Key Points
Brain ECs possess a functional TLR3/RIG-I system that is able to mount an effective IFN induction upon immune activation.
Brain ECs may be a key regulatory bystander, playing a crucial role in the BBB innate immunity against HIV infection.
Abstract
There is limited information about the role of blood-brain barrier (BBB) endothelial cells (ECs) in the central nervous system (CNS) and their innate immunity against HIV. We examined whether brain ECs can be immunologically activated to produce antiviral factors that inhibit HIV replication in macrophages. Human brain microvascular ECs expressed functional toll-like receptor 3 (TLR3) that could be activated by polyinosinic-polycytidylic acid (PolyI:C), resulting in the induction of endogenous interferon-β (IFN-β) and IFN-λ. The TLR3 activation of ECs also induced the phosphorylation of interferon regulatory transcription factor 3 (IRF3) and IRF7, the key regulators of IFN signaling pathway. When supernatant (SN) of PolyI:C-activated EC cultures was applied to infected macrophage cultures, HIV replication was significantly suppressed. This SN action of ECs on HIV was mediated through both IFN-β and IFN-λ because antibodies to their receptors could neutralize the SN-mediated anti-HIV effect. The role of IFNs in EC-mediated anti-HIV activity is further supported by the observation that treatment with SN from EC cultures induced the expression of IFN-stimulated genes (ISGs: ISG56, OAS-1, and MxA) in macrophages. These observations indicate that brain microvascular ECs may be a key regulatory bystander, playing a crucial role in the BBB innate immunity against HIV infection.
Introduction
The blood-brain barrier (BBB), consisting mainly of brain microvascular endothelial cells (ECs), astrocytes, and pericytes, separates circulating blood from the brain extracellular fluid in the central nervous system (CNS).1 The BBB is critical in maintaining CNS homeostasis and regulating the neuronal microenvironment. The brain ECs are sealed together with tight junctions and form the major structural and functional element of the BBB, which plays a key role in physiological processes such as blood supply, nutrient delivery, metabolic homeostasis, and immune cell trafficking. In addition, ECs also actively participate in the immunologic processes of the BBB, including cytokine-mediated inflammatory reactions. The BBB ECs are involved in regulating the influx of immune cells into the brain and in modifying immunologic reactions within the CNS.2
ECs represent a highly restrictive population in terms of immune activation at the BBB where they encounter a number of stimuli and immune cells, including HIV-infected cells. HIV infection has been largely known to compromise the BBB integrity and increase BBB permeability. A potential involvement of ECs in HIV infection is suggested by several observations, showing that ECs could be the target of HIV.3,4 An early study showed that HIV infected human brain capillary ECs through a CD4/galactosylceramide-independent mechanism.3 HIV gene expression has been found in ECs of the brains of AIDS patients.4,5 However, these findings are controversial and are not supported by the studies of others.6 Nevertheless, exposure to HIV or to HIV-infected cells could have a profound effect on the immune and barrier functions of ECs, even without clear evidence of productive infection.7,8 Coculture of brain ECs with HIV-infected macrophages was found to induce a number of proinflammatory and interferon (IFN)-inducible genes in comparison with ECs exposed to uninfected cells.9
Toll-like receptors (TLRs) specifically recognize pathogen-associated molecular patterns and play a critical role in eliciting host innate defense responses to viral infections. TLR3 together with TLR7 and TLR9 constitutes a powerful system to detect genetic material of viruses, with TLR3 implicated in the recognition of viral double-stranded RNA (dsRNA), TLR7 of single-stranded RNA, and TLR9 of cytosine phosphate guanine DNA, respectively.10 Several reports have demonstrated the expression of TLRs on ECs, such as human umbilical vein ECs, coronary artery ECs, dermal ECs, intestinal microvasular ECs, and pulmonary ECs.11-14 A recent study demonstrated that human brain ECs expressed TLR2, TLR3, TLR4, and TLR6.15 The expression of TLR3 in ECs is of importance, because TLR3 has a crucial role in virus-mediated innate immune responses, inducing both type I and type III IFNs.16 In addition to TLR3, retinoic acid-inducible gene I (RIG-I) has been identified as an important mediator of antiviral immunity, because it can detect viral genomic RNA during negative-strand RNA virus infection17 and trigger a type I IFN-mediated immune protection against viral infections.18 Thus, activating TLRs and/or RIG-I in ECs may be beneficial for CNS protection. Although most studies have focused on the interactions between HIV and the CNS immune cells, there is little information about whether the brain ECs participate in the BBB innate immunity against HIV infection of the CNS. Specifically, there is a lack of published data on whether the brain ECs possess functional TLR3/RIG-I signaling pathways and produce anti-HIV factors. Therefore, this study examined whether the brain ECs have the ability to mount a TLR3/RIG-I-mediated innate immunity that is effective in controling HIV infection of human macrophages.
Methods
Reagents
Rat tail collagen type I was purchased from BD Biosciences (San Diego, CA), fetal bovine serum was obtained from Sigma-Aldrich (St. Louis, MO), and penicillin-streptomycin solution was from Lonza (Walkersville, MD). All culture plasticware was obtained from Corning (Corning, NY). Unless otherwise specified, all other culture reagents were purchased from Invitrogen (Carlsbad, CA).
Cell culture
The human brain endothelial capillary cell line hCMEC/D3 has recently been developed as an excellent widely used model for studies of the human BBB.1,19,20 The cells were maintained in complete growth medium composed of EBM-2 and growth supplements (Lonza). Cells were cultured in an incubator at 37°C with 5% CO2 and 100% humidity. Medium was changed every 3 days.
Stimulation of ECs with TLR and RIG-I ligands
hCMEC/D3 cells seeded on collagen-coated multiwell plates were stimulated with polyinosinic-polycytidylic acid (PolyI:C) (TLR3), CL264 (TLR7), ODN2216 (TLR9), 5′ppp dsRNA (RIG-I), or 5′ppp dsRNA control (InvivoGen, San Diego, CA). LyoVec-treated cells were used as a vehicle control. For disruption of TLR3 function, cells were pretreated with 100 nM Bafilomycin A1 (Bafi A1; Tocris Bioscience, Ellisville, MO) for 1 hour before PolyI:C stimulation. After 16 hours, the culture medium was removed and fresh medium was added (to remove the ligands that may have direct anti-HIV activity in macrophages).21 Culture supernatant (SN) was collected at 48 hours poststimulation for an IFN-β and IFN-λ enzyme-linked immunosorbent assay (ELISA) and used to treat macrophages.
Quantitative real-time PCR
Quantitative real-time polymerase chain reaction (PCR) was described previously.22 The oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, IA), and sequences will be available upon request. All values were expressed as the increase relative to the expression of glyceraldehyde phosphate dehydrogenase (GAPDH) messenger RNA (mRNA). RNA was extracted with Tri-Reagent (Sigma-Aldrich) according to the manufacturer’s protocol.
RIG-I siRNA transfection
hCMEC/D3 cells were transfected with 100 nM silencing RNA (siRNA) for human RIG-I (Qiagen) or negative control siRNA according to the manufacturer’s protocol. The target sequence of siRNA for RIG-I was 5′-ACGGATTAGCGACAAATTTAA-3′. The negative control siRNA was 5′-AATTCTCCGAACGTGTCACGT-3′. To confirm efficient gene silencing, we performed quantitative real-time PCR with extracted total RNA 48 hours after transfection.
Immunofluorescence assay
Cells were treated with 0.1 µg/mL PolyI:C Rhodamine. The culture medium was then removed 16 hours later. Forty-eighty hours posttreatment, cells were fixed and stained with antibody to tight junction marker ZO-1 (Invitrogen; mouse; 1:100). Cells were stained with Hoechst 33342 and observed under a fluorescence microscope (Olympus IX71).
Western blotting
After PolyI:C stimulation of hCMEC/D3 cells, nuclear lysates were prepared by using the Nuclear Extraction Kit from Panomics (Santa Clara, CA) according to the manufacturer’s instructions. Equal amounts of protein lysates (20 µg) were separated on 4% to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis precast gels and transferred to an Immunobiolon-P membrane (Millipore, Eschborn, Germany). The blots were incubated with primary antibodies in 2% nonfat milk in phosphate-buffered saline with 0.05% Tween 20 (PBST) overnight at 4°C (interferon regulatory transcription factor 3 [IRF3], 1:2000; IRF7, 1:2000; IFN-stimulated gene factor 3 [ISGF3]γp48, 1:2500; histone H4, 1:1000; phospho-Erk1/2, 1:4000; phospho-JNK, 1:4000; phospho-STAT3, 1:4000; STAT3, 1:4000; GAPDH, 1:2000). Horseradish peroxidase–conjugated anti-rabbit immunoglobulin G (IgG), anti-goat IgG, and anti-mouse IgG were diluted at 1:2000 to ∼1:8000 in 2% nonfat milk PBST. Blots were developed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific). Densitometric analysis was performed by using ImageJ 1.44 software (National Institutes of Health).
ELISA
IFN-β and IFN-λ protein levels in hCMEC/D3 culture supernatant were measured by using ELISA (IFN-β: Invitrogen; IFN-λ1: eBioscience, San Diego, CA; IFN-λ2: BioLegends, San Diego, CA). Assays were performed according to the manufacturer’s instructions.
Macrophage treatment and HIV infection
Primary macrophages were derived from monocytes obtained from the Center for AIDS Research at the University of Pennsylvania School of Medicine.23 Macrophages were pretreated for 24 hours with SN from hCMEC/D3 cultures collected at 48 hours poststimulation. HIV isolate Jago, which is a macrophage-tropic isolate derived from cell-free cerebral spinal fluid from a patient with confirmed HIV-associated dementia24 (provided by Dr. Dennis Kolson, University of Pennsylvania) was used to infect macrophages. Briefly, 10 µL of Jago stock prepared in primary macrophages derived from the blood of healthy volunteers (HIV reverse transcriptase activity is 52 219 cpm) was added into a 48-well plate. After overnight incubation, the medium was removed and cells were washed 3 times with fresh Dulbecco’s modified Eagle medium. During the postinfection period, hCMEC/D3 SN was added to the macrophages where appropriate. At days 4, 8, 12, and 16 postinfection, half the medium was collected for detection of HIV reverse transcriptase activity.
HIV reverse transcriptase assay
Data analysis
Data were expressed as the mean ± standard deviation from at least 3 independent experiments, and statistical significance was measured by Student t test or one-way analysis of variance followed by the Newman-Keul test where appropriate. Statistical significance was defined as P < .05.
Results
PolyI:C-induced IFN expression in ECs
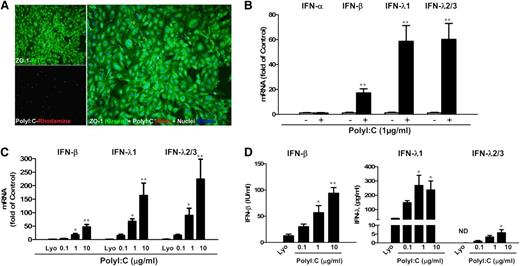
We first examined the expression of TLRs in human brain ECs (hCMEC/D3 and BB19). Both hCMEC/D3 cells and BB19 cells expressed TLR1, 2, 3, 4, and 6, with TLR4 being expressed at the highest level (supplemental Figure 1). We then investigated the effect of the ligands for TLR3, TLR7, and TLR9 on IFN expression in the ECs (hCMEC/D3 and primary HBMEC; supplemental Materials and Methods). Among the 3 ligands examined, only PolyI:C stimulation induced IFN expression in these ECs (supplemental Figures 2 and 3). As shown in Figure 1A, there was internalization of PolyI:C in hCMEC/D3 cells, which resulted in the induction of IFN-β and IFN-λ at both mRNA (Figure 1B) and protein (Figure 1D) levels. These effects of PolyI:C stimulation on IFN-β and IFN-λ expression in the ECs were dose-dependent (Figure 1C-D; supplemental Figure 4).
TLR3 activation induced IFN-β and IFN-λ expression. (A) hCMEC/D3 cells were stimulated with 0.1 µg/mL PolyI:C-Rhodamine for 16 hours and cultured for 48 hours poststimulation. Cells were fixed and stained with antibody to ZO-1 tight junction protein (mouse; 1:100; green). After nuclear counterstaining with Hoechst dyes, cells were observed under a fluorescence microscope. White arrows indicate the internalization of PolyI:C (magnification ×100). (B) Effect of PolyI:C on IFN expression of brain ECs. RNA extracted from the cells was subjected to real-time polymerase chain reaction (RT-PCR) for IFNs as indicated. (C,D) Dose-dependent effect of PolyI:C on IFN induction of hCMEC/D3 cells at (C) mRNA and (D) protein levels. Data were the mean ± standard deviation (SD) of 3 independent experiments. Asterisks indicate that the differences between the indicated groups are statistically significant (*P < .05; **P < .01). FITC, fluorescein isothiocyanate; Lyo, LyoVec (transfection reagent).
TLR3 activation induced IFN-β and IFN-λ expression. (A) hCMEC/D3 cells were stimulated with 0.1 µg/mL PolyI:C-Rhodamine for 16 hours and cultured for 48 hours poststimulation. Cells were fixed and stained with antibody to ZO-1 tight junction protein (mouse; 1:100; green). After nuclear counterstaining with Hoechst dyes, cells were observed under a fluorescence microscope. White arrows indicate the internalization of PolyI:C (magnification ×100). (B) Effect of PolyI:C on IFN expression of brain ECs. RNA extracted from the cells was subjected to real-time polymerase chain reaction (RT-PCR) for IFNs as indicated. (C,D) Dose-dependent effect of PolyI:C on IFN induction of hCMEC/D3 cells at (C) mRNA and (D) protein levels. Data were the mean ± standard deviation (SD) of 3 independent experiments. Asterisks indicate that the differences between the indicated groups are statistically significant (*P < .05; **P < .01). FITC, fluorescein isothiocyanate; Lyo, LyoVec (transfection reagent).
The role of TLR3 and RIG-I in PolyI:C-induced IFN expression
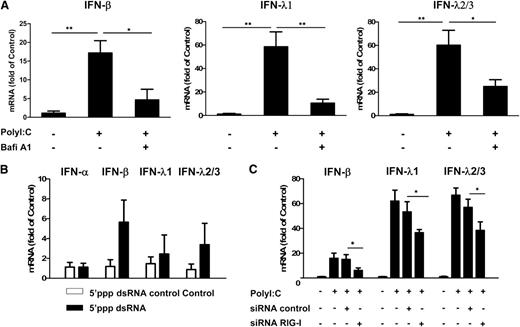
We next examined the role of TLR3 in PolyI:C-stimulated IFN expression. The pretreatment of hCMEC/D3 cells with Bafi A1, a strong inhibitor of the vacuolar type H(+)-ATPase that can disrupt the TLR3 function of cells, could largely but not completely block PolyI:C-induced expression of IFN-β and IFN-λ (Figure 2A). We then examined whether RIG-I, which also recognizes dsRNA, can induce IFNs in ECs. As shown in Figure 2B, stimulation of hCMEC/D3 cells with RIG-I ligand 5′ppp dsRNA also upregulated IFN-β and IFN-λ expression. In addition, PolyI:C-induced IFN-β and IFN-λ expression could be compromised by RIG-I siRNA, although control siRNA had little effect (Figure 2C).
Role of TLR3 and RIG-I in PolyI:C-mediated IFN induction. (A) Effect of disruption of TLR3 function by Bafi A1 on the induction of IFN-β and IFN-λ by PolyI:C stimulation. hCMEC/D3 cells were pretreated with 100 nM Bafi A1 for 1 hour and then stimulated with 1 µg/mL PolyI:C for 16 hours. (B) hCMEC/D3 cells were stimulated with 1 µg/mL 5′ppp dsRNA or 5′ppp dsRNA control for 16 hours, washed with plain medium 3 times, and then cultured for 48 hours poststimulation. (C) hCMEC/D3 cells were pretransfected with RIG-I siRNA or control siRNA for 24 hours prior to stimulation with 1 µg/mL PolyI:C. RNA was extracted and IFN expression was measured by quantitative real-time polymerase chain reaction (qRT-PCR). Data were the mean ± SD of 3 independent experiments. Asterisks indicate that the differences between the indicated groups are statistically significant (*P < .05; **P < .01).
Role of TLR3 and RIG-I in PolyI:C-mediated IFN induction. (A) Effect of disruption of TLR3 function by Bafi A1 on the induction of IFN-β and IFN-λ by PolyI:C stimulation. hCMEC/D3 cells were pretreated with 100 nM Bafi A1 for 1 hour and then stimulated with 1 µg/mL PolyI:C for 16 hours. (B) hCMEC/D3 cells were stimulated with 1 µg/mL 5′ppp dsRNA or 5′ppp dsRNA control for 16 hours, washed with plain medium 3 times, and then cultured for 48 hours poststimulation. (C) hCMEC/D3 cells were pretransfected with RIG-I siRNA or control siRNA for 24 hours prior to stimulation with 1 µg/mL PolyI:C. RNA was extracted and IFN expression was measured by quantitative real-time polymerase chain reaction (qRT-PCR). Data were the mean ± SD of 3 independent experiments. Asterisks indicate that the differences between the indicated groups are statistically significant (*P < .05; **P < .01).
PolyI:C-induced nuclear translocation of IRF3 and IRF7
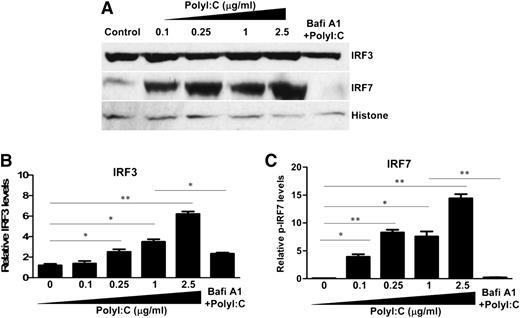
We next examined the effect of PolyI:C on the expression of IRF3 and IRF7, the key regulators of IFN expression. Although PolyI:C stimulation had no effect on the mRNA expression of IRF3 and IRF7 (data not shown), PolyI:C induced the nuclear translocation of IRF3 and IRF7 at posttranscriptional levels (Figure 3). Bafi A1 pretreatment of hCMEC/D3 cells significantly reduced PolyI:C-mediated nuclear translocation of IRF3 and IRF7 (Figure 3).
Effect of TLR3 activation on the nuclear translocation of IRF3 and IRF7. (A) hCMEC/D3 cells were pretreated with 100 nM Bafi A1 for 1 hour and then stimulated with the indicated concentrations of PolyI:C. Nuclear lysates were prepared, and expression of IRF3 and IRF7 was examined by immunoblot analysis. (B,C) Densitometry analysis of relative IRF3 and IRF7 levels compared with histone was performed by using ImageJ 1.44 software. The data are the average of at least 3 independent experiments and are expressed as mean ± SD. Asterisks indicate that the differences are statistically significant (*P < .05; **P < .01).
Effect of TLR3 activation on the nuclear translocation of IRF3 and IRF7. (A) hCMEC/D3 cells were pretreated with 100 nM Bafi A1 for 1 hour and then stimulated with the indicated concentrations of PolyI:C. Nuclear lysates were prepared, and expression of IRF3 and IRF7 was examined by immunoblot analysis. (B,C) Densitometry analysis of relative IRF3 and IRF7 levels compared with histone was performed by using ImageJ 1.44 software. The data are the average of at least 3 independent experiments and are expressed as mean ± SD. Asterisks indicate that the differences are statistically significant (*P < .05; **P < .01).
PolyI:C-stimulated hCMEC/D3 culture SN-inhibited HIV infection of macrophages
Brain ECs are the first line of brain cells to come into contact with circulating HIV-infected immune cells. We thus investigated the functional significance of TLR3 activation of brain ECs in the context of HIV infection of macrophages. As shown in Figure 4A, HIV Jago infection caused syncytia in macrophage cultures. In contrast, pretreatment of macrophages with SN from PolyI:C-stimulated hCMEC/D3 cultures greatly reduced HIV-induced syncytia in macrophages (Figure 4A). In addition, HIV reverse transcriptase activity and GAG gene expression were suppressed in macrophages pretreated with SN from PolyI:C-stimulated hCMEC/D3 cultures (Figure 4B-C). This inhibitory effect on HIV was dose dependent on the concentrations of PolyI:C used to activate hCMEC/D3 cells (Figure 4B-C) or on the percentage of SN added to macrophage cultures (Figure 4D).
Effect of SN from hCMEC/D3 cultures on HIV replication in macrophages. Macrophages were derived from human primary monocytes after 7-day culture and then pretreated with SN from control (Control/SN), LyoVec (LyoVec/SN) or PolyI:C (PolyI:C/SN) -stimulated hCMEC/D3 cultures for 24 hours. Macrophages were then infected with HIV Jago for overnight incubation, and after washing 3 times, cells were maintained in fresh medium for 8 days postinfection. (A) Morphologic observation of HIV-infected macrophages with no pretreatment, LyoVec/SN, or PolyI:C/SN pretreatment under phase contrast microscope (arrows indicate syncytium). (B,C) Intracellular HIV GAG gene expression in macrophages (B) or extracellular HIV reverse transcriptase (RT) activity in macrophage cultures (C) with 10% (volume to volume ratio [v/v]) of indicated SN pretreatments. For PolyI:C/SN, the SNs were collected from 0.1, 0.25, 1, or 2.5 μg/mL PolyI:C-stimulated hCMEC/D3 cultures. (D) HIV GAG gene expression at 8 days postinfection in macrophages pretreated with indicated volumes of 1 μg/mL PolyI:C-stimulated hCMEC/D3 cultures for 24 hours. Representative data from at least 5 donor macrophages were shown and were expressed as mean ± SD from triplicate wells. Asterisks indicate that the differences between indicated groups are statistically significant (*P < .05; **P < .01). GAPDH, glyceraldehyde phosphate dehydrogenase.
Effect of SN from hCMEC/D3 cultures on HIV replication in macrophages. Macrophages were derived from human primary monocytes after 7-day culture and then pretreated with SN from control (Control/SN), LyoVec (LyoVec/SN) or PolyI:C (PolyI:C/SN) -stimulated hCMEC/D3 cultures for 24 hours. Macrophages were then infected with HIV Jago for overnight incubation, and after washing 3 times, cells were maintained in fresh medium for 8 days postinfection. (A) Morphologic observation of HIV-infected macrophages with no pretreatment, LyoVec/SN, or PolyI:C/SN pretreatment under phase contrast microscope (arrows indicate syncytium). (B,C) Intracellular HIV GAG gene expression in macrophages (B) or extracellular HIV reverse transcriptase (RT) activity in macrophage cultures (C) with 10% (volume to volume ratio [v/v]) of indicated SN pretreatments. For PolyI:C/SN, the SNs were collected from 0.1, 0.25, 1, or 2.5 μg/mL PolyI:C-stimulated hCMEC/D3 cultures. (D) HIV GAG gene expression at 8 days postinfection in macrophages pretreated with indicated volumes of 1 μg/mL PolyI:C-stimulated hCMEC/D3 cultures for 24 hours. Representative data from at least 5 donor macrophages were shown and were expressed as mean ± SD from triplicate wells. Asterisks indicate that the differences between indicated groups are statistically significant (*P < .05; **P < .01). GAPDH, glyceraldehyde phosphate dehydrogenase.
IFN-β and IFN-λ are the key anti-HIV factors
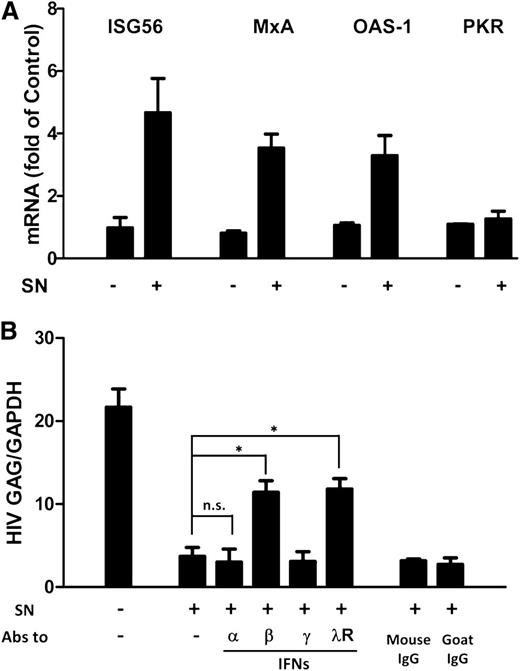
ISGs are a class of antiviral factors induced by IFNs. We next investigated whether ISGs were induced by PolyI:C-stimulated hCMEC/D3 cultures SN in macrophages. As shown in Figure 5A, the expression of ISG56, MxA, and OAS-1 was upregulated by the PolyI:C-stimulated hCMEC/D3 culture SN. To determine which IFN was responsible for activated hCMEC/D3 cell-mediated anti-HIV activity, we used neutralization antibodies against IFN-α, IFN-β or IFN-γ to pretreat the SN or antibody against IFN-λ receptor to pretreat macrophages, respectively. As shown in Figure 5B, preincubation of SN from PolyI:C-stimulated hCMEC/D3 cell cultures with antibody against IFN-α and IFN-γ had little effect on anti-HIV activity. In contrast, antibody to IFN-β significantly attenuated the anti-HIV activity of SN from PolyI:C-activated hCMEC/D3 cultures (Figure 5B). In addition, anti–interleukin-10 receptor β (IL-10Rβ) antibody pretreatment of macrophages significantly blunted the anti-HIV activity of the SN (Figure 5B).
Role of IFN-β and IFN-λ in PolyI:C-mediated anti-HIV activity. (A) Effect of PolyI:C-stimulated hCMEC/D3 culture SN on ISG expression of macrophages. HCMEC/D3 cells were stimulated with 1 µg/mL PolyI:C for 16 hours. Culture SN was collected for treatment of macrophages (10% v/v) for 6 hours. RNA was extracted, and the expression of ISGs (ISG56, MxA, OAS-1, and PKR) was measured by qRT-PCR. The results shown are mean ± SD of triplicate cultures, representative of 3 separate experiments. (B) Effect of neutralization antibodies (Abs) to IFNs or IFN-λ receptor on PolyI:C-stimulated hCMEC/D3 culture SN-mediated anti-HIV activity. PolyI:C-stimulated hCMEC/D3 culture SN was preincubated with anti-IFN-α (10 µg/mL), anti-IFN-β (10 µg/mL), or anti-IFN-γ (10 µg/mL) for 1 hour and then used to treat macrophages 24 hours prior to HIV Jago infection. For IFN-λ receptor pretreatment, the anti–IL-10Rβ neutralization antibody (10 µg/mL) was added to treat macrophages for 1 hour prior to the addition of SN. Eight days postinfection, RNA was extracted, and HIV GAG expression was measured by qRT-PCR. The results shown are mean ± SD of triplicate separate experiments. Asterisks indicate that the differences between the indicated groups are statistically significant (*P < .05; **P < .01). n.s., not significant.
Role of IFN-β and IFN-λ in PolyI:C-mediated anti-HIV activity. (A) Effect of PolyI:C-stimulated hCMEC/D3 culture SN on ISG expression of macrophages. HCMEC/D3 cells were stimulated with 1 µg/mL PolyI:C for 16 hours. Culture SN was collected for treatment of macrophages (10% v/v) for 6 hours. RNA was extracted, and the expression of ISGs (ISG56, MxA, OAS-1, and PKR) was measured by qRT-PCR. The results shown are mean ± SD of triplicate cultures, representative of 3 separate experiments. (B) Effect of neutralization antibodies (Abs) to IFNs or IFN-λ receptor on PolyI:C-stimulated hCMEC/D3 culture SN-mediated anti-HIV activity. PolyI:C-stimulated hCMEC/D3 culture SN was preincubated with anti-IFN-α (10 µg/mL), anti-IFN-β (10 µg/mL), or anti-IFN-γ (10 µg/mL) for 1 hour and then used to treat macrophages 24 hours prior to HIV Jago infection. For IFN-λ receptor pretreatment, the anti–IL-10Rβ neutralization antibody (10 µg/mL) was added to treat macrophages for 1 hour prior to the addition of SN. Eight days postinfection, RNA was extracted, and HIV GAG expression was measured by qRT-PCR. The results shown are mean ± SD of triplicate separate experiments. Asterisks indicate that the differences between the indicated groups are statistically significant (*P < .05; **P < .01). n.s., not significant.
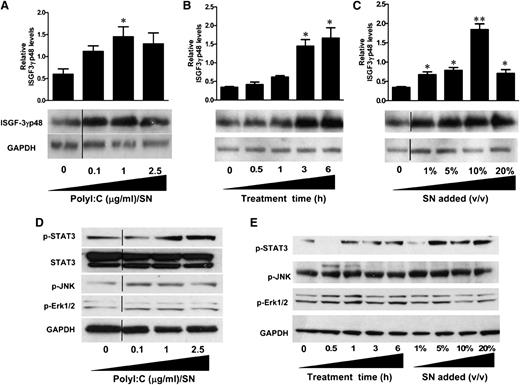
Induction of ISGF3 expression by hCMEC/D3 SN-activated macrophages
Because IFN-β and IFN-λ were found to be the key anti-HIV factors in PolyI:C-stimulated hCMEC/D3 culture SN, we next examined the effect on ISGF3 expression. As shown in Figure 6A, treatment of macrophages with PolyI:C-stimulated hCMEC/D3 culture SN significantly upregulated ISGF3 expression and exhibited dose dependence on the concentrations of PolyI:C (0.1, 1, and 2.5 µg/mL). The upregulation of ISGF3 by 1 µg/mL PolyI:C-stimulated hCMEC/D3 SN was also increased with the time period of SN treatment (0.5, 1, 3, and 6 hours) (Figure 6B). Additionally, ISGF3 induction was proportional to the dose of SN (1%, 5%, 10%, and 20%) used to treat macrophages (Figure 6C).
Effect of PolyI:C-stimulated hCMEC/D3 culture SN on the activation of ISGF3, STAT3, and MAPK in macrophages. (A) hCMEC/D3 cells were stimulated with 0.1, 1, and 2.5 µg/mL PolyI:C for 16 hours, then washed 3 times, and further incubated in fresh medium to 48 hours poststimulation. SN was collected to treat macrophages for 6 hours (10% v/v). (B) Macrophages were treated with 10% (v/v) of SN from 1 µg/mL PolyI:C-stimulated hCMEC/D3 culture for the indicated time periods. (C) Macrophages were treated with indicated percentage (v/v) of SN from 1 µg/mL PolyI:C-stimulated hCMEC/D3 culture for 6 hours. Protein was extracted and western blot was performed to examine the expression of ISGF-3γp48. Representative blots from 3 independent experiments ae shown. Densitometry analysis of the blot was performed by using ImageJ 1.44 software (National Institutes of Health) and plotted into graphs using data collected from triplicate experiments. Asterisks indicate that the differences between the SN-treated and control cells are statistically significant (*P < .05; **P < .01). (D) Macrophages were treated with 10% (v/v) of SN from indicated concentrations of PolyI:C stimulated hCMEC/D3 cultures for 6 hours. (E) Macrophages were treated with 10% (v/v) of SN from 1 μg/ml PolyI:C-stimulated hCMEC/D3 culture for indicated time points or with indicated percentage (v/v) of SN from 1 μg/ml PolyI:C stimulated hCMEC/D3 cutlure for 6 hours. Protein was extracted after treatment and western blot was performed to examine the expression of STAT3, p-STAT3, p-JNK, p-Erk1/2 amd GAPDH. Representative data from three times independent experiments was shown. Vertical lines have been inserted to indicate repositioned blot lanes.
Effect of PolyI:C-stimulated hCMEC/D3 culture SN on the activation of ISGF3, STAT3, and MAPK in macrophages. (A) hCMEC/D3 cells were stimulated with 0.1, 1, and 2.5 µg/mL PolyI:C for 16 hours, then washed 3 times, and further incubated in fresh medium to 48 hours poststimulation. SN was collected to treat macrophages for 6 hours (10% v/v). (B) Macrophages were treated with 10% (v/v) of SN from 1 µg/mL PolyI:C-stimulated hCMEC/D3 culture for the indicated time periods. (C) Macrophages were treated with indicated percentage (v/v) of SN from 1 µg/mL PolyI:C-stimulated hCMEC/D3 culture for 6 hours. Protein was extracted and western blot was performed to examine the expression of ISGF-3γp48. Representative blots from 3 independent experiments ae shown. Densitometry analysis of the blot was performed by using ImageJ 1.44 software (National Institutes of Health) and plotted into graphs using data collected from triplicate experiments. Asterisks indicate that the differences between the SN-treated and control cells are statistically significant (*P < .05; **P < .01). (D) Macrophages were treated with 10% (v/v) of SN from indicated concentrations of PolyI:C stimulated hCMEC/D3 cultures for 6 hours. (E) Macrophages were treated with 10% (v/v) of SN from 1 μg/ml PolyI:C-stimulated hCMEC/D3 culture for indicated time points or with indicated percentage (v/v) of SN from 1 μg/ml PolyI:C stimulated hCMEC/D3 cutlure for 6 hours. Protein was extracted after treatment and western blot was performed to examine the expression of STAT3, p-STAT3, p-JNK, p-Erk1/2 amd GAPDH. Representative data from three times independent experiments was shown. Vertical lines have been inserted to indicate repositioned blot lanes.
Activation of STAT3 and MAPK in macrophages
IFN-β and IFN-λ are the major factors responsible for the anti-HIV activity of the PolyI:C-stimulated hCMEC/D3 SN. These two types of IFNs have been shown to induce similar responses despite the distinct receptor systems. STAT3 and MAPK, including Erk1/2 and JNK, are important regulators of ISG expression. As shown in Figure 6D, TLR3 activation of hCMEC/D3 cells by PolyI:C induced the phosphorylation of STAT3 as well as MAPK kinases Erk1/2 and JNK. This induction was in a dose-dependent manner on the PolyI:C used to stimulate hCMEC/D3 cells (Figure 6D). Moreover, SN collected from PolyI:C-stimulated hCMEC/D3 cultures also exhibit time- (0.5, 1, 3, and 6 hours) and dose-dependent (1%, 5%, 10%, and 20%) induction of STAT3, Erk1/2, and JNK phosphorylations (Figure 6E).
Discussion
The BBB is well-known for its role as a physical barrier in separating blood and the brain and in controlling the trafficking of HIV and/or HIV-infected cells into the CNS. In the BBB, ECs are the first line of cells that come in contact with HIV or HIV-infected cells. Although it is unclear whether ECs play a role in protecting the CNS from HIV infection, the brain ECs display distinct antimicrobial properties, including antiviral responses.26 We demonstrated, for the first time, that human brain ECs express functional TLR3 that could be activated, which results in the induction of both IFN-β and IFN-λ. Interestingly, TLR7 and TLR9 stimulation with their ligands had little effect on IFN induction (supplemental Figures 2 and 3), suggesting that TLR3 is a key player in the immunologic activation of human brain ECs. The lack of TLR7 and TLR9 activation in the brain ECs could be due to the fact that these cells do not express the receptors for these TLRs. A recent study demonstrated that human cerebral ECs express TLR3 but not TLR7 and TLR9.15 We found that the brain ECs had detectable expression of TLR1, 2, 3, 4, and 6, but very low to undetectable levels of TLR5, 7, 8, 9, and 10 (supplemental Figure 1). In addition to TLR3, the RIG-I signaling of the ECs also induced the expression of IFN-β and IFN-λ. Our further investigation of the mechanisms for the induction of IFNs by ECs indicated that there was an upregulation of IRF3 and IRF7 in the ECs activated by PolyI:C.
Given the antiviral activity of IFNs in the innate host immunity, we examined whether PolyI:C-activated brain ECs exhibit anti-HIV activity in macrophages. We found that PolyI:C-mediated immune activation of ECs resulted in a potent anti-HIV effect in infected macrophages, which was evidenced by the observation that SN from TLR3-activated EC cultures could inhibit HIV replication in macrophages. IFN-β and IFN-λ in SN appeared to be responsible for the anti-HIV effect, because the antibodies to IFN-β and IFN-λ receptors could block the inhibitory effect of EC SN on HIV. IFN-λ has been shown to have antiviral activities against a number of viruses, including HIV.27-31 IFN-λ expression has been identified in a tissue-dependent fashion,32,33 and cells that respond to IFN-λ closely depend on the expression of the IL-28Rα subunit.34,35 We found that hCMEC/D3 cells express a nondetectable mRNA level of IL-28Rα (supplemental Figure 6), suggesting that TLR3 signaling of brain EC-induced IFN-λ had little autocrine action. In addition, it was reported that hCMEC/D3 cells were nonresponsive to IFN-α treatment, because IFN-α could not activate STAT1 or STAT3.36 Although the mechanism(s) for the lack of the autocrine action of IFN-α and IFN-λ from the activated brain ECs remain to be determined, it is likely that IFNs produced by the ECs can bind to their receptors in macrophages (supplemental Figure 6) and induce ISGF3 expression, phosphorylation of STAT3, and activation of MAPK pathway, leading to the induction of anti-HIV ISGs, such as ISG56, MxA, and OAS-1.
To determine whether the TLR3-mediated antiviral effect is limited to the brain ECs, we also examined the effect of PolyI:C on IFN expression in human lymphatic microvascular endothelial cells derived from the lung (HMVEC-L). HMVEC-L has been reported to express multiple functional TLRs, including TLR3, and can be immunologically activated.37,38 We found that PolyI:C treatment induced IFN-β and IFN-λ expression in HMVEC-L cells (supplemental Figure 7B-C). In addition, SN from PolyI:C-stimulated HMVEC-L cultures also significantly inhibited HIV replication in primary macrophages (supplemental Figure 7D-E). Given the importance of the brain and lung ECs in protecting the encapsulated tissue, the fact that TLR3 signaling of these cells could induce the production of IFNs is highly significant.
It is well known that plasmacytoid dendritic cells and monocytes/macrophages are the major producers of type I IFNs upon viral infections.39-41 However, their numbers and ability to produce type I IFNs are significantly reduced by HIV infection.42-46 Thus, an IFN-mediated antiviral response mounted by nonimmune cells such as brain ECs is likely to be beneficial for CNS protection. Although two early studies reported that brain ECs could be infected with HIV,3,47 our results showed that hCMEC/D3 cells were not susceptible to HIV infection (supplemental Figure 5), which is consistent with the majority of evidence.6 As a non-HIV target cell in the CNS, it is unlikely that the ability of ECs to mount an IFN-mediated anti-HIV response would be compromised by HIV infection. However, as the first line of cells in the BBB, the brain ECs have to encounter a number of stimuli and immune cells. Thus, the immune activation of these cells is inevitable, which leads to the activation of IFN signaling pathways, producing IFNs. We and others have shown that IFNs are produced not only by the immune cells but also by the nonimmune cells in the CNS, such as neurons and astrocytes.48-51 In contrast to PolyI:C induction of both IFN-α and IFN-β in neurons or astrocytes, TLR3 signaling of ECs induced only IFN-β expression. This finding is consistent with the report by Starace et al showing that PolyI:C induced IFN-β but not IFN-α in mouse Sertoli cells.48 These observations along with the findings in this study support the notion that ECs and other nonimmune cells in the CNS could act as important bystanders in mounting effective antiviral responses that restrict HIV infection and replication in the target cells in the CNS.
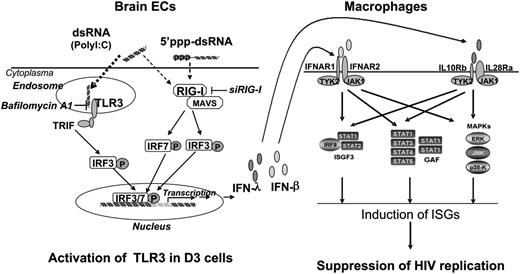
In summary, we have provided experimental evidence that human brain ECs possess a functional TLR3/RIG-I system that can mount an effective innate immune response and produce anti-HIV factors (Figure 7). Future animal and clinical studies are necessary to determine whether the BBB EC-mediated anti-HIV activity is beneficial for CNS protection. In addition, further research into the fundamental and immunologic functions of ECs is key to understanding the BBB innate immunity against viral infections. These studies will be critical not only for our basic understanding of EC-mediated control of HIV infection but also for the design and development of TLR3/RIG-I-based intervention and treatment strategies for neuroAIDS.
Hypothetic anti-HIV mechanism of TLR3 signaling of ECs. Stimulation of brain ECs with dsRNA activates TLR3 and/or RIG-I pathways, which facilitates phosphorylation and translocation of IRF3 and IRF7, initiating the transcription of IFN-β and IFN-λ in the ECs. When released from the ECs, IFN-β and IFN-λ bind to their receptors in macrophages and activate ISGF3, STAT3, and MAPK, inducing anti-HIV ISGs in macrophages. GAF, gamma-interferon activation factor; JAK1, Janus kinase 1; MAVS, mitochondrial antiviral-signaling protein; P, phosphorylation; TRIF, TIR-domain-containing adapter-inducing interferon-β; TYK2, Tyrosine kinase 2.
Hypothetic anti-HIV mechanism of TLR3 signaling of ECs. Stimulation of brain ECs with dsRNA activates TLR3 and/or RIG-I pathways, which facilitates phosphorylation and translocation of IRF3 and IRF7, initiating the transcription of IFN-β and IFN-λ in the ECs. When released from the ECs, IFN-β and IFN-λ bind to their receptors in macrophages and activate ISGF3, STAT3, and MAPK, inducing anti-HIV ISGs in macrophages. GAF, gamma-interferon activation factor; JAK1, Janus kinase 1; MAVS, mitochondrial antiviral-signaling protein; P, phosphorylation; TRIF, TIR-domain-containing adapter-inducing interferon-β; TYK2, Tyrosine kinase 2.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This investigation was supported by grants from the National Institutes of Health (DA012815, DA027550, DA022177) to W.H.
Authorship
Contribution: J.L. and W.H. conceived and designed the experiments; J.L., Y.W., X.W., L.Y., and Y.Z. performed the experiments; J.L., Y.W., and W.H. analyzed the data; X.W. and Y.P. contributed reagents, materials, and analysis tools; and J.L. and W.H. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wenzhe Ho, Department of Pathology and Laboratory Medicine, Temple University School of Medicine, 843 MERB, 3500 N Broad St, Philadelphia, PA 19140; e-mail: wenzheho@temple.edu.

![Figure 4. Effect of SN from hCMEC/D3 cultures on HIV replication in macrophages. Macrophages were derived from human primary monocytes after 7-day culture and then pretreated with SN from control (Control/SN), LyoVec (LyoVec/SN) or PolyI:C (PolyI:C/SN) -stimulated hCMEC/D3 cultures for 24 hours. Macrophages were then infected with HIV Jago for overnight incubation, and after washing 3 times, cells were maintained in fresh medium for 8 days postinfection. (A) Morphologic observation of HIV-infected macrophages with no pretreatment, LyoVec/SN, or PolyI:C/SN pretreatment under phase contrast microscope (arrows indicate syncytium). (B,C) Intracellular HIV GAG gene expression in macrophages (B) or extracellular HIV reverse transcriptase (RT) activity in macrophage cultures (C) with 10% (volume to volume ratio [v/v]) of indicated SN pretreatments. For PolyI:C/SN, the SNs were collected from 0.1, 0.25, 1, or 2.5 μg/mL PolyI:C-stimulated hCMEC/D3 cultures. (D) HIV GAG gene expression at 8 days postinfection in macrophages pretreated with indicated volumes of 1 μg/mL PolyI:C-stimulated hCMEC/D3 cultures for 24 hours. Representative data from at least 5 donor macrophages were shown and were expressed as mean ± SD from triplicate wells. Asterisks indicate that the differences between indicated groups are statistically significant (*P < .05; **P < .01). GAPDH, glyceraldehyde phosphate dehydrogenase.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/15/10.1182_blood-2012-08-450353/4/m_2934f4.jpeg?Expires=1769090626&Signature=pobH6ToYa88xpMgl~fACedzKnF1DZV7f8k81L9xImYAWoRAwDq2dumQEXwyQJ55suwmauVkXpKS0qbPvtrz3LKIM7rmI9MxKG8vbpZW0uH~SjB9bb3O36DFWbob1bRfLBLqxTJGpXepWEt0Jy2B9jrc2TQLXEh93fncv3G34gzwjaVyh0iq6aORXcqqBrkv9OQXzqf2GqPHqxJqMblvPdvg-~hEF~zd4lecOeOLvx-fxFC1FeTiqXFiTyMDV8NrOa6W8HTs4I7KIVMH1EHgPB5upD-l6IPms7k1I9kpvvvaHXdSlI917DjSRkeosqt03IEjKo64OYImUWrLof1f4hg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal