Key Points
Aryl hydrocarbon receptor (AhR) mediates the ERK-dependent maintenance of the immature state of monocyte-derived dendritic cells (MDDCs).
MEK-ERK regulates antigen capture, lymph node homing, and the acquisition of maturation-associated genes in MDDCs.
Abstract
Dendritic cells (DCs) promote tolerance or immunity depending on their maturation state, which is enhanced or accelerated upon MEK-ERK signaling pathway inhibition. We have determined the contribution of MEK-ERK activation to the profile of gene expression of human immature monocyte-derived dendritic cells (MDDCs) and peripheral blood myeloid DCs. ERK inhibition altered the expression of genes that mediate Chemokine (C-C motif) ligand 19 (CCL19)–directed migration (CCR7) and low-density lipoprotein (LDL) binding (CD36, SCARB1, OLR1, CXCL16) by immature DCs. In addition, ERK upregulated CCL2 expression while impairing the expression of DC maturation markers (RUNX3, ITGB7, IDO1). MEK-ERK–regulated genes exhibited an overrepresentation of cognate sequences for the aryl hydrocarbon receptor (AhR) transcription factor, whose transcriptional and DNA-binding activities increased in MDDCs upon exposure to the MEK1/2 inhibitor U0126. Therefore, the MEK-ERK signaling pathway regulates antigen capture, lymph node homing, and acquisition of maturation-associated genes, and its contribution to the maintenance of the immature state of MDDCs and myeloid DCs is partly dependent on the activity of AhR. Since pharmacologic modulation of the MEK-ERK signaling pathway has been proposed as a potential therapeutic strategy for cancer, our findings indicate that ERK inhibitors might influence antitumor responses through regulation of critical DC effector functions.
Introduction
Dendritic cells (DCs) are the essential link between innate and adaptive immune responses.1 In the steady-state, tissue-resident immature DCs capture extracellular material and contribute to peripheral tolerance.1,2 By contrast, upon detection of danger-associated molecular patterns by germ line–encoded pathogen-recognition receptors, immature DCs increase their expression of major histocompatibility complex (MHC) and costimulatory molecules and migrate toward peripheral lymph nodes in a chemokine receptor 7 (CCR7) –dependent manner.1,2 This maturation event endows DCs with the ability to promote the generation of primary immune responses.1 Thus, DC maturation constitutes an ideal target for modulation of immune responses in autoimmune diseases, transplant rejection, or cancer.2
Whereas the critical role of nuclear factor kappa B (NFκB) activation in DC maturation is firmly established, various signaling mechanisms function as modulators of the DC maturation process.3 ERK and p38MAPK signaling pathways have been shown to oppositely contribute to DC maturation.4,5 p38MAPK activation is a requisite for increased NFκB recruitment to the regulatory regions of genes whose expression is modulated during DC maturation, and its inhibition reduces costimulatory molecule expression, interleukin-12p70 (IL-12p70) production, and T-cell stimulatory ability.6 Conversely, the MEK-ERK signaling pathway prevents an optimal DC maturation.4 In response to maturation stimuli, inhibition of MEK-ERK activation results in higher expression of MHC-II, costimulatory and adhesion molecules (CD83, CD86, and CD49d),5 higher allostimulatory activity, elevated secretion of IL-12p70 and IL-12p40,5,7 and decreased proinflammatory cytokine production.4 These functions point to a negative regulatory role of the MEK-ERK signaling pathway on DC maturation.
DC maturation is strongly impaired in patients with tumors, in whom the low number and the immature state of DCs are responsible for cancer-associated immunosuppression.8 Tumor cells, myeloid-derived suppressor cells, and tumor-associated macrophages create a potent immunosuppressive environment that prevents DC maturation.8 In this regard, DCs in rapidly growing tumors usually exhibit an immature phenotype, and the majority of DCs remain immature within the tumors, whereas mature DCs reside in the surrounding areas.9 DC-based cancer immunotherapy is currently considered a means to overcome the immunosuppression that takes place in patients with tumors.10,11 Small-molecule inhibitors of ERK and p38MAPK signaling pathways are also being evaluated in clinical trials for the treatment of pathologic disorders.12-14 Therefore, on the basis of their effects on DC maturation, it is conceivable that pharmacologic MEK inhibitors might diminish the tumor-imposed halt on DC maturation, thus overcoming immunosuppression.
