the measurement of gene expression need to be developed.
We thought that assembling a larger dataset of T-NHL patients was mandatory to discovering reliable biomarkers for the recognition of ALK− ALCL patients. Stringent quality controls of the arrays in public datasets, diagnostic verifications, and detection of high-level expression of known markers, suggestive of high percentage of neoplastic cells, were the strategies applied to increase the statistical significance of analysis. The application of prediction analysis to this large T-NHL dataset identified a simple and very robust classifier for ALK− ALCL (overall error rate, 4.1%), confirming our previous observations.20 The aim of the present study was to identify a small group of genes the expression of which predicts the diagnosis of ALK− ALCL and that can be readily measured in the daily practice. The prediction model generated by gene-expression profiling analysis was validated by RT-qPCR in unrelated groups of patients. The results of this evaluation allowed the design of a model consisting of 3 genes (TNFRSF8, BATF3, and TMOD1) that distinguish ALK− ALCL from PTCL-NOS with overall accuracy near 97%. Our analysis clearly indicated that the 3 genes included in the classifier are superior to GZMB and PRF1 in the distinction of the 2 classes.
Because CD30/TNFRSF8 is the conventional (although not exclusive) marker for ALCL, it was not unexpected that TNFRSF8 scored at the top of the 3-gene model. However, its expression does not have an independent predictive power, indicating that BATF3 and TMOD1 contributions are also required. We verified that including more genes did not enhance the performance of the predictive model significantly. On the contrary, a 3-gene classifier resulted in a robust and practical model that was further simplified by the generation of an easy mathematical procedure to convert RT-qPCR values into a single output to discriminate the 2 phenotypes. Moreover, our data suggest that the predictor discriminates ALK− ALCL from CD30+ PTCL-NOS, despite the high levels of CD30/TNFRSF8 expression in these patients, in all likelihood thanks to the pivotal contribution of BATF3 and TMOD1 expression.
The 3-gene model described herein could be easily applied to FFPE sections without the need for shifting or scaling of the expression data to match the values of frozen samples. Although RNA isolated from FFPE is considered a poor material for gene-expression analysis, we have shown that optimization strategies such as limited amplicon size and gene-specific retrotranscription can effectively overcome its limitations. Our experience allowed the generation of biologic relevant data from at least 80% of FFPE samples, supporting the results obtained on frozen samples and suggesting the possibility of applying RT-qPCR protocols to routine clinical settings.
Overall, our present findings support the hypothesis that ALCLs, independent of the presence of ALK fusion proteins, are closely related and may derive from a different set of progenitors compared with PTCL-NOS. In addition, the lack of an exclusive signature for ALK− ALCL leads us to speculate that this “provisional entity” could be an heterogeneous group with distinct pathogenetic defects yet to be identified. We foresee that massively parallel DNA and RNA sequencing technologies will be pivotal to a definitive classification and to an effective use of targeted therapeutics in T-NHL.11,33
In conclusion, the results of the present study led to the identification of novel diagnostic markers for more objective differential diagnosis within T-NHL. Specifically, the application of RT-qPCR protocols to FFPE tissues will allow the possibility of developing simple and cost-effective molecular diagnostic tools and reducing error and ambiguity in the stratification of T-NHL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Appendix
The members of the European T-Cell Lymphoma Study Group are: Barreca A, Cuccuru G, Inghirami G, Medico E, Mereu E, Pellegrino E, Spaccarotella E, Scarfò I, Piva R, Fornari A, Ferreri C, Novero D, Chilosi M, Zamó A, Facchetti F, Lonardi S, De Chiara A, Fulciniti F, Doglioni C, Ponzoni M, Agnelli L, Neri A, Todoerti K, Agostinelli C, Piccaluga PP, Pileri S, Falini B, Tiacci E, Van Loo P, Tousseyn T, De Wolf-Peeters C, Geissinger E, Muller-Hermelink HK, Rosenwald A, Piris MA, Rodriguez ME, Bertoni F, Kwee I, and Boi M.
Acknowledgments
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (grants IG-8675, 10007, and IG-4569); Regione Piemonte; Compagnia di San Paolo, Torino (Progetto Oncologia); Fondazione Italiana Ricerca sul Cancro; Oncosuisse (grant KLS-02403-02-2009); Fondazione per la Ricerca e la Cura sui Linfomi (Lugano, Switzerland); and the Nelia et Amadeo Barletta Foundation (Lausanne, Switzerland).
Authorship
Contribution: L.A., F.B., G.I., and R.P., designed the study, interpreted the data, and wrote the manuscript; E.M., E.P., and E.B. performed the experiments; L.A., E.M., E.P., T.L., I.K., A.N., F.B., G.I., and R.P. interpreted the biologic data; M.P., A.Z., J.I., P.P.P., W.C.C., and S.P. provided well-characterized study materials; and P.P.P., S.P., and G.I. reviewed the pathology.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for T.L. is Istituto Italiano di Tecnologia (IIT), Genova, Italy.
Correspondence: Roberto Piva and Giorgio Inghirami, Department of Molecular Biotechnology and Health Sciences, Center for Experimental Research and Medical Studies, University of Torino, Via Santena 5, Torino 10126 Italy; e-mail: roberto.piva@unito.it or giorgio.inghirami@unito.it.
References
Author notes
L.A. and E.M. contributed equally to this work.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal