Key Points
We demonstrate that vitronectin plays an important role in tumor growth.
We show that the urokinase receptor can promote tumor growth through its interaction with vitronectin.
Abstract
Extensive evidence implicates the urokinase plasminogen activator receptor (uPAR) in tumor growth, invasion, and metastasis. Recent studies have substantiated the importance of the interaction between uPAR and the extracellular matrix protein vitronectin (VN) for the signaling activity of the receptor in vitro, however, the possible relevance of this interaction for the activity of uPAR in tumor growth and metastasis has not been assessed. We generated a panel of HEK293 cell lines expressing mouse uPAR (muPARWT), an uPAR mutant specifically deficient in VN binding (muPARW32A), and a truncation variant (muPARΔD1) deficient in both VN and uPA binding. In vitro cells expressing muPARWT display increased cell adhesion, spreading, migration, and proliferation associated with increased p130Cas and MAPK signaling. Disruption of VN binding or ablation of both VN and uPA binding specifically abrogates these activities of uPAR. When xenografted into SCID (severe combined immunodeficiency) mice, the expression of muPARWT, but not muPARW32A or muPARΔD1, accelerates tumor development, demonstrating that VN binding is responsible for the tumor-promoting activity of uPAR in vivo. In an orthotopic xenograft model using MDA-MB-231 cells in RAG1−/−/VN−/− mice, we document that host deficiency in VN strongly impairs tumor formation. These 2 lines of in vivo experimentation independently demonstrate an important role for VN in tumor growth even if the uPAR dependence of the effect in the MDA-MB-231 model remains to be ascertained.
Introduction
The urokinase-type plasminogen activator (uPA) and its cellular receptor (uPAR) enhance cell adhesion, migration, proliferation, and invasion by accelerating extracellular proteolysis and by modulating the activity of a variety of signaling receptors.1 There is extensive evidence in the literature supporting an important role of the uPA system in a variety of pathological conditions and, in particular, in cancer progression.2 Great effort has been made to develop drugs that inhibit the individual component of the uPA system. However, the multifunctional properties of a protein like uPAR require the precise knowledge of which receptor function to interfere with and possibly also which function not to interfere with. This type of knowledge is difficult to extract from wholesale knockdown or overexpression studies in which all uPAR functions are either reduced or augmented. Although overexpression studies have consistently provided support for an important role of uPA and uPAR,3 they have failed to identify which uPAR functions/interactions are critical for the receptor’s activity in disease.
The proteolytic functions of uPA/uPAR were the first to be identified and are the best described. Briefly, the concomitant binding of pro-uPA and plasminogen (Plg) to the cell surface mediated by uPAR and the recently identified Plg-RKT4 result in a reciprocal zymogen activation cascade, leading to the generation of active uPA and plasmin.5 The proteolytic activities of uPA and plasmin allow cells to degrade the surrounding extracellular matrix either directly or through the activation of other proteases from the matrix metalloproteinase family and thus enhance the capability of the cell to invade the surrounding tissue.6 In addition, uPA and plasmin are caable of activating latent growth factors, such as transforming growth factor β and hepatocyte growth factor, thus modulating cell migration, proliferation, and differentiation.7
Subsequent to the identification of uPAR as the cellular binding site for uPA, it became clear that overexpression of uPAR, or the treatment of uPAR-expressing cells with catalytically inactive uPA variants, induces a variety of cellular responses that are dependent on cell type, including changes in adhesion, migration, and proliferation and commonly known as the nonproteolytic functions of the receptor. Compilation of the published data on the nonproteolytic functions of uPAR yields a complex scenario in which the interactions with a variety of different ligands and coreceptors are required to explain the receptor’s many biological activities.1,8 In line with this, uPAR’s signaling activity relies on an association with a wide variety of transmembrane receptors such as tyrosine kinase receptors, G-protein–coupled receptors, and integrin adhesion receptors.1,8 Among the many interactions implicated in the signaling activity of uPAR, the direct molecular binding to the extracellular matrix protein vitronectin (VN) has recently emerged as crucial for most of the nonproteolytic receptor functions.1 Indeed, binding of VN is required for uPAR activity to induce cell adhesion, spreading, migration, and invasion in vitro.9,10 The effect of the uPAR–VN interaction on cell morphology and migration involves integrin-dependent regulation of Rho guanosine triphosphatases activity through the p130-Cas adaptor molecule.10,11 Moreover, uPAR-mediated VN adhesion initiates intracellular signaling, enhancing mitogen-activated protein kinase (MAPK) activation in different cell lines.9,12 The activation of these migratory and mitogenic signaling pathways upon VN engagement suggests that the adhesive function of uPAR could promote tumor growth and metastasis dissemination in vivo. However, despite the strong evidence supporting a central role for VN binding in the biological activity of uPAR in vitro, the possible relevance of this interaction for tumor growth and metastasis in vivo has not been investigated.
It has previously been shown that overexpression of both human and mouse uPAR in HEK293 cells promotes metastatic dissemination in SCID (severe combined immunodeficiency) mice,13 suggesting that the uPAR function responsible for this effect must be different from uPA binding, which displays pronounced species specificity.14 Because the critical residues for the interaction with VN are well conserved between human and mouse uPAR,14 we hypothesized that this interaction might be responsible for the tumorigenic activity of uPAR.