To determine the extent of the contribution of the MEK-ERK signaling pathway to the maturation state of DCs, we have determined the ERK-dependent gene expression profile in immature human monocyte-derived dendritic cells (MDDCs). Our results indicate that the MEK-ERK signaling axis regulates DC functions that are critical in the initiation and regulation of immune response, and they provide evidence for the involvement of the aryl hydrocarbon receptor (AhR) transcription factor in the ERK-dependent gene expression profile of immature MDDCs.
Methods
Dendritic cell generation and maturation
Human peripheral blood mononuclear cells were isolated from buffy coats from normal donors over a Lymphoprep (Nycomed Pharma, Oslo, Norway) gradient. Monocytes were purified from peripheral blood mononuclear cells by magnetic cell sorting using CD14 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). Monocytes (>95% CD14+ cells) were cultured at 0.7 × 106 cells/mL for 5 days in RPMI 1640 supplemented with 10% fetal calf serum (complete medium) at 37°C in a humidified atmosphere with 5% CO2 containing 1000 U/mL granulocyte macrophage colony-stimulating factor (GM-CSF) (ImmunoTools; Friesoythe, Germany) and 1000 U/mL IL-4 (ImmunoTools) to generate immature MDDCs, with cytokine addition every 2 days. Macrophages were generated following the same scheme but using 10 ng/mL M-CSF (ImmunoTools). For maturation, MDDCs were treated with 10 ng/mL lipopolysaccharide (LPS; E. coli 055:B5; Sigma). For gene profiling experiments, immature MDDCs were exposed to p38MAPK inhibitors SB203580 (13 μM; Enzo Life Sciences, Farmingdale, NY) and BIRB0796 (0.1 μM)15 or MEK inhibitors U0126 (2.5 μM) and PD98059 (40 μM; Calbiochem, Nottingham, UK).16,17 A final dose of GM-CSF and IL-4 was added to the culture 1 hour after treatment with inhibitors, and cells were collected after 4, 10, or 24 hours. Peripheral blood CD1c+ DCs were isolated by using the CD1c (BDCA-1)+ Dendritic Cell Isolation Kit (Miltenyi Biotec). Chemokine (C-C motif) ligand 2 (CCL2) was measured by using a commercially available enzyme-linked immunosorbent assay (BD OptEIATM Human MCP-1 ELISA Set; BD Bioscience).
Cell transfection and reporter gene assays
MDDCs (2 × 106 cells) were nucleofected with 10 μg of a constitutively active (MEK-A) or a double negative (MEK-E) form of MEK, provided by Dr Pura Muñoz-Canoves (Universidad Pompeu Fabra, Barcelona, Spain), using the Human Dendritic Cell Nucleofector Kit V (Lonza, Basel, Switzerland). As a control, MDDCs were nucleofected with pCDNA3.1 empty vector. After nucleofection, cells were cultured for 24 hours in complete medium containing GM-CSF and IL-4. In reporter gene assays, MDDCs (2 × 106 cells) or monocytes (107 cells) were nucleofected with 975 ng of a xenobiotic responsive element (XRE) –dependent construct (XRE-TATA-luc) and 25 ng of a construct expressing the Renilla luciferase from a constitutive promoter (Cignal XRE Reporter Kit; SABiosciences). Where indicated, cells were transfected in parallel with a negative control TATA-luc construct. After 24 hours, cells were treated for 12 hours with dimethylsulfoxide (DMSO), U0126 (2.5 μM), or tetrachlorodibenzo-p-dioxin (TCDD; 10 nM) and lysed. Firefly and Renilla luciferase activities were determined by using the Dual-Luciferase Reporter Assay System (Promega). HepG2 cells were transfected by using Superfect (Qiagen, Valencia, CA).
Gene expression profiling and quantitative reverse transcriptase polymerase chain reaction
RNA was isolated from three independent preparations of MDDCs exposed to either DMSO or U0126 for 4, 10, or 24 hours by using AllPrep DNA/RNA/Protein Mini Kit (Qiagen). Labeled RNA was used as a hybridization probe on Whole Human Genome Microarrays (Agilent Technologies, Palo Alto, CA). A threshold of 1 was assigned to raw signals and quantiles were normalized.18 After removing probes with low signals and questionable quality, a moderated t test, as implemented in the Bioconductor limma package,19 was used to calculate the P values for each gene comparing conditions. P values were adjusted by using Benjamini-Hochberg method to account for multiple testing. Adjusted P values smaller than .05 were considered significant. Gene ontology (GO) analysis of microarray results was performed by using the Web-based DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/home.jsp). Significantly changed genes were compared with a background set of the total complement of genes that were represented on the microarray. GO terms were considered significant if they had a Benjamini-corrected P value lower than .05. All microarray data have been deposited in the Gene Expression Omnibus under accession number GSE39745. For quantitative reverse transcriptase polymerase chain reaction, total RNA was retrotranscribed, and complementary DNA was quantified by using the Universal ProbeLibrary (Roche Diagnostics). Triplicate assays were performed, and results were normalized according to the expression levels of GAPDH RNA and expressed by using the ΔΔCT method for quantization.
Western blot
Affinity-purified rabbit monoclonal antibodies from Cell Signaling Technology (Danvers, MA) were used to detect ERK1/2, phospho-ERK1/2 (Thr202/Tyr204), p38MAPK, and phospho-p38MAPK (Thr180/Tyr182) (D3F9). A polyclonal antibody against AhR (N-19) and a monoclonal anti–glyceraldehyde phosphate dehydrogenase (GAPDH) antibody (6C5) were from Santa Cruz Biotechnology (Santa Cruz, CA), and the Ab-11 cocktail (Thermo-Scientific, Fremont, CA) was used to detect THBS1.
Low-density lymphocyte uptake assay
MDDCs from 5 independent donors were treated with DMSO or U0126 for 24 hours, washed twice with phosphate-buffered saline, and exposed to 1 μg/mL of acetylated low-density lymphocyte (LDL) -PE/Cy5 (acLDL; Invitrogen, Carlsbad, CA) in serum-free RPMI 1640 for 30 to 180 minutes at 37°C. After washing, acLDL uptake was evaluated by flow cytometry.
Chemotaxis assays
Chemotaxis in response to CCL19 was determined in MDDCs previously exposed to DMSO or U0126 for 24 hours; 1 × 105 cells were washed, diluted in 100 µL of RPMI 1640 medium containing 0.1% bovine serum albumin, and seeded in the upper chamber of a 5-μm pore size 24-well Transwell chamber (Costar; Cambridge, MA). The lower chamber contained 600 µL of the same medium supplemented or not with CCL19 (100 ng/mL). After 2.5 hours, MDDCs that migrated into the lower chamber were measured.
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed on MDDCs treated with either DMSO or U0126 for 10 hours by using the Magna ChIP G Chromatin Immunoprecipitation Kit (Millipore), according to the manufacturer’s recommendations. For PCR detection of the AhR-binding sequence within the CYP1B1 gene promoter, the oligonucleotides 5′-ATATGACTGGAGCCGACTTTCC-3′ and 5′- GGCGAACTTTATCGGGTTGA-3′ were used, which amplify the −790/−890 CYP1B1 promoter region. Five micrograms of an anti-AhR RPT9 antibody (Abcam) was used for immunoprecipitation, and an irrelevant immunoglobulin G-1 mouse monoclonal antibody was used as a negative control.
Results
Identification of genes regulated by MEK-ERK1/2 in immature human MDDCs
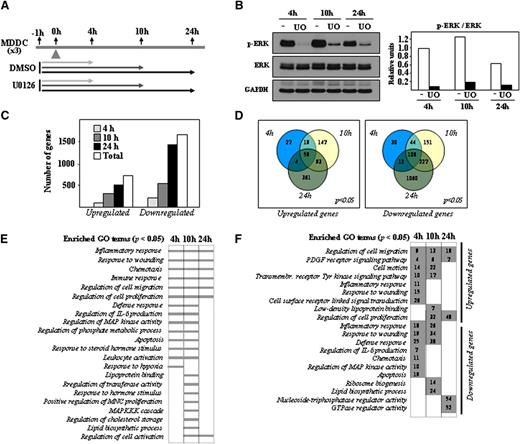
MEK-ERK and p38MAPK differentially contribute to the phenotype and effector functions of LPS-treated mature MDDCs.4,5 To determine the ERK-dependent gene expression profile in immature MDDCs, cells were exposed to the MEK1/2-specific inhibitor U0126, and gene expression changes were analyzed after 4, 10, or 24 hours by using DMSO-treated MDDCs as a control (Figure 1A). ERK1/2 phosphorylation was inhibited throughout the whole period of analysis, with no significant changes in the level of ERK1/2 protein (Figure 1B). Transcriptional profiling revealed a higher percentage of downregulated genes at all time points after U0126 treatment. After 4 hours in the presence of U0126, 199 genes were upregulated and 194 genes were downregulated (Figure 1C). These numbers increased to 318 and 530 after 10 hours and to 517 and 1407 after 24 hours (Figure 1C), whereas 59 genes were downregulated and 108 were upregulated at all time points after U0126 treatment (Figure 1D). Therefore, gene transcription in immature MDDCs is actively controlled by the MEK-ERK1/2 axis. Besides modulation of genes involved in “Regulation of MAP kinase activity”, GO terms significantly enriched at all time points after treatment with U0126 included “Inflammatory Response,” “Immune Response,” “Response to wounding,” “Regulation of cell migration,” and “Chemotaxis” (Figure 1E and supplementary Table 1), whereas “Response to hypoxia” and “Lipoprotein binding” terms were connected only to genes whose expression was modified by U0126 after 4 or 10 hours (Figure 1E and supplementary Tables 2 and 3). The “Lipoprotein binding” function was enriched among the genes upregulated by U0126 after 10 hours (Figure 1F and supplementary Table 3), whereas the “Lipid biosynthesis” process was exclusively enriched in genes downregulated by U0126 at the same time point (Figure 1F and supplementary Table 3). Supporting the GO analysis, the list of the genes whose expression was more profoundly affected by U0126 included numerous genes involved in immune/inflammatory responses, cell migration, and lipoprotein binding (Table 1). Importantly, U0126 treatment significantly reduced the expression of well-known targets of ERK (FOS, MYC, DUSP6). Therefore, inhibition of the MEK-ERK signaling axis in immature MDDCs alters the expression of genes that mediate critical effector functions of immature MDDCs as well as functions related to sterol and lipid metabolism.
Determination of the U0126-dependent transcriptomic profile of immature MDDCs. (A) Schematic representation of the profiling experiments, with indication of the treatment and time points for sample collection. (B) Effect of U0126 (UO) or DMSO (-) treatment on ERK1/2 phosphorylation in immature MDDCs, as determined by western blot. The level of total ERK1/2 and GAPDH protein were determined in parallel. The histogram illustrates the ratio p-ERK/ERK for each sample, as determined by densitometric analysis of the western blot. One of 3 representative experiments is presented. (C) Quantification of the number of genes whose expression is significantly altered (adjusted P < .05) at all time points (Total) and after the indicated duration of treatment with U0126. (D) Comparison of the number of genes whose expression is significantly (adjusted P < .05) upregulated (left panel) or downregulated (right panel) after the indicated time of exposure to U0126. (E) GO analysis for molecular functions significantly (adjusted P < .05) enriched within the sets of genes whose expression is altered by U0126 exposure at the indicated times. (F) Number of genes whose expression is significantly (adjusted P < .05) upregulated or downmodulated by U0126 at the indicated times and for the indicated GO terms.
