In this issue of Blood, Hu et al1 reported that expression of CD30 by diffuse large B-cell lymphoma (DLBCL) cells is associated with better outcome.
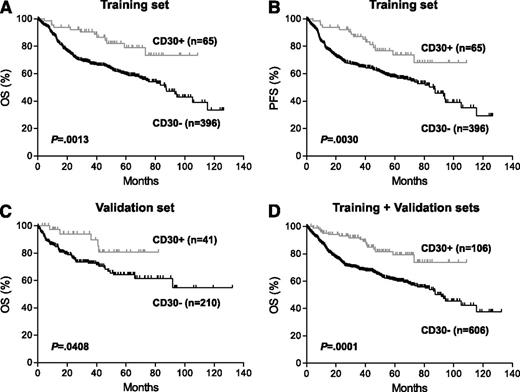
Prognostic impact of CD30 expression in de novo DLBCL. (A-B) Overall survival (OS) (A) and progression-free survival (PFS) (B) of patients with CD30+ vs CD30– DLBCL in the training set. (C) OS of patients with CD30+ vs CD30– DLBCL in the validation set. These patients were part of an independent cohort of 442 patients with available survival information (supplemental Table 1). (D) OS of patients with CD30+ vs CD30– DLBCL in combined training and validation sets. See Figure 2 in the article by Hu et al that begins on page 2715.
Prognostic impact of CD30 expression in de novo DLBCL. (A-B) Overall survival (OS) (A) and progression-free survival (PFS) (B) of patients with CD30+ vs CD30– DLBCL in the training set. (C) OS of patients with CD30+ vs CD30– DLBCL in the validation set. These patients were part of an independent cohort of 442 patients with available survival information (supplemental Table 1). (D) OS of patients with CD30+ vs CD30– DLBCL in combined training and validation sets. See Figure 2 in the article by Hu et al that begins on page 2715.
DLBCL is the most common type of non-Hodgkin lymphoma, and with modern immunochemotherapy, more than half of patients are cured.2 DLBCL is in fact a heterogeneity entity, and it can be divided into subgroups that are biologically and clinically distinct; however, even within these subgroups, there is still marked biological and clinical heterogeneity.3
One of the most consistent predictors of outcome is the International Prognostic Index (IPI) with a proposed revision to R-IPI2 for patients treated with Rituximab, cyclophosphamide, Hydroxydaunorubicin, Oncovin (Vincristine), Prednisone, R-CHOP. Many attempts have been made to find biomarkers that can improve our outcome prediction beyond the IPI. However, the literature has been littered with numerous prognostic markers, especially single markers that additional studies have failed to validate. Many factors contribute to the inconsistency of the findings. One major factor relates to the assay used, which may not be sufficiently robust or standardized to be readily reproducible. Biomarkers are frequently assayed by immunohistochemistry (IHC) on formalin-fixed, paraffin-embedded tissues. The tissues are often not processed uniformly, and the IHC assays are generally not standardized regarding the antibodies used, the antigen retrieval methods, the detection system, and the instruments employed. The scoring is usually subjective, and diverse criteria are used by different investigators.
Another problem is the heterogeneity of the patient population included in the different studies that were also frequently underpowered for analysis. The influence of the targeted population on the results obtained is illustrated by the different prognostic implication of BCL2 expression between the germinal center B-cell–like (GCB) and activated B-cell–like (ABC) subtypes of DLBCL: in the pre-rituximab era, BCL2 predicts for worse survival in the ABC, but not in the GCB, subtype, whereas with the addition of rituximab, the reverse is true.4 Studying BCL2 without taking the GCB/ABC subtypes into consideration may produce inconsistent findings.
Another important consideration is the complexity of the biological system. Tumor behavior is influenced by multiple factors, and the study of a single marker does not take into consideration other important modifiers. This is illustrated by a recent study that examined the expression of both MYC and BCL2 in DLBCL and demonstrated the importance of assaying both parameters for outcome prediction.5 Even with both markers included, there are layers of complexity that needed to be further explored. Not all MYCs are created equal, and it has been demonstrated that MYC with T58A mutation is a more potent oncogene. The mutant is associated with diminished activation of BCL2-like Protein 11, and can give rise to tumors without p19ARF and TP53 inactivation in a murine model.6 Thus, adding MYC mutation analysis may make the MYC/BCL2 model more robust. Although abnormalities in the tumor are important, we must not forget the important role of host/tumor interactions. Gene expression signatures derived from the microenvironment have been shown to be predictive of outcome,7 whereas abnormal expression of major histocompatibility complex molecules may impair immune surveillance and thus contribute to poorer survival.8
In this issue of Blood, Hu and coworkers1 reported from the retrospective study of a large series of DLCBL that expression of CD30 by the lymphoma cells is associated with better outcome in patients treated with R-CHOP therapy (see figure). CD30 expression is a hallmark of T/null anaplastic large cell lymphoma, T/natural killer cell lymphoma, nasal type, and Hodgkin lymphoma, but it may also be expressed in DLBCL including B anaplastic large cell lymphoma, primary mediastinal large cell lymphoma (PMBCL), and DLBCL-associated Epstein-Barr virus infection. The authors appropriately excluded the latter 2 categories from this study. They also examined the impact of some possible confounding variables such as GCB vs ABC subtype and MYC and BCL2 alterations on the findings of this study. In such a multi-institutional, retrospective study, it is not likely that patients would be treated entirely uniformly, but the large number of cases studied added credibility to the findings. CD30 (TNFRSF8) belongs to the tumor necrosis factor receptor superfamily. Its role in DLBCL is unclear, and the mechanistic basis for its association with better prognosis remains to be defined. Gene expression profiling (GEP) suggested differences in a number of pathways between the CD30-positive and CD30-negative groups and may indicate some possible directions for future investigations. However, to consider this group of CD30+ DLBCL as a distinct entity is premature at this point. The findings of this study should be validated in future independent studies. It is important that PMBCL be carefully excluded, possibly by using defined molecular signatures from previous GEP studies, as PMBCL can present as extramediastinal lesions.
With the limitations of single-marker IHC studies as discussed previously, should we still continue with this type of investigation? There are likely to be a number of biomarkers representing major biological determinants that could influence outcome even in the presence of other confounding factors. Their measurement could have immediate impact on patient management. Preferably, multiple such markers can be identified and assayed as a panel to determine if the panel is more robust and have greater predictive power compared with individual markers. A multimarker approach has been attempted in a number of studies: LIM Domain Only 2 (LMO2)/TNFRSF9,9 MYC/BCL2,5 and cell of origin/microenvironment.10 However, this could well be a transitional stage. We should move toward a more comprehensive, mechanism-based evaluation. The goal of our future studies should not only be finding reliable predictors of outcome, but the predictors should inform us of the mechanisms responsible for adverse outcome and the options available to improve it. The tools for this modern investigation, such as gene and micro RNA expression profiling and next-generation sequencing, are available. It is possible to construct a comprehensive picture for each tumor that will help us understand its strengths and vulnerabilities and its response to therapeutic perturbations. However, this information needs to be coupled with well-designed, large clinical studies, and a partnership of multiple disciplines and institutions is essential. The authors of this paper should be applauded for organizing such a large multi-institutional effort that makes their study feasible.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal