Key Points
TFPI inhibits CD26 activity in murine and human HSPCs via GPC3 resulting in better transwell migration as well as BM homing.
GPC3−/− mice show increased CD26 activity that leads to poor HSC retention in BM and increased circulating HSPCs.
Abstract
Directional migration determines hematopoietic stem/progenitor cell (HSPC) homing, which depends upon the interaction between the chemokine CXCL12 and its receptor CXCR4. CD26 is a widely expressed membrane-bound ectopeptidase that cleaves CXCL12 thereby depleting its chemokine activity. We identified tissue-factor pathway inhibitor (TFPI) as a biological inhibitor of CD26 in murine and human HSPCs. We observed low-level TFPI expression in endothelial cells in the bone marrow (BM), which did not increase following radiation injury. Treatment of HSPCs with TFPI in vitro led to enhanced HSPC migration toward CXCL12, as well as homing and engraftment in the BM upon transplantation. We found that Glypican-3 (GPC3), a heparan sulfate proteoglycan expressed on murine as well as human HSPCs, mediated this effect. TFPI did not affect CD26 activity, migration, or homing of GPC3−/− HSPCs, while it affected GPC1−/− HSPCs similar to wild-type HSPCs. Moreover, proliferation of GPC3−/− but not GPC1−/− BM HSPCs was significantly increased, which was associated with a decrease in the primitive HSC pool in BM and an increase in proportion of the circulating HSPCs in the peripheral blood. Hence, we present a novel role for TFPI and GPC3 in regulating HSC homing as well as retention in the BM.
Introduction
Hematopoietic stem cells (HSCs) are responsible for maintaining all blood cells throughout the lifetime of an individual, and are used clinically to treat various malignant and nonmalignant disorders.1 However, for some HSC grafts, for instance from umbilical cord blood (UCB), limited numbers of HSCs restrict their application to pediatric patients.2 Expanding HSCs in vitro or improving their homing efficiency would overcome this hurdle.3 As the HSC niche regulates HSC function in vivo, it is believed that additional insights in the regulation of HSCs by their niche may identify novel ways to manipulate HSCs and enhance their clinical use.4
A number of niche factors regulate HSC function by interacting with their respective receptors expressed on HSCs.5 These molecular interactions also play important roles in homing of the transplanted HSCs, which adhere to the vasculature through integrins and pass through the endothelium following rolling mediated by selectins.6 Directional migration of hematopoietic stem/progenitor cells (HSPCs) is mediated in large part by interaction of cell-surface–expressed CXCR4 with a gradient of CXCL12 or stroma-derived factor-1α expressed in the bone marrow (BM) niche.7,8 Loss of CXCR4 or annexin 2, involved in the presentation of CXCL12 to HSCs, severely reduces the number of HSCs in BM of adult mice.9,10 Incubation of murine or human HSPCs with anti-CXCR4 antibodies significantly reduces their homing and engraftment ability,7 while infusion of CXCR4-selective antagonists induces an increase in circulating HSPCs.11
CD26, a serine protease, cleaves an N-terminal dipeptide from CXCL12 thereby depleting its chemotactic activity.12-14 CD26-deficient or CD26 inhibitor–treated mouse BM as well as human UCB-derived HSPCs display enhanced migration toward CXCL12, which is translated in improved engraftment.15-17
During a screen of stromal feeders from fetal sites of hematopoiesis, used to mimic the hematopoietic niche, we found that transcripts for Tfpi were >20-fold higher in murine stromal cells that supported long-term repopulating (LTR) HSCs in noncontact cultures.18 Tissue-factor (TF) pathway inhibitor (TFPI) mediates the coagulation cascade. TFPI is a serine protease inhibitor that contains 3 Kunitz-type domains, 2 of which bind to factor VIIa and Xa.19 Although there was no evidence for a role of TFPI in hematopoiesis, other molecules involved in coagulation such as uPA and uPAR have been shown to affect HSC homeostasis.20
Here, we report that TFPI acts as a biological inhibitor of CD26 in murine BM as well as human UCB-derived HSPCs. Decrease in CD26 activity led to better chemotactic activity of HSPCs resulting in enhanced homing and engraftment potential. We further demonstrate that TFPI binds to heparan sulfate proteoglycan Glypican-3 (GPC3), which itself is known to inhibit CD26 activity in hepatocarcinoma cells.21,22 As GPC3 plays a role in inactivating CD26 in HSPCs, loss of this receptor caused increased proliferation and decreased retention of HSPCs in the BM, as well as decreased homing and engraftment of HSPCs.
Materials and methods
Animals
Six- to 8-week-old C57BL/6J-CD45.2 (Centre d’Elevage R. Janvier, Le Genest-St Isle, France), B6.SJL-PTPRCA-CD45.1 (Charles River Laboratories, Raleigh, NC), GPC3−/− (gift from Prof Jorge Filmus), GPC1−/− (gift from Prof Guido David, Department of Molecular and Developmental Genetics, VIB, K.U.Leuven), and Rag1−/− (gift from Prof Georges Coremans, Faculty of Medicine, UZ Leuven) mice were bred and maintained in the animal facility at KU Leuven. During the experiments, mice were maintained in isolator cages, fed with autoclaved acidified water, and irradiated food ad libitum. All experiments were approved by the institutional ethics committee.
Murine and human hematopoietic progenitor cell sorting
B6.SJL-PTPRCA-CD45.1 mice were used to collect BM cells, from which were enriched for lin− fraction using the EasySep hematopoietic progenitor cell enrichment kit (Stem Cell Technologies, Vancouver, Canada). Subsequently, fluorescence-activated cell sorting (FACS)–based purification of the c-kit+lin−Sca-1+ (KLS) cell fraction was performed on a FACSVantage sorter (Becton Dickinson, Mountain View, CA). Human umbilical cord blood (hUCB) products collected at room temperature and unsuitable for banking were supplied by the Leuven cord blood bank (UZ Leuven, Gasthuisberg, Belgium). All studies were conducted in accordance with the Declaration of Helsinki and carried out with approval of the medical ethics committee of the UZ Leuven, Gasthuisberg. Details of the methods followed to isolate HSPC fractions are provided in supplemental Methods (available on the Blood website).
Sorting of cellular components of the HSC niche
BM cells were isolated by flushing the long bones of the hind limbs followed by crushing of the bones and treatment with 3 mg/mL collagenase type 1 (Sigma-Aldrich, St. Louis, MO). Following magnetic-activated cell sorting–based depletion of Lin+/CD45+ cells (lineage depletion kit and CD45 microbeads; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany), the cells were stained with an anti-mouse lineage allophycocyanin (APC) cocktail (BD Biosciences, San Jose, CA), CD45-PerCPCy5.5, CD31 phycoerythrin (PE), Sca-1 PECy7 (all from eBioscience, San Diego, CA), and CD51-fluorescein isothiocyanate (FITC) (Biolegend, San Diego, CA). Endothelial (lin−CD45−CD31+), osteoblastic (lin−CD45−CD31−CD51+Sca-1−), and mesenchymal stem cell (lin−CD45−CD31−CD51+Sca-1+) subpopulations were sorted as reported earlier.23 Some lin+ cells were also sorted as negative control for the HSC niche cell populations.
Cell culture
The in vitro HSPC culture system was adapted from Zhang et al.24 Sorted murine KLS cells (50 cells per well), were cultured in U-bottom 96-well plates (BD Biosciences) in 100 μL of Stemspan (Stem Cell Technologies) supplemented with 100 ng/mL murine thrombopoietin (mTPO) and 50 ng/mL murine stem cell factor (mSCF), with or without 100 ng/mL recombinant mouse (rm)TFPI (all from R&D Systems, Minneapolis, MN). Recombinant human SCF, Tpo, and TFPI (all from R&D Systems) were used to culture hUCB-derived Lin−CD34+ (50 000 cells per well in ultra-low attachment 24-well plates). Murine cells were cultured for 5 days, while human HSPCs were cultured for 2 days at 37°C with 5% CO2.
Serial transplantation
Culture progeny of 200 KLS cells from CD45.1 mice was transplanted along with 1 × 106 competitor CD45.2 BM cells into lethally irradiated (10 Gy) C57BL/6J-CD45.2 mice. From GPC3−/− mice, 0.5 × 106 BM cells were mixed with 0.5 × 106 competitor CD45.1 BM cells and injected into lethally irradiated B6.SJL-PTPRCA-CD45.1 mice. Peripheral blood chimerism analysis was performed every 4 weeks. After 12 weeks, primary recipients were sacrificed, BM was harvested, and 1 × 106 cells were grafted in secondary lethally irradiated CD45.2 mice. After 3 months, chimerism in secondary recipients was evaluated.
Flow cytometry
Chimerism and lineage analysis of the BM-derived cells was performed by flow cytometry. Lineage-specific antibodies used were PE-conjugated Mac-1 and Gr-1 for myeloid cells, APC-conjugated B220 for B cells, and PE-conjugated anti-CD4 and anti-CD8 for T cells. For chimerism analysis, FITC-conjugated anti-CD45.1 and PerCPCy5.5-conjugated anti-CD45.2 antibodies were used. All antibodies were procured from BD Pharmingen (San Diego, CA). To detect binding of TFPI on HSPCs, anti-mouse TFPI antibody was used from R&D Systems. Flow cytometric analysis for primitive HSCs and hematopoietic progenitors was performed using anti-mouse CD48 APC and CD150 PECy7 (eBioscience) along with KLS cell staining (as for sorting).
GPC3 expression in UCB-derived hematopoietic progenitors was analyzed by using anti-human GPC3-PE antibody (R&D Systems) along with anti-human CD45-APC, anti-human lineage cocktail-FITC (all from BD Biosciences), and anti-human CD26-FITC (Abcam, Cambridge, United Kingdom). Similar strategy was used for analyzing human HSPCs homed in mouse BM. The cells were analyzed by flow cytometry using FACSCanto (BD Biosciences). Suitable isotype controls for each antibody were used in all experiments.
PKH-26 labeling
For adhesion and migration experiments, cultured KLS cells were labeled with the PKH-26 membrane dye,25 according to the manufacturer’s instructions (Sigma-Aldrich). The cells were harvested and washed thoroughly to remove any protein content. Cells were resuspended at a density of 1 × 107/mL diluent C. The cell suspension was mixed with an equal volume of 4mM PKH26 dye (in diluent C) for 5 minutes at room temperature. An equal volume of fetal bovine serum (FBS) was added to stop the reaction and the cells were washed with medium containing 10% FBS.
In vitro migration assays
In vitro transwell migration assays were performed as described previously with slight modifications.26 PKH-26–labeled KLS cell progeny (5000-50 000) in 200 μL of chemotaxis buffer (RPMI 1640, 1% FBS [Gibco BRL, Grand Island, NY], and antibiotics) were added to the upper chambers of a 6.5-mm, 3-μm pore-size transwell (Costar, Cambridge, MA). Chambers were incubated at 37°C, 5% CO2 for 3 hours after which cell migration toward CXCL12 in lower chamber was analyzed. The percentage of migration was determined by calculating the percentage of input cells migrated into the lower chamber.
Cell-cycle analysis
For cell-cycle analysis, cells were fixed by adding ethanol (precooled at −20°C) dropwise in the suspension while vortexing. RNase treatment (Ambion, Austin, TX) of the cells was performed followed by propidium iodide (10 μg/mL; Sigma-Aldrich) staining.27 Chicken erythrocytes were used as standard. Flow cytometer (FACSCanto; Becton Dickinson) based detection of DNA content was analyzed, and the percentage of cells in the sub-G0 fraction as well as G2/M phase of the cell cycle was plotted for different samples.
Quantitative RT-PCR
Total RNA was prepared using the RNA Isolation kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. DNase treatment of RNA was performed using the Turbo DNase kit (Ambion). The purity and the concentration of RNA were assessed using a ND-1000 spectrophotometer (NanoDrop Technologies Rockland, DE). RNA (100 ng-1 µg) from each sample was used to synthesize cDNA using the Superscript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. For cDNA preparation from HSC niche cells, the Cell-to-cDNA II kit (Ambion) was used. Quantitative reverse transcription–polymerase chain reaction (qRT-PCR) was carried out using TaqMan SYBR Green Universal Mix PCR buffer (Applied Biosystems, Foster City, CA). The PCRs were carried out in a Mastercycler Realplex (Eppendorf, Hamburg, Germany) using the following program: 2 minutes at 50°C, 1 minutes at 95°C and 40 cycles of 30 seconds at 95°C and 30 seconds at 60°C. Amplified products from qRT-PCR performed on HSC niche components were also electrophoresed on agarose gel (1.5% in tris-base acetic acid EDTA buffer; Sigma Aldrich). The list of primers used is given in supplemental Table 1.
DPP-IV activity assay
The analyses of the dipeptidyl peptidase IV (DPP-IV) (CD26) activity were performed by DPP-IV Glo protease assay (Promega, Madison, WI) on 100 000 hUCB-derived cultured lin−CD34+ or 50 000 KLS progeny per well in 96-well flat-bottom plates (BD Biosciences) following the manufacturer’s instructions. Diprotin A (100 ng/mL; Sigma-Aldrich), an inhibitor of CD26 activity, was used as a positive control. Recombinant human DPP-IV (R&D Systems) was used as positive control for all experiments. Chemiluminiscence was detected using a microplate luminometer, Luminoskan Ascent (Thermo Fisher Scientific Inc, Waltham, MA).
Immunostaining
Following incubation with recombinant TFPI for 3 hours, the wild-type (WT) or GPC3−/− mice BM-derived KLS cells were used for immunostaining to detect TFPI binding on the cell surface. TFPI expression in BM endothelial cells was examined by staining BM sections with anti-mouse TFPI and CD31 antibodies. Details of the methods used are provided in supplemental Methods. Immunostaining was analyzed by using an AxioImager microscope (Zeiss, Jena, Germany) fitted with an AxioCam MRc5 digital camera used to capture images. Images were prepared using AxioVision AC software (Zeiss) and assembled in Adobe Photoshop.
Coimmunoprecipitation and western blotting
Coimmunoprecipitation (Co-IP) for TFPI and GPC3 was performed using a Co-IP kit (Thermo Scientific Pierce, Rockford, IL) following the manufacturer’s protocol. The samples were analyzed for pulldown of GPC3 (TFPI Co-IP) or TFPI (GPC3 Co-IP) by immunoblotting using standard protocols described in supplemental Methods.
Homing experiments
Statistical analysis
Data are shown as mean ± SE. Statistical analysis was performed using a 2-tailed Student t test. P values < .05 were considered statistically significant.
Results
TFPI increases the migration and homing potential of murine as well as human HSPCs by inhibiting CD26 activity
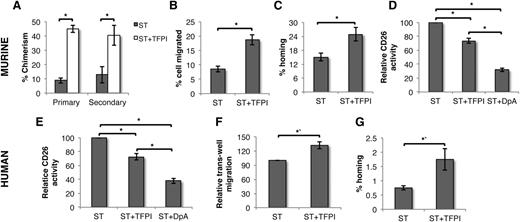
We first examined the effect of TFPI on the LTR potential of murine BM-derived KLS cells. Competitive repopulation experiments were performed following culture of KLS cells in serum-free medium in the presence of SCF and TPO with or without TFPI. Chimerism resulting from TFPI-treated cells, both in primary and secondary recipients, was twofold to threefold higher compared with KLS progeny from cultures without TFPI (Figure 1A). We did not see any difference in the in vivo multilineage differentiation potential (supplemental Figure 1a). TFPI treatment did not alter the total number of cells (supplemental Figure 1b), which was also reflected by unchanged cell-cycle status (supplemental Figure 1c). Methylcellulose colony forming assays did not show any change in the frequency of colony forming cells (CFCs) following TFPI treatment (supplemental Figure 1d). These results suggested that enhanced long-term engraftment following TFPI treatment may not be caused by HSC expansion, but could be due to more efficient homing.
TFPI improves LTR potential of HSPCs by inhibiting CD26 activity. (A) Progeny of 200 CD45.1 murine KLS cells from cultures with ST with or without TFPI was injected along with 1 × 106 CD45.2 BM cells in lethally irradiated mice. Peripheral blood chimerism derived from CD45.1 cells in primary recipients was analyzed after 3 months (n = 12); 1 × 106 BM cells from primary recipients were injected into lethally irradiated secondary recipient and peripheral blood chimerism from CD45.1 cells was analyzed after 3 months (n = 12). (B) Fifty thousand cells obtained from KLS cells cultured for 5 days with or without TFPI were allowed to migrate through a 3-μm pore-size membrane toward CXCL12 (n = 5). Percentage of cells migrated was assessed. (C) One hundred thousand cells obtained from KLS cells cultured for 5 days with or without TFPI were injected in lethally irradiated hosts (n = 8). The fraction of transplanted Colony forming unit cells that homed into the BM after 16 hours of transplantation was assessed. (D) CD26 activity in KLS cells cultured for 5 days were compared by CD26 enzyme assay (n = 8). Error bars represent SEM. (E) CD26 activity of human UCB derived lin−CD34+, cultured for 2 days with or without TFPI (n = 5). (F) One hundred thousand cells obtained from human lin−CD34+ cells cultured for 2 days with or without TFPI were allowed to migrate through a 3-μm pore-size membrane toward CXCL12 (n = 5). (G) One hundred thousand human lin−CD34+ cells cultured for 2 days with or without TFPI were transplanted in lethally irradiated Rag1−/− preinjected with anti-NK1.1 antibodies. After 16 hours, mice were sacrificed and human HSPCs homed in the BM were quantified by flow cytometric detection of human CD45+lin−CD34+ cells. Total number of HSPCs homed was compared with the number of HSPCs injected and the proportion of homed cells was plotted for different conditions (n = 8, P = .039). Error bars represent SEM; *P < .05; ST, SCF TPO.
TFPI improves LTR potential of HSPCs by inhibiting CD26 activity. (A) Progeny of 200 CD45.1 murine KLS cells from cultures with ST with or without TFPI was injected along with 1 × 106 CD45.2 BM cells in lethally irradiated mice. Peripheral blood chimerism derived from CD45.1 cells in primary recipients was analyzed after 3 months (n = 12); 1 × 106 BM cells from primary recipients were injected into lethally irradiated secondary recipient and peripheral blood chimerism from CD45.1 cells was analyzed after 3 months (n = 12). (B) Fifty thousand cells obtained from KLS cells cultured for 5 days with or without TFPI were allowed to migrate through a 3-μm pore-size membrane toward CXCL12 (n = 5). Percentage of cells migrated was assessed. (C) One hundred thousand cells obtained from KLS cells cultured for 5 days with or without TFPI were injected in lethally irradiated hosts (n = 8). The fraction of transplanted Colony forming unit cells that homed into the BM after 16 hours of transplantation was assessed. (D) CD26 activity in KLS cells cultured for 5 days were compared by CD26 enzyme assay (n = 8). Error bars represent SEM. (E) CD26 activity of human UCB derived lin−CD34+, cultured for 2 days with or without TFPI (n = 5). (F) One hundred thousand cells obtained from human lin−CD34+ cells cultured for 2 days with or without TFPI were allowed to migrate through a 3-μm pore-size membrane toward CXCL12 (n = 5). (G) One hundred thousand human lin−CD34+ cells cultured for 2 days with or without TFPI were transplanted in lethally irradiated Rag1−/− preinjected with anti-NK1.1 antibodies. After 16 hours, mice were sacrificed and human HSPCs homed in the BM were quantified by flow cytometric detection of human CD45+lin−CD34+ cells. Total number of HSPCs homed was compared with the number of HSPCs injected and the proportion of homed cells was plotted for different conditions (n = 8, P = .039). Error bars represent SEM; *P < .05; ST, SCF TPO.
Homing of HSCs depends on their ability to migrate through endothelium and subsequently attach to the BM niche.30 We therefore assessed the in vitro adhesion of KLS progeny cultured with or without TFPI to BM stromal cells as well as their chemotactic response toward CXCL12 using a transwell system. Adhesion of TFPI-treated cells to BM stroma remained unchanged (supplemental Figure 2a), consistent with the finding that expression of important adhesion receptors in cultured KLS cells was not influenced by TFPI (supplemental Figure 2b). However, a significantly greater proportion of TFPI-treated cells migrated toward CXCL12 (Figure 1B). To further these in vitro observations to in vivo context, we performed short-term homing experiments (Figure 1C). KLS cells cultured with or without TFPI were harvested after 5 days and were transplanted intravenously into lethally irradiated animals. After 16 hours of transplantation, BM cells from the recipients were harvested and the total number of CFCs was enumerated and compared with the number of CFCs injected. As lethal irradiation depleted the animals of their BM HSPCs, CFCs obtained thus represent the homed donor population. Results clearly showed that TFPI-treated HSPCs homed significantly better (P = .02, n = 8; Figure 1C). Although TFPI-treated cells migrated significantly more toward recombinant CXCL12, levels of CXCR4 gene as well as protein expression were similar between cells cultured with or without TFPI (supplemental Figure 3a-b left), suggesting that TFPI affects the chemotactic response of HSPCs toward CXCL12, not by increasing CXCR4 expression.
As chemokine activity of CXCL12 can be regulated by the proteolytic activity of CD26, we analyzed the expression (supplemental Figure 3a-b right) and activity (Figure 1D) of CD26 in murine HSPCs. TFPI did not affect the expression of CD26 in murine KLS cells, however, the protease activity of CD26 in cultured murine KLS cells was inhibited significantly by TFPI. The inorganic inhibitor diprotin A, a known inhibitor of CD26 activity,31 was used as positive control in most experiments. The effect of TFPI on CD26 activity was less pronounced than the effect of diprotin A. The effect of TFPI on CD26 activity of KLS cells could be seen as early as 3 hours (supplemental Figure 4a), which was also reflected in increased migration (supplemental Figure 4b). This early effect of TFPI on the HSPCs was corroborated by homing assays performed using KLS cells incubated with or without TFPI for 3 hours (supplemental Figure 5).
Our observations that TFPI inhibited CD26 activity in murine HSPCs thereby affecting their in vitro migration and in vivo BM homing potential led us to explore the effect of TFPI in human HSPCs. We used UCB-derived HSPCs to assess the effect of TFPI on their migration and homing capacity. UCB-derived lin−CD34+ were sorted and cultured in the presence of SCF and TPO with or without TFPI. After 48h, the cells were harvested and CD26 activity, transwell migration and in vivo BM homing assays were performed. We found that TFPI treatment led to decreased CD26 activity in HSPCs cultured in the presence of TFPI (Figure 1E). As in murine BM-derived HSPCs, this also resulted in improved migration toward CXCL12 (Figure 1F). A significant increase in homing potential of the TFPI-treated lin−CD34+ cells was also observed (Figure 1G). Homed human HSPCs in recipient mice BM were identified using flow cytometry–based detection of CD45+lin−CD34+ cells.
BM endothelial cells express low levels of TFPI that remain unaffected following radiation injury
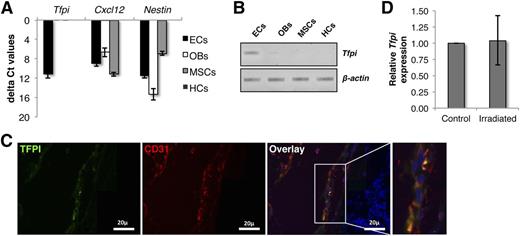
To understand the physiological relevance of TFPI, we examined its expression in different cellular components of the HSC niche. We sorted endothelial cells (ECs; lin−CD45−CD31+), osteoblastic cells (OBs; lin−CD45−CD31−CD51+Sca-1−) and mesenchymal stem cell (MSCs; lin−CD45−CD31−CD51+Sca-1+) subpopulations from the crushed bones from mice and analyzed the expression of Tfpi by qRT-PCR for Tfpi, Cxcl12, and Nestin (Figure 2A) (gating strategy is shown in supplemental Figure 6). As expected, we observed the highest expression of Cxcl12 in OBs while MSCs expressed the highest levels of Nestin. ECs in various organs have been reported to be the major source of TFPI.32 Albeit low, we detected Tfpi transcripts in ECs, but not in the OB- or MSC-sorted cell fractions (semiquantitative RT-PCR in Figure 2B). We confirmed these findings by immunostaining demineralization bone sections for TFPI and CD31+ (Figure 3C). We demonstrate TFPI staining of CD31+ ECs. We also assessed whether Tfpi levels are influenced by radiation injury. Mice were lethally irradiated and Tfpi expression was quantified in lin−CD45− BM cells harvested from irradiated and nonirradiated collagenase I and crushed long bones (Figure 2D). Levels of Tfpi were similar in irradiated and nonirradiated BM lin−CD45− cells.
TFPI is expressed in BM endothelial cells. (A) Murine BM cells were isolated by first flushing the long bones from the hind limbs, crushing the bones treated with collagenase I. Lin+CD45+ cells were depleted by magnetic-activated cell sorter columns and cells subsequently stained with antibodies against lineage, CD45, CD31, CD51, and Sca-1 antigens. ECs (lin−CD45−CD31+), OBs (lin−CD45−CD31−CD51+Sca-1−), MSCs (lin−CD45−CD31−CD51+Sca-1+), and HCs (lin+) were FACS sorted and qRT-PCR was performed for Tfpi, Cxcl12, and Nestin. HC, hematopoietic cell. (B) Resulting cDNAs were also identified by DNA gel electrophoresis. (C) Expression of TFPI protein in the BM was assessed by immunostaining decalcified long bones with antibodies against CD31 and TFPI. (D) Effect of lethal radiation on Tfpi expression was examined by comparing its expression in lin−CD45− cells from control and irradiated long bones.
TFPI is expressed in BM endothelial cells. (A) Murine BM cells were isolated by first flushing the long bones from the hind limbs, crushing the bones treated with collagenase I. Lin+CD45+ cells were depleted by magnetic-activated cell sorter columns and cells subsequently stained with antibodies against lineage, CD45, CD31, CD51, and Sca-1 antigens. ECs (lin−CD45−CD31+), OBs (lin−CD45−CD31−CD51+Sca-1−), MSCs (lin−CD45−CD31−CD51+Sca-1+), and HCs (lin+) were FACS sorted and qRT-PCR was performed for Tfpi, Cxcl12, and Nestin. HC, hematopoietic cell. (B) Resulting cDNAs were also identified by DNA gel electrophoresis. (C) Expression of TFPI protein in the BM was assessed by immunostaining decalcified long bones with antibodies against CD31 and TFPI. (D) Effect of lethal radiation on Tfpi expression was examined by comparing its expression in lin−CD45− cells from control and irradiated long bones.
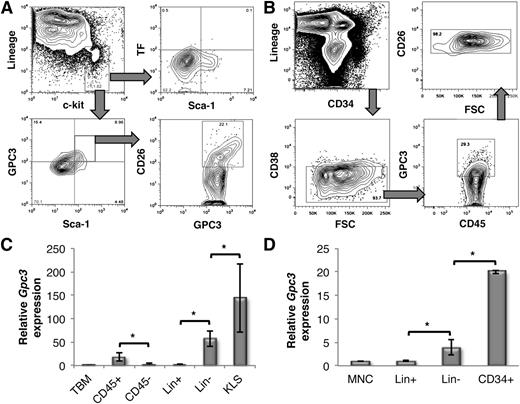
GPC3 is expressed on the cell surface of murine as well as human HSPCs. (A) Expression of TF, GPC3, and CD26 on KLS cells. Lin−c-kit+ BM MNCs were gated (top left) and the cell-surface expression of TF (top right) and GPC3 (bottom left) was determined in combination with Sca-1. GPC3-expressing KLS cells were further gated to determine CD26 expression (bottom right). (B) Freshly isolated MNCs from human UCB samples were analyzed for coexpression of CD26 (top right) and GPC3 (bottom right) in lin−CD34+ cells (top left) gated for CD38− cells (bottom left). (C) qRT-PCR analysis of Gpc3 expression in different subpopulations of mouse BM (n = 4). (D) qRT-PCR analysis of Gpc3 expression in different subpopulations human UCB-derived MNCs (n = 5). Error bars represent SEM; *P < .05.
GPC3 is expressed on the cell surface of murine as well as human HSPCs. (A) Expression of TF, GPC3, and CD26 on KLS cells. Lin−c-kit+ BM MNCs were gated (top left) and the cell-surface expression of TF (top right) and GPC3 (bottom left) was determined in combination with Sca-1. GPC3-expressing KLS cells were further gated to determine CD26 expression (bottom right). (B) Freshly isolated MNCs from human UCB samples were analyzed for coexpression of CD26 (top right) and GPC3 (bottom right) in lin−CD34+ cells (top left) gated for CD38− cells (bottom left). (C) qRT-PCR analysis of Gpc3 expression in different subpopulations of mouse BM (n = 4). (D) qRT-PCR analysis of Gpc3 expression in different subpopulations human UCB-derived MNCs (n = 5). Error bars represent SEM; *P < .05.
Murine as well as human HSCs express GPC3 but not TF as a potential TFPI-binding partner
As TF is the classical receptor for TFPI,19 we examined its expression on murine KLS cells. FACS analysis demonstrated that TF is not expressed on murine BM-derived KLS cells (Figure 3A top panel). TFPI is also known to bind to proteoglycans such as GPC3.21 To assess whether TFPI binds to murine BM and human UCB-derived HSPCs through GPC3, we first examined its expression on these cells by flow cytometry (Figure 3A-B). A significant proportion of murine BM-derived KLS cells and UCB-derived lin−CD45+CD34+CD38− cells expressed GPC3. Importantly, a significant fraction of HSPCs that expressed GPC3 coexpressed CD26. We further compared the expression of Gpc3 in different subpopulations of murine BM and human UCB-derived mono-nuclear cells (MNCs). qRT-PCR experiments demonstrated significantly higher expression of Gpc3 transcripts in HSPCs than either the nonhematopoietic or mature hematopoietic cells in murine BM as well as hUCB-derived MNCs (Figure 3C-D).
TFPI binds to GPC3 in murine BM-derived HSPCs
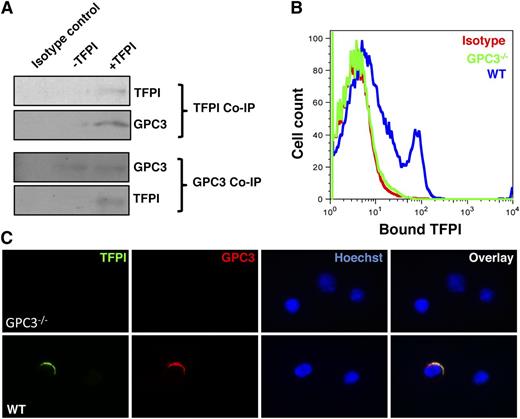
As GPC3, a potential binding partner for TFPI, was expressed on HSPCs, we tested whether TFPI indeed binds to HSPCs through GPC3. To achieve this, we used lin− BM murine cells and incubated them for 3 hours with or without TFPI. Following this, we performed Co-IP experiments using antibodies against TFPI and GPC3, and performed immunoblots to detect TFPI and GPC3, respectively (Figure 4A). Results clearly showed coprecipitation of GPC3 along with TFPI, and TFPI was coprecipitated along with GPC3. These experiments showed that TFPI binds to GPC3 expressed on the HSPCs. We further confirmed these results using GPC3−/− mice where we performed flow cytometry (Figure 4B) and immunostaining-based (Figure 4C) based detection of TFPI binding on the cell surface of HSPCs. Using FACS, we could detect binding of TFPI to Lin− BM cells from WT mice but not GPC3−/− mice. To confirm the binding of TFPI to the HSC fraction of lin− cells, we sorted KLS cells from WT and GPC3−/− mice, and incubated them with/without TFPI for 3 hours, followed by immunostaining for TFPI and GPC3 (Figure 4C). Consistent with the FACS results, TFPI did not bind to KLS cells from GPC3−/− mice (Figure 4C top panel), while binding to WT KLS cells was clearly observed (Figure 4C bottom panel).
TFPI binds to GPC3 expressed on the cell surface of murine as well as human HSPCs. (A) Co-IP experiments were performed for TFPI (top panel) or GPC3 (bottom panel) using mouse BM-derived lin− cells incubated with or without TFPI for 3 hours. Western blotting was performed to detect GPC3 or TFPI in the immunoprecipitated protein, respectively. (B) Lin− BM cells from WT or GPC3−/− mice were used for flow cytometric detection of TFPI bound to HSPCs. The cells were incubated with or without TFPI for 3 hours and stained with specific anti-TFPI antibodies. TFPI binding to the HSPCs was detected by performing flow cytometry. (C) WT and GPC3−/− mouse BM-derived KLS cells were incubated for 3 hours with or without TFPI. Immunostaining was performed to detect binding of TFPI. All results are representative of at least 3 experiments.
TFPI binds to GPC3 expressed on the cell surface of murine as well as human HSPCs. (A) Co-IP experiments were performed for TFPI (top panel) or GPC3 (bottom panel) using mouse BM-derived lin− cells incubated with or without TFPI for 3 hours. Western blotting was performed to detect GPC3 or TFPI in the immunoprecipitated protein, respectively. (B) Lin− BM cells from WT or GPC3−/− mice were used for flow cytometric detection of TFPI bound to HSPCs. The cells were incubated with or without TFPI for 3 hours and stained with specific anti-TFPI antibodies. TFPI binding to the HSPCs was detected by performing flow cytometry. (C) WT and GPC3−/− mouse BM-derived KLS cells were incubated for 3 hours with or without TFPI. Immunostaining was performed to detect binding of TFPI. All results are representative of at least 3 experiments.
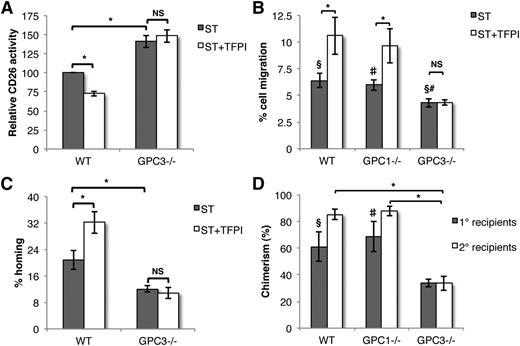
TFPI does not affect CD26 activity, migration, and homing potential of GPC3−/− HSPCs
We next evaluated the effect of TFPI on the CD26 activity in GPC3−/− KLS cells in comparison with WT cells (Figure 5A). TFPI reduced the CD26 activity of WT KLS cells while there was no significant change in the CD26 activity of GPC3−/− KLS cells. Consistent with published results,22 GPC3−/− KLS cells invariably showed higher CD26 activity than WT cells (n = 5, P = .006). These results confirmed that TFPI affects CD26 activity of HSPCs via binding to GPC3. Consistent with the notion that the effect of TFPI on CD26 activity translates in improved migration to CXCL12 and homing into the BM, GPC3−/− cells migrated less toward CXCL12 than WT cells, and migration of GPC3−/− cells was unaffected by exposure to TFPI (Figure 5B). Likewise, GPC3−/− cells homed significantly less to BM, and homing was unaffected by culture with TFPI (Figure 5C). The poor homing potential of GPC3−/− BM-derived HSPCs was also reflected in significantly lower LTR potential in both primary and secondary recipients (Figure 5D). To ensure that the effect was GPC3 specific, we also included KLS cells from GPC1−/− mice. No difference in migration potential or LTR ability between HSPCs from WT and GPC1−/− HSPCs was observed (Figure 5B,D).
Higher CD26 activity, decreased migration, and engraftment potential of TFPI unresponsive GPC3−/− HSPCs. (A) WT or GPC3−/− mouse BM-derived KLS cells were cultured in the presence of SCF and Tpo with or without TFPI. After 5 days, the cells were harvested and CD26 enzyme activity was assessed (n = 5). (B) KLS cells from WT, GPC1−/−, and GPC3−/− mouse BM were treated with or without TFPI, and migration toward CXCL12 assessed (n = 5). (C) Proportion of transplanted progenitors homed into the recipient BM after 16 hours of transplantation was compared between TFPI-treated and untreated KLS cells from WT and GPC3−/− mice by Colony forming unit-cell assays (n = 8). (D) A total of 0.5 × 106 BM cells from WT, GPC1−/−, and GPC3−/− mice together with 0.5 × 106 competitor CD45.1 BM cells were transplanted in lethally irradiated hosts and donor-derived chimerism in peripheral blood was analyzed after 3 months (n = 12). One million BM cells from primary recipients were injected in lethally irradiated secondary recipients (n = 12). Donor-derived chimerism in secondary recipients was analyzed after 3 months of transplantation. Error bars represent SEM. *§#, P < .05; §, WT vs GPC3−/−; #, GPC1−/− vs GPC3−/−.
Higher CD26 activity, decreased migration, and engraftment potential of TFPI unresponsive GPC3−/− HSPCs. (A) WT or GPC3−/− mouse BM-derived KLS cells were cultured in the presence of SCF and Tpo with or without TFPI. After 5 days, the cells were harvested and CD26 enzyme activity was assessed (n = 5). (B) KLS cells from WT, GPC1−/−, and GPC3−/− mouse BM were treated with or without TFPI, and migration toward CXCL12 assessed (n = 5). (C) Proportion of transplanted progenitors homed into the recipient BM after 16 hours of transplantation was compared between TFPI-treated and untreated KLS cells from WT and GPC3−/− mice by Colony forming unit-cell assays (n = 8). (D) A total of 0.5 × 106 BM cells from WT, GPC1−/−, and GPC3−/− mice together with 0.5 × 106 competitor CD45.1 BM cells were transplanted in lethally irradiated hosts and donor-derived chimerism in peripheral blood was analyzed after 3 months (n = 12). One million BM cells from primary recipients were injected in lethally irradiated secondary recipients (n = 12). Donor-derived chimerism in secondary recipients was analyzed after 3 months of transplantation. Error bars represent SEM. *§#, P < .05; §, WT vs GPC3−/−; #, GPC1−/− vs GPC3−/−.
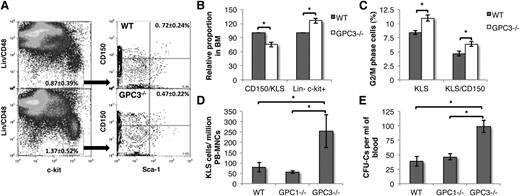
HSC maintenance in the BM of GPC3−/− mice is altered
As decreased levels of CXCL12 in the BM are associated with increased mobilization and proliferation of HSCs,33 we examined the retention of HSCs within the BM of GPC3−/− mice, which exhibited higher CD26 activity than the cells from WT animals (Figure 6A). FACS analysis demonstrated that the number of CD48−CD150+ KLS cells (HSCs) was significantly lower in GPC3−/− BM than WT BM (n = 6, P = .006), whereas the number of more committed Lin−c-kit+ cells (progenitors) was higher in GPC3−/− BM than WT BM (n = 6, P = .04) (Figure 6A-B). Moreover, significantly more KLS as well as CD48−CD150+KLS cells from GPC3−/− mice were in the G2M phase of the cell cycle compared with WT mice (Figure 6C). We did not observe any significant change in the lineage-committed subpopulations in GPC3−/− BM cells (supplemental Figure 7). To determine whether higher CD26 activity in GPC3−/− mice BM also led to mobilization of HSPCs into circulation, we performed FACS analysis on peripheral blood cells from WT, GPC1−/−, and GPC3−/− mice (Figure 6D). We found no differences in the circulating KLS cell number between WT and GPC1−/− mice, while there was a significant increase in KLS cells in the peripheral blood of GPC3−/− mice. This was also reflected in the increased number of CFCs in GPC3−/− but not in GPC1−/− mice compared with WT mice (Figure 6E).
Impaired HSC retention of primitive HSCs in GPC3−/− BM resulting in a higher proportion of circulating HSPCs. (A) Flow cytometric analysis to assess the proportion of primitive HSCs and hematopoietic progenitors in WT (top panel) and GPC3−/− (bottom panel) mice BM. Lin−c-kit+ cells (hematopoietic progenitors) were gated and analyzed for Sca-1+CD150+ cells (primitive HSCs). (B) The proportion of hematopoietic progenitors and primitive HSCs in WT and GPC3−/− BM was compared. (C) Cell-cycle analysis of gated Lin− (left) and KLS (right) cells from BM of WT and GPC3−/− cells. (D) Comparison of the number of KLS cells in peripheral blood of WT, GPC1−/−, and GPC3−/− mice by flow cytometry. (E) Comparison of the circulating hematopoietic progenitors in WT, GPC1−/−, and GPC3−/− mice by CFC assay. n = 8-12; *P < .05.
Impaired HSC retention of primitive HSCs in GPC3−/− BM resulting in a higher proportion of circulating HSPCs. (A) Flow cytometric analysis to assess the proportion of primitive HSCs and hematopoietic progenitors in WT (top panel) and GPC3−/− (bottom panel) mice BM. Lin−c-kit+ cells (hematopoietic progenitors) were gated and analyzed for Sca-1+CD150+ cells (primitive HSCs). (B) The proportion of hematopoietic progenitors and primitive HSCs in WT and GPC3−/− BM was compared. (C) Cell-cycle analysis of gated Lin− (left) and KLS (right) cells from BM of WT and GPC3−/− cells. (D) Comparison of the number of KLS cells in peripheral blood of WT, GPC1−/−, and GPC3−/− mice by flow cytometry. (E) Comparison of the circulating hematopoietic progenitors in WT, GPC1−/−, and GPC3−/− mice by CFC assay. n = 8-12; *P < .05.
Discussion
That the CXCR4-CXCL12 axis plays an important role in HSPC homing, mobilization, and maintenance has been very extensively demonstrated. Insights into how this axis is regulated would be important in our understanding of HSC biology and can lead to designing improved engraftment and mobilization regimens. Among various chemokine-modifying proteases, CD26 plays a crucial role in regulating activity of CXCL12.34 CD26 cleaves amino-terminal dipeptide in proteins with proline at the penultimate position. CD26 is expressed in many solid tumors such as hepatocarcinoma cells, as well as various cell types, including HSCs. As CD26 decreases the active CXCL12 concentration gradient, loss of its expression or activity in murine HSPCs as well as human UCB-derived HSPCs improved homing.15,17 Various inorganic inhibitors of CD26 have been synthesized while biological regulators of CD26 activity are largely unknown. Davoodi et al22 demonstrated that GPC3 binds to CD26 on hepatocarcinoma cells, and causes its inactivation.
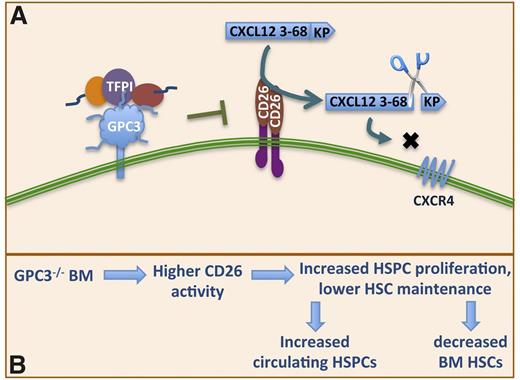
GPC3 is also expressed on various embryonic mesodermal cells.35,36 Loss of GPC3 causes a number of growth factor–dependent defects, as it serves as coreceptor for growth factors, including transforming growth factor family members,37,38 fibroblast growth factors,39 Wnts40 among others. We here demonstrate that GPC3 and CD26 are coexpressed on HSPCs, and as in hepatocarcinoma cells, GPC3 inactivates CD26 activity, whereas this was not the case for another glypican GPC1. Engraftment of LTR HSPCs from GPC3−/− mice was significantly impaired, whereas this was not the case for GPC1−/− BM. This may in part be explained by the fact that homing of GPC3−/− HSPCs is decreased because of higher CD26 activity, and in part because the number primitive HSCs in the BM is decreased, whereas the frequency of circulating HSPCs in GPC3−/− mice was significantly higher than the WT mice. All of these findings indicate poor HSC retention/maintenance potential of GPC3−/− BM consistent with higher CD26 activity that weakens the CXCL12-CXCR4 chemotactic axis. Therefore, our studies demonstrate for the first time a role for GPC3 in maintenance, engraftment, and mobilization of HSPCs (Figure 7). As GPC3 is involved in various cellular processes as well as regulating the local concentration of various growth factors, we cannot rule out the involvement of mechanisms other than alteration of CD26 activity in these observations.
Model representing the mechanism of TFPI action on HSCs. (A) Higher CD26 activity in GPC3−/− BM leads to lower maintenance of HSPCs, which leads to their entry into cell cycle and mobilization leading to increased circulating HSPCs. (B) TFPI through binding with GPC3 inhibits CD26 activity, enabling HSPCs to respond to the concentration gradient of CXCL12 thereby affecting HSPC homing into the BM positively.
Model representing the mechanism of TFPI action on HSCs. (A) Higher CD26 activity in GPC3−/− BM leads to lower maintenance of HSPCs, which leads to their entry into cell cycle and mobilization leading to increased circulating HSPCs. (B) TFPI through binding with GPC3 inhibits CD26 activity, enabling HSPCs to respond to the concentration gradient of CXCL12 thereby affecting HSPC homing into the BM positively.
The second, perhaps even more surprising, observation was that TFPI, heretofore only known to play a role in blood coagulation, is a natural ligand that enhances GPC3-mediated inhibition of CD26. We demonstrated that culture of HSPCs with TFPI enhances homing and engraftment. As only a single other report has demonstrated that TFPI may be able to bind to GPC3, we initially attempted to identify the typical receptor for TFPI, namely TF, on HSPCs. HSPCs did not show any expression of TF. However, we could conclusively demonstrate using FACS, immunostaining, and Co-IP that TFPI directly binds to GPC3 on HSPCs. TFPI affected the activity of CD26 in WT and GPC1−/− HSPCs, but not in GPC3−/− HSPCs, demonstrating that the effect of TFPI on CD26 activity was mediated by interaction of TFPI with GPC3 specifically. Inactivation of CD26 in response to TFPI lead to significantly improved migration, homing, as well as long-term repopulation of HSPCs. This effect was seen as early as 3 hours following incubation of HSPCs with TFPI, and persisted for at least 5 days of HSPC culture. These results might not be solely due to alterations of the CXCL12-CXCR4 axis as a recent report by Broxmeyer et al41 showed that CD26 can alter the activity of multiple other cytokines important in adult hematopoiesis. TFPI is expressed in endothelial cells of various organs and its expression as well as release into the circulation is regulated by physiological status of the cells.42 Under homeostatic conditions, we found low-level expression of TFPI in endothelial cells of the BM, which did not change significantly following radiation injury, pointing toward little physiological relevance of TFPI in adult hematopoiesis in vivo. During development, placenta has been reported as the major site of TFPI expression,43 so it might be insightful to examine its role in developmental hematopoiesis.
In conclusion, we show that TFPI inhibits CD26 via GPC3, resulting in better homing and long-term reconstitution. Loss of GPC3 was associated with decreased numbers of primitive HSCs in the BM and also resulted in increased numbers of circulating hematopoietic progenitors. Hence, this pathway is not only important in adult HSC maintenance in the BM but could also be used to manipulate HSCs in vitro to achieve better homing and engraftment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Jorge Filmus for providing GPC3−/− mice, Prof Guido David for providing GPC1−/− mice, Vik Vanduppen for excellent assistance with FACS sorting, and Rangarajan Sambathkumar for critical review for accurateness of the manuscript.
This work was supported by an Fonds Wetenschappelijk Onderzoek – Vlaanderen (FWO) grant (1.2.665.11.N.00) (S.K.), FWO funding (G085111N), National Institutes of Health NIH-PO1-CA-65493-06, Odysseus funding, Centre of Excellence and Geconcerteerde Onderzoeksacties/11/012 funding from KU Leuven, and the Vanwayenberghe fund (C.M.V.).
Authorship
Contribution: S.K. designed and performed the experiments, analyzed the data, and wrote the manuscript; L.M. bred GPC3−/− mice, genotyped, and performed in vitro assays; C.J. performed flow cytometry staining and in vitro assays with human UCB cells; S.S. performed FACS analyses and some of the engraftment studies; S.M.B. performed some chimerism experiments; and C.M.V. supervised all the research and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.M.B. is Howard Hughes Medical Institute and Department of Pathology, New York University School of Medicine, New York, NY.
Correspondence: Satish Khurana,Stem Cell Institute, KU Leuven, O&N4, Bus 804, Herestraat 49, 3000 Leuven, Belgium; e-mail: satish.khurana@med.kuleuven.be; and Catherine M. Verfaillie, Stem Cell Institute, K.U.Leuven, O&N4, Bus 804, Herestraat 49, 3000 Leuven, Belgium; e-mail: catherine.verfaillie@med.kuleuven.be.
References
Author notes
L.M. and C.J. contributed equally to this work.