To assess the possible role of the uPAR–VN interaction in tumor growth and metastasis, we performed a limited structure–function analysis of uPAR in vitro and in vivo using 293 Flp-In cells expressing murine uPAR (muPAR) and variants thereof that were carefully designed based on the extensive knowledge about the human counterpart. The expression of muPAR in vitro was found to increase VN-dependent cell adhesion, migration, and proliferation, consistent with previous observations using human uPAR. The expression of muPAR in 293 cells in vivo resulted in accelerated tumor formation and growth. This effect was lost upon specific disruption of VN binding to the receptor. In independent experiments of orthotopic tumor growth, host deficiency in VN abrogated tumor formation by MDA-MB-231 cells. This finding provides further evidence for the importance of VN in tumor growth, even if the uPAR dependence remains to be examined in this model. Our data, for the first time, provide experimental evidence that VN plays an important role in the growth of solid tumors and that this activity is, at least in part, mediated by its interaction with uPAR.
Methods
Materials
The VN(1–66)/Fc fusion protein was generated as previously described.9
Expression vectors construction
A mouse uPAR cDNA (muPAR115 ) was amplified with primers mupKpn/muPARre and cloned KpnI/NotI in pcDNA5/FRT (Invitrogen, Carlsbad, CA), generating the expression vector pFRT-muPAR. The muPARΔD1 deletion mutant was generated by coamplification (primers mupKpn/muPARre) of 2 polymerase chain reaction products generated using primer pairs mupKpn/mD1rev and mD1/muPARre and cloned as described for full-length mouse uPAR. This procedure deletes residues 1–84 of the mature receptor (ie, domain 1 [D1]), generating a truncated D2D3 receptor that has the same N terminal (Gly85) as that generated by proteolytic cleavage of the linker region by uPA or plasmin.16 The mouse uPARW32A variant was generated by site-directed mutagenesis using oligonucleotides mW32A/mW32Ar. The correct sequences of the complete coding regions of all constructs were verified by sequencing.
Oligonucleotide sequences included the following:
mupKpn: 5′-gaagatctcggtaccgatctcaatatgggactcccaaggc-3′
muPARre: 5′-atatagtttagcggccgcatcaggtccagaggagga-3′
mD1rev: 5′-gccctgagggaaagcacggccgccctgggaggctgggacacaggtagt-3′
mD1: 5′-tgtgtcccagcctcccagggcggccgtgctttccctcagggccgtta-3′
mW32A: 5′-accgtgcttcgggaagcgcaagatgatagagag-3′
mW32Ar: 5′-ctctctatcatcttgcgcttcccgaagcacggt-3′
Cell culture
HEK293 Flp-In T-Rex cells (Invitrogen) were cultured in Dulbecco’s modified Eagle medium (DMEM; BioWhittaker, Radnor, PA), supplemented with 10% fetal calf serum (Hyclone, Logan, UT), 100 U/mL penicillin, 100 U/mL streptomycin, 5 mM glutamine (EuroClone, Milan, Italy), 100 μg/mL zeocin (Invitrogen), and 5 μg/mL blasticidin (Invitrogen) at 37°C in 5% carbon dioxide (CO2). Parental HEK293 Flp-In T-Rex cells were transfected with cytosolic green fluorescent protein (GFP) expression vector (pEGFP-N1; Clontech Corp, Mountain View, CA) and subjected to single cell cloning in selective medium containing 1 mg/mL G418. A clone displaying homogenous and stable GFP expression was selected and further transfected with pcDNA5/FRT–based expression vector, containing full-length muPAR, muPARW32A, and muPARDΔ1 or with empty vector (mock), together with the Flp-recombinase expression vector pOG44 (Invitrogen) using FuGENE 6 (Roche Corp, Basel, Switzerland). Pools of stable transfectants were selected in medium containing 150 µg/mL hygromycin B.
Cell proliferation assay
Cells were seeded at 3 × 103 cells/well in 96-well tissue culture plates and grown at 37°C. Plates were removed at different time points after seeding (4, 24, 48, 72, and 96 hours), fixed in paraformaldehyde, and stored in phosphate-buffered saline (PBS) at 4°C until the end of the experiment. Cells were quantified by measuring the GFP fluorescence using a PerkinElmer EnVision plate reader (Waltham, MA) with a fluorescein-optimized filter set and bottom reading. Background fluorescence, measured in wells receiving no cells, was subtracted. To normalize for variability in seeding density, the readings at 24, 48, 72, and 96 hours were normalized to the 4-hour time point.
FACS analysis
Cell surface expression of the different muPAR variants was analyzed by flow cytometry using a monoclonal antibodies anti-uPAR antibody (AK17) at 5 µg/mL. Bound antibody was detected using a secondary Cy5-labeled antibody (Jackson ImmunoResearch Laboratories) and analyzed by flow cytometry (FACS [fluorescence-activated cell sorter] Calibur; BD Biosciences, Franklin Lakes, NJ).
Cell lysis and western blotting
Cells were washed with PBS and lysed directly on the culture dish in ice-cold radioimmunoprecipitation assay buffer (50 mM Tris, pH 8.0, 150 mM sodium chloride (NaCl), 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) supplemented with a protease inhibitor cocktail (complete-EDTA-free; Roche), 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 1 mM sodium fluoride, and 1 mM sodium ortovanadate followed by a brief sonication. The total protein content was determined using the DC-protein assay (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin (BSA) as standard. Equal amounts of total protein were separated by SDS polyacrylamide gel electrophoresis (SDS-PAGE) and probed as indicated. Polyclonal antibodies against total and phosphorylated ERK1/2 and pY410-p130Cas and secondary antibodies against anti-mouse horseradish peroxidase and anti-rabbit horseradish peroxidase were used (Cell Signaling Technology, Danvers, MA).
Cell adhesion assay
Cells were seeded at the concentration of 3 × 104/well in MaxiSorp 96-well plates (Nunc, Roskilde, UK), coated with different substrates, and allowed to adhere for 30 minutes at 37°C. After washing with warm DMEM, adherent cells were fixed, stained with crystal violet, and quantified by measuring the absorbance at 540 nm. Coatings were as follows: poly-L-lysine 100 µg/mL, fibronectin 10 µg/mL (Roche), and purified VN(1-66) and VN(1-66)RAD at 5 µg/mL. All measurements were done in triplicate, and specific binding was calculated by subtracting the nonspecific binding observed in BSA-coated wells. Specific adhesion is presented as percent of total cell adhesion measured on poly-L-lysine.
Differential interference contrast microscopy
Cells were plated in serum containing medium on cover glass chambers (Laboratory-TekII, Ashland, MA) and allowed to adhere for 24 hours. Cells were fixed in 4% paraformaldehyde, rinsed with PBS, and subjected to differential interference contrast (DIC) microscopy. DIC analysis was performed using an inverted Olympus IX81 microscope. Cells were viewed through a high-aperture 60× objective lens (UIS2 60× TIRFM PlanApo N, NA 1.45; Olympus, Tokyo, Japan). Images were acquired using a Hamamatsu Orca-ER digital camera with the software Metamorph 7.5.6.0. Cell area was measured with ImageJ 1.42q.
Migration assay
Time-lapse live-cell imaging was performed at 37°C in 5% CO2 with an inverted Olympus IX80 microscope equipped with an incubation chamber (Okolab, Ottaviano, Italy) to control CO2 and temperature. Cells were plated in serum containing medium in 12-well plates (Nunc) at the confluence of 1 × 105 cell/well. Cells were viewed through 10× (µPlan FLN 10× Ph1, N.A. 0.30; Olympus) objective lenses, and pictures were taken every 5 minutes for 5 hours. The acquisition system included a digital camera (Orca ER; Hamamatsu, Hamamatsu City, Japan) and System Control Software Olympus ScanR. Cell migration speed was quantified with ImageJ 1.42q using the plug-in “manual tracking.” In each experiment, 20 randomly chosen cells were tracked, and their average migration speed throughout the experiment was calculated.
Xenograft experiments
Anesthetized 8-week-old C.B-17/IcrCrl-scid-BR mice (Charles River Laboratories, Wilmington, MA) were inoculated in the fourth mammary fat pad with 1 × 106 living HEK293-GFP cells, suspended in 50 μL of 1:1 PBS and Matrigel (Sigma-Aldrich, St. Louis, MO). The development of palpable tumors was monitored every 2 to 3 days, and tumor volume was estimated by caliper measurement and the formula V = (length × width2 )/2. Mice were killed when the tumor size ≥2 cm3. Mice deficient in VN (B6.129S2(D2)-Vtntm1Dgi/J) and RAG1−/− (B6.129S7-Rag1tm1Mom/J) were obtained from Charles River Laboratories and interbred to homozygosis for both alleles. The 8-week-old RAG1−/− and RAG1−/−/VN−/− mice were xenografted with 1 × 106 MDA-MB-231 cells as described for the SCID mice, and the development of palpable tumors was monitored every 2 to 3 days. Mice were maintained in HEPA-filtered IVC units, and all experiments were performed according to the guidelines for care and use of laboratory animals approved by the institutional ethical animal care committee.
Apoptosis assay.
Adherent cells were detached and washed in annexin buffer (N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid 10 mM, NaCl 150 mM, magnesium chloride 1 mM, calcium chloride 3.6 mM, potassium chloride 5 mM). Then 5 × 105 cells were incubated for 1 hour at room temperature with a biotinylated anti-annexinV antibody (Sigma-Aldrich). Cells were washed with annexin buffer and incubated for 1 hour with APC-labeled streptavidin. Stained cells were then washed with annexin buffer and resuspended in PBS. Propidium iodide (PI) (Sigma-Aldrich) was added prior to acquisition. Samples were acquired with FACS CANTO II (Beckton Dickinson, Franklin Lakes, NJ) and analyzed with FACS DIVA software (Beckton Dickinson).
BrdU assay.
Semiconfluent cells were pulsed with 5-bromo-2'-deoxyuridine (BrdU; 33 μM; Sigma-Aldrich) for 1 hour, detached, and washed extensively in PBS. Next 3 × 106 cells were resuspended in PBS and fixed in 75% ethanol. Cells were then incubated for 25 minutes in denaturing solution (2 M hydrogen chloride) and 3 volumes of sodium borate (0.1 M) were added for 2 minutes. Cells were washed with 1% BSA in PBS and incubated with an anti-BrdU antibody (Beckton Dickinson) for 1 hour at room temperature. Cells were then washed and incubated with DyLight 649–conjugated anti-mouse immunoglobulin (Jackson Labs, Suffolk, UK) for 1 hour. After washing, cells were incubated overnight with propidium iodide (2.5 μg/mL) and RNase A (250 μg/mL). Samples were acquired using a FACS CANTO II flow cytometer and analyzed with the FACS DIVA software. For the cell cycle kinetics experiments, cells were pulsed with BrdU, extensively washed, and stained at the indicated time point after the initial pulse.
Statistical analyses.
One-way analysis of variance with the Dunnett post hoc test was used to test the null hypothesis that expression of the different receptor variants had no effect on ERK1/2-activation, p130-Cas substrate domain phosphorylation, and BrdU incorporation. Differences between the Kaplan-Meier curves were analyzed by repeated log-rank tests comparing the curves of the different receptor variants with the control (mock) curve. Differences in tumor formation between the different mouse genotypes were analyzed using repeated χ2 tests.
Results
Generation and characterization of mouse uPAR variants expressed in HEK293 Flp-In cells
To address directly the relative contribution of uPA and VN binding to the activity of uPAR in experimental tumor growth and metastasis, we exploited structure–function data obtained in alanine scans of human uPAR in which the physical and functional hot spots for VN binding have been mapped to a handful of residues located in domain 1 and in the domain 1/2 connecting region.9,17 Residues critical for VN binding in human uPAR are conserved in mouse uPAR.14 Among several tested substitutions, we selected the muPAR W32A substitution because it was found to disrupt VN binding efficiently without affecting uPA binding (Figure 1 and data not shown). In contrast to the effect of the W32A substitution on VN binding, even the most efficient single alanine substitutions in uPAR reduce uPA binding only moderately.18 To generate a receptor effectively devoid of uPA-binding activity, we therefore applied a more radical approach that consisted of the deletion of the entire domain 1. The resulting deletion variant (muPARΔD1) was predicted to be deficient in both uPA and VN binding and was a physiologically relevant form of uPAR because it is identical to the natural product of uPA or plasmin-catalyzed receptor cleavage.16
Characterization of HEK293 cells expressing mouse uPAR variants. (A) muPAR variants are expressed at comparable levels in transfected 293 cells. Cell surface expression profiles of mock (black), muPAR (red), muPARW32A (blue), and muPARΔD1 (green) transfected cells were analyzed by flow cytometry using a monoclonal antibody against mouse uPAR. (B) Western blot analysis illustrating the molecular weights of the different mouse uPAR variants. Equal amounts of cell extracts were fractionated by nonreducing SDS-PAGE and probed with the same antibody used in FACS analysis with an anti-GFP antibody as control. (C) W32A substitution or deletion of the entire domain 1 impairs the activity of muPAR in promoting cell adhesion to VN. Cells expressing the different receptor variants were seeded in wells coated with VN(1-66), VN(1-66)RAD, or FN and allowed to adhere for 30 minutes at 37°C. After washing, adherent cells were fixed and quantified. Specific cell adhesion is shown as percent of cell binding to poly-D-lysine. Data represents the mean ± standard error of the mean of independent experiments (n = 3).
Characterization of HEK293 cells expressing mouse uPAR variants. (A) muPAR variants are expressed at comparable levels in transfected 293 cells. Cell surface expression profiles of mock (black), muPAR (red), muPARW32A (blue), and muPARΔD1 (green) transfected cells were analyzed by flow cytometry using a monoclonal antibody against mouse uPAR. (B) Western blot analysis illustrating the molecular weights of the different mouse uPAR variants. Equal amounts of cell extracts were fractionated by nonreducing SDS-PAGE and probed with the same antibody used in FACS analysis with an anti-GFP antibody as control. (C) W32A substitution or deletion of the entire domain 1 impairs the activity of muPAR in promoting cell adhesion to VN. Cells expressing the different receptor variants were seeded in wells coated with VN(1-66), VN(1-66)RAD, or FN and allowed to adhere for 30 minutes at 37°C. After washing, adherent cells were fixed and quantified. Specific cell adhesion is shown as percent of cell binding to poly-D-lysine. Data represents the mean ± standard error of the mean of independent experiments (n = 3).
Stable pools of transfected cells were generated by homologous recombination using a GFP-positive clone of HEK293 Flp-In cells (see Methods). As predicted from the nature of this isogenic cell system, the vast majority of transfectants expressed the modified receptors on the cell surface at highly comparable levels when evaluated by FACS analysis (Figure 1A). Analysis of cell lysates by SDS-PAGE and immunoblotting confirmed that the apparent molecular weights of muPAR and muPARW32A are very similar and that muPARΔD1 displays increased electrophoretic mobility as a result of the deleted domain 1 (Figure 1B). The apparent lower expression of muPARΔD1 observed by western blotting is likely to be the result of faster membrane sorting or a reduced binding of this (smaller) uPAR variant to the nitrocellulose filters. Because the Flp-In system used in this study ensures equal transcriptional pressure for the different receptor variants and because we find comparable numbers of these on the cell surface by FACS analysis, the reason for the different intensities in western blotting was not further investigated.
Expression of human uPAR in HEK293 cells is described to enhance cell adhesion to VN. To confirm the predicted activity of the different mouse receptor variants, we measured cell adhesion to a recombinant fragment of VN in which the respective contribution of uPAR and integrins is dissected by mutation of the RGD-motif9 (Figure 1C). As previously reported for the human receptor,9 the expression of muPAR enhances RGD-independent cell adhesion on VN. On the contrary, cells expressing muPARW32A or muPARΔD1 do not display adhesion properties significantly different from those of mock-transfected cells on both of these VN variants. All cell lines showed comparable integrin-mediated cell adhesion to FN.
muPAR–VN interaction induces cell spreading and migration
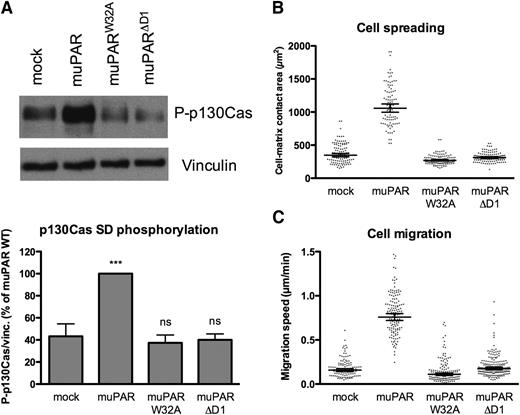
The expression of human uPAR in 293 cells and the consequent increase in VN adhesion induce marked changes in cell morphology, including extensive lamellipodia formation and cell spreading.9 To determine if the murine uPAR variants have a similar activity, we quantified the spreading of cells seeded in complete serum-containing medium (containing a large amount of VN) by differential interference contrast microscopy (Figure 2B). Expression of muPAR increases cell/matrix contact areas by ∼2.5-fold, while cells expressing muPARW32A or muPARΔD1 display spreading similar to that of mock-transfected cells. In vitro uPAR has been reported to promote tumor cell migration and invasion through activation of the p130Cas/DOCK180/Rac pathway;10 we consistently found that expression of muPAR induces robust phosphorylation of the substrate domain of p130Cas (Figure 2A). Furthermore, the muPAR-induced increase in cell adhesion, spreading, and p130Cas activation are associated with a marked increase in 2D cell migration on serum-coated surfaces (Figure 2C). These receptor activities are also dependent upon the direct interaction with VN because no significant p130Cas activation or enhanced cell migration are observed in cells expressing muPARW32A or muPARΔD1.
Mouse uPAR promotes VN-dependent p130Cas activation, cell spreading, and migration. (A) muPAR induces VN-dependent p130Cas substrate domain phosphorylation. Cells were incubated overnight in serum-supplemented medium in tissue culture dishes, washed, and lysed in SDS-PAGE sample buffer. Equal volumes of lysates were analyzed by western blotting using an antibody specific for p130Cas phosphorylated on Tyr410. The blots were stripped and then reprobed for VN as loading control. The lower panel shows the quantification of independent experiment (n = 5). (B) muPAR-induced cell spreading requires VN binding to the receptor. Cells in serum-supplemented medium were seeded sparsely on glass cover slides and left to adhere overnight. After fixation, DIC microscopy images were recorded and the cell-matrix contact areas quantified using ImageJ software. Each dot represents a single cell. Bars indicate the geometric mean with 95% confidence intervals. The data represent cell-matrix contact areas of randomly selected cells (n = 93–122) pooled from 2 independent experiments. (C) The interaction between uPAR and VN promotes migration of 293 cells on serum-coated surfaces. Cells grown in serum-supplemented medium were monitored by phase-contrast time-lapse microscopy. The migration speed of individual cells was quantified by manual cell tracking using the ImageJ software. Each dot represents the migration speed of a single cell. Bars indicate the geometric mean with 95% confidence intervals. The data represent migration speeds of randomly selected cells (n = 93–138) pooled from 2 independent experiments.
Mouse uPAR promotes VN-dependent p130Cas activation, cell spreading, and migration. (A) muPAR induces VN-dependent p130Cas substrate domain phosphorylation. Cells were incubated overnight in serum-supplemented medium in tissue culture dishes, washed, and lysed in SDS-PAGE sample buffer. Equal volumes of lysates were analyzed by western blotting using an antibody specific for p130Cas phosphorylated on Tyr410. The blots were stripped and then reprobed for VN as loading control. The lower panel shows the quantification of independent experiment (n = 5). (B) muPAR-induced cell spreading requires VN binding to the receptor. Cells in serum-supplemented medium were seeded sparsely on glass cover slides and left to adhere overnight. After fixation, DIC microscopy images were recorded and the cell-matrix contact areas quantified using ImageJ software. Each dot represents a single cell. Bars indicate the geometric mean with 95% confidence intervals. The data represent cell-matrix contact areas of randomly selected cells (n = 93–122) pooled from 2 independent experiments. (C) The interaction between uPAR and VN promotes migration of 293 cells on serum-coated surfaces. Cells grown in serum-supplemented medium were monitored by phase-contrast time-lapse microscopy. The migration speed of individual cells was quantified by manual cell tracking using the ImageJ software. Each dot represents the migration speed of a single cell. Bars indicate the geometric mean with 95% confidence intervals. The data represent migration speeds of randomly selected cells (n = 93–138) pooled from 2 independent experiments.
Promotion of cell growth by uPAR requires VN binding
In addition to its activity in promoting cell migration and invasion, the expression of uPAR has been shown to determine the balance between dormancy and growth of cancer cell lines by regulating the activation state of ERK1/2.19 We have shown that uPAR signaling to ERK1/2 requires the direct binding of the receptor to VN,9 suggesting that this interaction may also be responsible for the pro-proliferative activity of uPAR. Consistent with this hypothesis, we find that the expression of muPAR in 293 cells results in increased levels of phosphorylated ERK1/2 (Figure 3A) as well as increased cell growth quantified by GFP fluorescence (Figure 3B). This receptor activity is again dependent upon the interaction with VN because cells expressing muPARW32A or muPARΔD1 display levels of ERK1/2 activation and cell growth comparable to that of mock-transfected cells.
muPAR expression in 293 cells induces VN-dependent ERK1/2 activation and cell growth. (A) muPAR expression induces VN-dependent ERK1/2 activation. Semiconfluent cells were serum starved for 4 hours prior to cell lysis and immunoblot analysis of ERK1/2 activation. Blots were probed for phosphorylated (Thr202/Tyr204) ERK1/2, stripped, and reprobed for total ERK1/2. Representative blots are shown, and the quantification of independent (n = 5) experiments is shown (bar graph). (B) uPAR promotes VN-dependent cell growth of HEK293 cells. Cells were seeded in 96-well tissue culture plates. Cell growth was quantified by measuring the relative increase in GFP-fluorescence. (C) Effect of uPAR–VN interaction on cell proliferation. Semiconfluent cells were pulsed with BrdU for 1 hour and stained with an anti-BrdU antibody and PI. BrdU incorporation and DNA content were analyzed by flow cytometry. The graph represents the mean of percentages of cells in S phase according to BrdU signal and DNA content (n = 4). (D) Effect of uPAR–VN interaction on apoptosis. Cells were stained with an annexin V antibody and PI prior to analysis by flow cytometry. Representative dot plots of 3 independent experiments are shown. The global cell population was divided into 4 subpopulations according to the PI and annexin V signal intensity. The percentage of double-positive apoptotic cells is shown.
muPAR expression in 293 cells induces VN-dependent ERK1/2 activation and cell growth. (A) muPAR expression induces VN-dependent ERK1/2 activation. Semiconfluent cells were serum starved for 4 hours prior to cell lysis and immunoblot analysis of ERK1/2 activation. Blots were probed for phosphorylated (Thr202/Tyr204) ERK1/2, stripped, and reprobed for total ERK1/2. Representative blots are shown, and the quantification of independent (n = 5) experiments is shown (bar graph). (B) uPAR promotes VN-dependent cell growth of HEK293 cells. Cells were seeded in 96-well tissue culture plates. Cell growth was quantified by measuring the relative increase in GFP-fluorescence. (C) Effect of uPAR–VN interaction on cell proliferation. Semiconfluent cells were pulsed with BrdU for 1 hour and stained with an anti-BrdU antibody and PI. BrdU incorporation and DNA content were analyzed by flow cytometry. The graph represents the mean of percentages of cells in S phase according to BrdU signal and DNA content (n = 4). (D) Effect of uPAR–VN interaction on apoptosis. Cells were stained with an annexin V antibody and PI prior to analysis by flow cytometry. Representative dot plots of 3 independent experiments are shown. The global cell population was divided into 4 subpopulations according to the PI and annexin V signal intensity. The percentage of double-positive apoptotic cells is shown.
The increased growth of muPAR-expressing cells is the results of multiple effects on the cell cycle. In particular, muPAR-expressing cells display a significant increase in the number of cells in S phase as determined by BrdU incorporation (Figure 3C). Concomitantly, the fraction of propidium iodide/Annexin V double positive cells is decreased upon muPAR expression, indicating a mild reduction in apoptosis (Figure 3D). Expression of muPAR also accelerates cell cycle progression since the relative movement of the peek of BrdU-positive cells is faster in cells expressing muPAR (supplemental Figure 1). Importantly, cells expressing muPARW32A or muPARΔD1 show levels of DNA synthesis and apoptosis that are similar to those of mock-transfected cells, demonstrating conclusively that the interaction with VN is responsible of this uPAR activity. Further studies will be required to clarify how these minor changes in cell cycle synergize to cause the strong effects on cell proliferation seen at later time points after seeding.
uPAR accelerates tumor formation by HEK293 cells in SCID mice
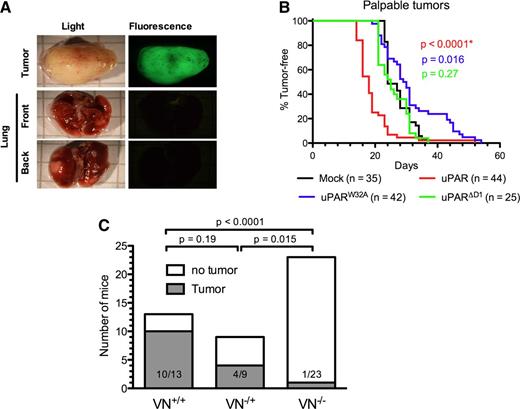
To evaluate the biological activity of the different muPAR variants in promoting tumor growth and metastasis, we xenografted the different cell lines into the mammary fat pad of SCID mice and monitored the appearance of palpable tumors and their growth by caliper measurement. When the primary tumor mass was ≥2 cm3, the animals were sacrificed and carefully inspected by fluorescence microscopy for the presence of GFP-positive macrometastasis in the lungs. In contrast to a previous study,13 we failed to identify any GFP-positive foci on the surface of the lungs in any of the animals independent of the uPAR variant expressed by the grafted cells (Figure 4A and data not shown). The lack of identifiable lung foci is unlikely to be caused by a loss of GFP expression because the primary tumors were homogenously and brightly GFP positive in all animals. Experiments aimed at promoting the expansion of possible micrometastasis into visible macrometastasis by surgical removal of the primary tumor (sized ∼1 cm3) and extended observation times also failed to evidence colonization to the lungs (data not shown).
Role of VN and the uPAR–VN interaction in xenograft tumor growth. (A) Absence of macro-metastasis foci in the lungs of SCID mice xenografted with HEK293 cells expressing mouse uPAR. Primary tumors and lungs were collected from killed mice, fixed in PFA, and imaged using an inverted fluorescence microscope. Representative images of primary tumors (upper panel) and front and back views of lungs (lower panels) are shown. (B) Expression of uPAR reduces tumor latency. Anesthetized 8-week-old SCID mice were inoculated in the fourth mammary fat pad with mock (black), muPAR (red), muPARW32A (blue), and muPARΔD1 (green) transfected cells. The appearance of tumors was monitored by palpation. Kaplan-Meier curves show the fraction of tumor-free mice. The total number of mice analyzed with each cell line is indicated and includes complete data from 3 to 5 independent experiments. (C) VN deficiency impairs MDA-MB-231 tumor formation in immunodeficient RAG−/− mice. MDA-MB-231 cells were injected into the fourth mammary fat pad of RAG−/−/VN+/+, RAG−/−/VN−/−, or RAG−/−/VN+/− mice and tumor formation monitored. The graph shows data pooled from 2 independent experiments and indicates the number of tumor-bearing mice and total number of mice injected. At the end of the observational period (4 months), none of the remaining mice (RAG−/−/VN−/−, n = 22; RAG−/−/VN+/+, n = 3; and RAG−/−/VN+/−, n = 5) had palpable tumors.
Role of VN and the uPAR–VN interaction in xenograft tumor growth. (A) Absence of macro-metastasis foci in the lungs of SCID mice xenografted with HEK293 cells expressing mouse uPAR. Primary tumors and lungs were collected from killed mice, fixed in PFA, and imaged using an inverted fluorescence microscope. Representative images of primary tumors (upper panel) and front and back views of lungs (lower panels) are shown. (B) Expression of uPAR reduces tumor latency. Anesthetized 8-week-old SCID mice were inoculated in the fourth mammary fat pad with mock (black), muPAR (red), muPARW32A (blue), and muPARΔD1 (green) transfected cells. The appearance of tumors was monitored by palpation. Kaplan-Meier curves show the fraction of tumor-free mice. The total number of mice analyzed with each cell line is indicated and includes complete data from 3 to 5 independent experiments. (C) VN deficiency impairs MDA-MB-231 tumor formation in immunodeficient RAG−/− mice. MDA-MB-231 cells were injected into the fourth mammary fat pad of RAG−/−/VN+/+, RAG−/−/VN−/−, or RAG−/−/VN+/− mice and tumor formation monitored. The graph shows data pooled from 2 independent experiments and indicates the number of tumor-bearing mice and total number of mice injected. At the end of the observational period (4 months), none of the remaining mice (RAG−/−/VN−/−, n = 22; RAG−/−/VN+/+, n = 3; and RAG−/−/VN+/−, n = 5) had palpable tumors.
Despite the apparent absence of lung metastasis, the expression of uPAR had remarkable effects on the latency and growth of the primary tumors (Figure 4B). All cell lines formed primary tumors with complete penetrance, but the expression of muPAR significantly shortened the latency period as compared with mock-transfected cells (Figure 4B). On the contrary, the expression of muPARW32A moderately prolonged latency, while the expression of muPARΔD1 was without any measurable activity at all. These data demonstrate that the activity of uPAR in reducing tumor latency is mediated specifically by its interaction with VN.
The differential effect of muPAR and muPARW32A on tumor growth in the 293/SCID model not only documents that the interaction with VN is critical for the growth-promoting activity of uPAR, it also identifies an as yet unrecognized importance for VN in tumor growth. To directly exploit the possible role of VN in the growth of xenografted tumors, we intercrossed immunodeficient RAG1−/− with VN−/− mice to generate the double-deficient RAG1−/−/VN−/− strain. Initially we wanted to compare the growth of 293 cells expressing wild-type muPAR and muPARW32A in these animals. However, independent of host VN status, the 293 cell lines failed to form palpable tumors in the RAG1−/− background (data not shown). As an alternative approach, we turned to the highly aggressive breast adenocarcinoma cell line MDA-MB-231 because this line expresses high levels of uPAR and uPA,20 displays uPAR-dependent tumor formation,21 and grows in the RAG1−/− strain.22 The pooled data from 2 independent experiments are shown in Figure 4C. In the first experiment 70% (7/10) of the VN-proficient mice formed tumors, while no tumors (0/10) were detected in VN-deficient mice. In the second experiment, where VN-haploinsufficient mice were included, 100% (3/3) of the VN-proficient, 44% (4/9) of the VN-haploinsufficient, and 8% (1/13) of the VN-deficient mice developed tumors. These experiments clearly document that host VN is required for efficient tumor formation by MDA-MB-231 cells xenografted in RAG1−/− mice and indicate that this effect may be dose dependent. Although the MDA-MB-231 xenograft model documents a strong dependence for host VN, the importance of uPAR in this context has yet to be established.
Discussion
VN was identified as the circulating factor responsible for the cell attachment-promoting activity of serum23 required by most cultured tumor cell lines to escape cell death by anoikis. Despite the evident importance of VN for cell growth in vitro, the role of this protein for tumor growth in vivo has not been tested, presumably discouraged by the surprising finding that VN-deficient mice display normal development and fertility.24 In contrast, extensive evidence exists in support of an important role for uPAR in tumor growth, despite the fact that this protein seems to have very limited importance in normal physiological processes.25,26
In separate lines of evidence, we document that the acceleration of tumor growth caused by overexpression of uPAR in 293 cells is mediated by its interaction with VN and that host VN deficiency impairs tumor formation by MDA-MB-231 mammary carcinoma cells in an orthotopic xenograft model. These data document for the first time an important role for VN in tumor growth and demonstrate that the interaction between uPAR and VN is responsible for the tumor growth–promoting activity of uPAR.
Through a focused structure–function analysis of mouse uPAR, we first show that uPAR promotes cell migration and proliferation in vitro by virtue of its interaction with VN. These data confirm, and extend, previous studies conducted with human uPAR9,10 that demonstrated that the direct interaction with VN is crucial for the evolutionary conserved signaling activity of uPAR. Second, we document that the VN-dependent signaling activity of uPAR is paralleled by a highly significant reduction of tumor latency in xenografted mice, demonstrating that the tumor growth–promoting activity of uPAR in vivo is mediated by its interaction with VN. Our evidence for the central importance of VN in the biological activity of uPAR is based on the head-to-head comparison between wild-type muPAR and a muPAR variant that carries the single amino acid substitution W32A. The muPAR W32A mutant clearly displays a strong impairment in VN binding, and the cause for this is understood to be based on the structural level for both human and mouse uPAR.14 Is it possible for the W32A substitution to perturb other important functions of the receptor? We believe this is very unlikely because this uPAR variant has been shown to display normal uPA binding,9,17 falls outside of the chemotactic epitope located in the D1/D2 linker region (residues 88-9227 ), and is located in a different domain (D1) than all of the uPAR hot spots previously implicated in uPAR–integrin interactions.28-30 Our data clearly show that binding of VN to uPAR is required. However, is it also sufficient? In this study we specifically addressed the role of VN, and our results are therefore not informative with respect to the involvement of other uPAR interactions. The involvement of other downstream transducers is, however, very likely because uPAR/VN signaling has been shown to involve integrins10 and it has been shown recently that uPAR signaling via EGFR12 and GPCRs31 is closely connected to VN binding.
Interestingly we found that 293 cells transfected with VN-deficient uPAR (muPARW32A) displayed a significant delay in tumor formation compared with cells expressing receptors deficient in both VN and uPA binding (muPARΔD1) or mock-transfected cells. If we assume that the only functional difference between these variants is the inability of the latter to bind uPA, our findings would imply that binding of uPA to uPAR might in fact slow tumor growth. Because the difference in tumor formation by muPARW32A and muPARΔD1 cells is modest and the molecular structure of these variants very different, further experimentation is required to determine conclusively the role of uPA in the studied process.
Even if our data are consistent with other investigations on the role of uPAR in cancer progression, there are some discrepancies with a previous study in which it was found that overexpression of uPAR in HEK293 cells promotes metastatic dissemination, but not primary tumor growth.13 The experimental approach used in that study is very similar to the one applied here, and differences between the cell lines are the most likely cause of the discordance. The cells used by Jo et al13 were from American Type Culture Collection (Manassas, VA), while we used a commercial subclone of these (HEK293 Flp-In T-Rex from Invitrogen). In the study of Jo et al13 and here, these cells were further subcloned to express the different receptor variants or to render them GFP positive. It is thus very possible that these discordant results could be due to clonal differences. Importantly, however, both studies were carefully designed to exclude the possibility that clonal differences could affect the conclusions drawn from the results.
Finally, we show that host VN deficiency strongly impairs tumor formation by MDA-MB-231 cells in an orthotopic xenograft model. In this model the effect of VN deficiency is much greater than the effect of uPAR in the HEK293 model. This strongly suggests that it cannot be accounted for alone by an interaction between VN and tumor cell–expressed uPAR. Knockdown of uPAR in MDA-MB-231 cells has been shown to reduce tumor growth in a similar orthotopic xenograft model.21 However, in this case, the effect is not comparable to that observed with VN-deficient animals. VN is a multifunctional protein that interacts with various proteins known to play roles in tumor growth, including PAI-1 and integrins collagen. The striking effect of VN deficiency on tumor growth likely reflects a sum of different processes. Further studies are clearly required to determine the critical VN interactions.
How do VN and its interaction with uPAR promote tumor growth? Our in vitro data show that the interaction between uPAR and matrix VN promote cell proliferation by potentiation of adhesion-dependent proliferative signaling. However, it remains to be determined whether this is also the mechanism in vivo. In humans, VN is found in both healthy and cancerous breast tissue but with markedly different distribution.32 In ductal cancer in situ VN colocalize with collagen IV in the basement membrane and in adjacent periductal zones.32,33 This location coincides with areas where a prominent expression of uPA, PAI-1, and uPAR is observed by myofibroblasts located in the vicinity of microinvasive lesions,34 supporting a possible functional link between the biological activity of these molecules and the onset of invasive tumor growth. Expression of uPAR by malignant epithelial cells is the exception in ductal breast cancer,35 and further experiments are required to determine if the importance of VN is restricted to uPAR-positive tumors cells or if it also acts on the stromal compartment.
In conclusion, our data, for the first time, document a direct role for VN and for the interaction between uPAR and VN in the growth of xenografted human cancers cells. Together with the fact that both uPAR and VN are dispensable for normal development and fertility, our data identifies VN and possibly the interaction between uPAR and VN as promising novel drug targets for the treatment of cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
N.S. was supported by research grants from the Italian Association for Cancer Research.
Authorship
Contribution: V.P., G.M.S.F., and N.S. planned, conducted, and analyzed the experimentation presented in this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nicolai Sidenius, Italian Federation for Cancer Research (FIRC) Institute of Molecular Oncology, Via Adamello 16, 20139, Milan, Italy; e-mail: nicolai.sidenius@ifom.eu.
References
Author notes
V.P. and G.M.S.F. contributed equally to this study.