Determination of the U0126-dependent transcriptomic profile of immature MDDCs. (A) Schematic representation of the profiling experiments, with indication of the treatment and time points for sample collection. (B) Effect of U0126 (UO) or DMSO (-) treatment on ERK1/2 phosphorylation in immature MDDCs, as determined by western blot. The level of total ERK1/2 and GAPDH protein were determined in parallel. The histogram illustrates the ratio p-ERK/ERK for each sample, as determined by densitometric analysis of the western blot. One of 3 representative experiments is presented. (C) Quantification of the number of genes whose expression is significantly altered (adjusted P < .05) at all time points (Total) and after the indicated duration of treatment with U0126. (D) Comparison of the number of genes whose expression is significantly (adjusted P < .05) upregulated (left panel) or downregulated (right panel) after the indicated time of exposure to U0126. (E) GO analysis for molecular functions significantly (adjusted P < .05) enriched within the sets of genes whose expression is altered by U0126 exposure at the indicated times. (F) Number of genes whose expression is significantly (adjusted P < .05) upregulated or downmodulated by U0126 at the indicated times and for the indicated GO terms.
Top 25 upregulated and downregulated genes in U0126-treated MDDCs at 4, 10, and 24 hours posttreatment
| . | 4 hours . | 10 hours . | 24 hours . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene symbol . | Log2 fold change . | Adjusted p . | Gene symbol . | Log2 fold change . | Adjusted p . | Gene symbol . | Log2 fold change . | Adjusted P . | |
| Upregulated genes | PLAT | 2.39 | 1.69 E-07 | PLAT | 3.39 | 1.78 E-10 | ANGPT2 | 5.48 | 0.012988 |
| SHF | 1.95 | 7.79 E-06 | PTGER3 | 3.06 | 1.44 E-08 | RBMXL3 | 5.09 | 0.043762 | |
| AHRR | 1.83 | 0.011751 | ATP1B2 | 2.71 | 3.64 E-08 | NR4A3 | 4.85 | 0.011564 | |
| ROR2 | 1.70 | 0.014002 | TTC9 | 2.61 | 2.89 E-12 | RALGAPA1 | 4.38 | 0.020438 | |
| TTC9 | 1.67 | 1.50 E-08 | SHF | 2.50 | 7.10 E-08 | ATP1B2 | 4.06 | 2.16 E-10 | |
| LRG1 | 1.64 | 1.69 E-07 | AHRR | 2.23 | 1.53 E-11 | A4GALT | 3.97 | 0.000855 | |
| ATP1B2 | 1.59 | 0.000199 | CYP1B1 | 2.21 | 1.39 E-09 | TMPRSS6 | 3.71 | 1.95 E-05 | |
| GAD1 | 1.48 | 4.16 E-07 | GAD1 | 2.21 | 2.31 E-10 | GSTT1 | 3.41 | 7.67 E-10 | |
| RASAL1 | 1.38 | 0.0001908 | SYTL3 | 2.13 | 2.36 E-05 | TTC9 | 3.10 | 7.46 E-13 | |
| ZNF395 | 1.38 | 1.69 E-07 | ROR2 | 2.06 | 0.001021 | MUC1 | 2.78 | 1.96 E-09 | |
| RARG | 1.36 | 0.0005075 | CD163L1 | 2.01 | 5.72 E-05 | CD163L1 | 2.76 | 2.74 E-06 | |
| ARL4C | 1.33 | 0.0491981 | CSRP2 | 1.94 | 3.14 E-09 | XPNPEP2 | 2.71 | 0.000513 | |
| CXCL2 | 1.33 | 0.0020045 | RARG | 1.85 | 4.36 E-06 | ARL4C | 2.64 | 5.30 E-05 | |
| TIPARP | 1.24 | 7.11 E-07 | MUC1 | 1.82 | 4.68 E-07 | PTGER3 | 2.64 | 8.52 E-07 | |
| PDGFB | 1.20 | 0.0109397 | THBS1 | 1.78 | 1.05 E-05 | SLC26A1 | 2.62 | 0.04403 | |
| PTGER2 | 1.17 | 3.49 E-07 | ARL4C | 1.77 | 0.002025 | CYP1B1 | 2.49 | 9.26 E-10 | |
| NINL | 1.16 | 0.0108421 | TIPARP | 1.69 | 1.96 E-09 | CSRP2 | 2.46 | 2.81 E-10 | |
| PRDM1 | 1.14 | 0.000317 | LPAR6 | 1.64 | 4.57 E-09 | HES1 | 2.43 | 2.46 E-05 | |
| THBS1 | 1.14 | 0.0048596 | ANK3 | 1.61 | 0.020469 | PTP4A3 | 2.41 | 1.80 E-05 | |
| SYTL3 | 1.13 | 0.0417727 | RASAL1 | 1.60 | 1.27 E-05 | ITGB7 | 2.41 | 4.05 E-10 | |
| CYP1B1 | 1.10 | 0.000145 | ITGB7 | 1.59 | 9.34 E-08 | SHF | 2.41 | 6.22 E-07 | |
| NCF1 | 1.08 | 0.0274784 | GSG1 | 1.59 | 0.002054 | RPS4Y1 | 2.41 | 8.68 E-05 | |
| XYLT1 | 1.08 | 0.0172593 | NCF1 | 1.54 | 0.000415 | CCDC108 | 2.38 | 0.031671 | |
| MUC1 | 1.02 | 0.0020864 | PLA2G16 | 1.52 | 1.31 E-09 | BDH2 | 2.29 | 0.009805 | |
| LPAR6 | 1.01 | 2.03 E-05 | ZNF395 | 1.51 | 1.36 E-08 | EPHB2 | 2.28 | 8.15 E-09 | |
| Downregulated genes | SPRED2 | −1.50 | 1.66 E-05 | HIVEP3 | −1.57 | 1.55 E-03 | GATAD1 | −2.57 | 3.01 E-08 |
| KMO | −1.50 | 9.46 E-08 | ASPHD2 | −1.57 | 3.05 E-09 | NDP | −2.60 | 1.19 E-08 | |
| CDC42EP2 | −1.54 | 3.71 E-05 | KMO | −1.59 | 1.53 E-07 | TDG | −2.65 | 2.81 E-10 | |
| CCR2 | −1.55 | 3.61 E-04 | LEPREL1 | −1.61 | 1.00 E-08 | MYO1D | −2.74 | 1.77 E-08 | |
| ASPHD2 | −1.56 | 1.04 E-08 | ARAP3 | −1.61 | 6.63 E-08 | PDLIM7 | −2.77 | 5.34 E-10 | |
| EGR1 | −1.56 | 9.69 E-06 | SLAMF8 | −1.64 | 8.99 E-08 | EBPL | −2.77 | 1.21 E-12 | |
| CLDN1 | −1.57 | 3.34 E-06 | PLA2G4A | −1.64 | 1.76 E-07 | MTERFD1 | −2.81 | 2.61 E-10 | |
| TAGAP | −1.61 | 1.53 E-08 | KCNN4 | −1.66 | 1.74 E-07 | PPIAL4A | −2.84 | 1.61 E-04 | |
| PLA2G4A | −1.65 | 3.49 E-07 | EXT1 | −1.70 | 2.85 E-05 | STEAP3 | −2.92 | 1.91 E-10 | |
| FNIP2 | −1.69 | 8.25 E-08 | DMPK | −1.70 | 6.05 E-06 | EBAG9 | −2.94 | 1.34 E-11 | |
| KCNN4 | −1.69 | 3.23 E-07 | ADORA2B | −1.72 | 8.57 E-03 | FAM26F | −2.96 | 1.56 E-12 | |
| DAB2 | −1.69 | 2.95 E-06 | CCR2 | −1.74 | 3.98 E-02 | CHCHD10 | −3.00 | 1.81 E-09 | |
| CDR2 | −1.70 | 3.63 E-04 | SLC4A7 | −1.76 | 1.81 E-03 | TMEM41B | −3.02 | 9.03 E-12 | |
| SPRY2 | −1.71 | 1.45 E-04 | CLDN1 | −1.81 | 1.53 E-07 | EPCAM | −3.03 | 1.80 E-11 | |
| PTX3 | −1.73 | 7.12 E-09 | ENTPD1 | −1.86 | 3.14 E-09 | GRHPR | −3.08 | 9.66 E-11 | |
| TRIB2 | −1.81 | 4.97 E-10 | TRIB2 | −1.95 | 4.13 E-11 | IGSF6 | −3.16 | 5.83 E-10 | |
| MYC | −1.83 | 2.08 E-08 | ZC3H8 | −1.97 | 9.34 E-08 | CLK1 | −3.22 | 6.68 E-10 | |
| ADORA2B | −1.86 | 8.93 E-03 | PPAP2B | −2.04 | 1.73 E-10 | PATZ1 | −3.39 | 8.81 E-13 | |
| FLT1 | −1.88 | 1.69 E-07 | FOS | −2.20 | 1.00 E-08 | MTFMT | −3.41 | 1.21 E-12 | |
| PPAP2B | −2.08 | 4.25 E-10 | FLT1 | −2.32 | 5.60 E-09 | DNM1L | −3.43 | 1.42 E-13 | |
| CCL7 | −2.18 | 9.23 E-07 | IL1R2 | −2.43 | 1.00 E-08 | PNKD | −3.44 | 2.71 E-14 | |
| FOS | −2.26 | 1.62 E-08 | EHD1 | −2.43 | 2.21 E-12 | CCR2 | −3.61 | 7.47 E-05 | |
| CCL2 | −2.46 | 1.79 E-06 | PTX3 | −2.47 | 3.75 E-12 | TMEM167A | −3.77 | 2.79 E-13 | |
| EHD1 | −2.81 | 1.34 E-13 | CCL2 | −2.61 | 3.05 E-07 | G3BP2 | −3.83 | 2.71 E-14 | |
| DUSP6 | −3.80 | 1.34 E-13 | DUSP6 | −3.05 | 4.66 E-12 | MRPL33 | −3.88 | 2.49 E-13 | |
| . | 4 hours . | 10 hours . | 24 hours . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene symbol . | Log2 fold change . | Adjusted p . | Gene symbol . | Log2 fold change . | Adjusted p . | Gene symbol . | Log2 fold change . | Adjusted P . | |
| Upregulated genes | PLAT | 2.39 | 1.69 E-07 | PLAT | 3.39 | 1.78 E-10 | ANGPT2 | 5.48 | 0.012988 |
| SHF | 1.95 | 7.79 E-06 | PTGER3 | 3.06 | 1.44 E-08 | RBMXL3 | 5.09 | 0.043762 | |
| AHRR | 1.83 | 0.011751 | ATP1B2 | 2.71 | 3.64 E-08 | NR4A3 | 4.85 | 0.011564 | |
| ROR2 | 1.70 | 0.014002 | TTC9 | 2.61 | 2.89 E-12 | RALGAPA1 | 4.38 | 0.020438 | |
| TTC9 | 1.67 | 1.50 E-08 | SHF | 2.50 | 7.10 E-08 | ATP1B2 | 4.06 | 2.16 E-10 | |
| LRG1 | 1.64 | 1.69 E-07 | AHRR | 2.23 | 1.53 E-11 | A4GALT | 3.97 | 0.000855 | |
| ATP1B2 | 1.59 | 0.000199 | CYP1B1 | 2.21 | 1.39 E-09 | TMPRSS6 | 3.71 | 1.95 E-05 | |
| GAD1 | 1.48 | 4.16 E-07 | GAD1 | 2.21 | 2.31 E-10 | GSTT1 | 3.41 | 7.67 E-10 | |
| RASAL1 | 1.38 | 0.0001908 | SYTL3 | 2.13 | 2.36 E-05 | TTC9 | 3.10 | 7.46 E-13 | |
| ZNF395 | 1.38 | 1.69 E-07 | ROR2 | 2.06 | 0.001021 | MUC1 | 2.78 | 1.96 E-09 | |
| RARG | 1.36 | 0.0005075 | CD163L1 | 2.01 | 5.72 E-05 | CD163L1 | 2.76 | 2.74 E-06 | |
| ARL4C | 1.33 | 0.0491981 | CSRP2 | 1.94 | 3.14 E-09 | XPNPEP2 | 2.71 | 0.000513 | |
| CXCL2 | 1.33 | 0.0020045 | RARG | 1.85 | 4.36 E-06 | ARL4C | 2.64 | 5.30 E-05 | |
| TIPARP | 1.24 | 7.11 E-07 | MUC1 | 1.82 | 4.68 E-07 | PTGER3 | 2.64 | 8.52 E-07 | |
| PDGFB | 1.20 | 0.0109397 | THBS1 | 1.78 | 1.05 E-05 | SLC26A1 | 2.62 | 0.04403 | |
| PTGER2 | 1.17 | 3.49 E-07 | ARL4C | 1.77 | 0.002025 | CYP1B1 | 2.49 | 9.26 E-10 | |
| NINL | 1.16 | 0.0108421 | TIPARP | 1.69 | 1.96 E-09 | CSRP2 | 2.46 | 2.81 E-10 | |
| PRDM1 | 1.14 | 0.000317 | LPAR6 | 1.64 | 4.57 E-09 | HES1 | 2.43 | 2.46 E-05 | |
| THBS1 | 1.14 | 0.0048596 | ANK3 | 1.61 | 0.020469 | PTP4A3 | 2.41 | 1.80 E-05 | |
| SYTL3 | 1.13 | 0.0417727 | RASAL1 | 1.60 | 1.27 E-05 | ITGB7 | 2.41 | 4.05 E-10 | |
| CYP1B1 | 1.10 | 0.000145 | ITGB7 | 1.59 | 9.34 E-08 | SHF | 2.41 | 6.22 E-07 | |
| NCF1 | 1.08 | 0.0274784 | GSG1 | 1.59 | 0.002054 | RPS4Y1 | 2.41 | 8.68 E-05 | |
| XYLT1 | 1.08 | 0.0172593 | NCF1 | 1.54 | 0.000415 | CCDC108 | 2.38 | 0.031671 | |
| MUC1 | 1.02 | 0.0020864 | PLA2G16 | 1.52 | 1.31 E-09 | BDH2 | 2.29 | 0.009805 | |
| LPAR6 | 1.01 | 2.03 E-05 | ZNF395 | 1.51 | 1.36 E-08 | EPHB2 | 2.28 | 8.15 E-09 | |
| Downregulated genes | SPRED2 | −1.50 | 1.66 E-05 | HIVEP3 | −1.57 | 1.55 E-03 | GATAD1 | −2.57 | 3.01 E-08 |
| KMO | −1.50 | 9.46 E-08 | ASPHD2 | −1.57 | 3.05 E-09 | NDP | −2.60 | 1.19 E-08 | |
| CDC42EP2 | −1.54 | 3.71 E-05 | KMO | −1.59 | 1.53 E-07 | TDG | −2.65 | 2.81 E-10 | |
| CCR2 | −1.55 | 3.61 E-04 | LEPREL1 | −1.61 | 1.00 E-08 | MYO1D | −2.74 | 1.77 E-08 | |
| ASPHD2 | −1.56 | 1.04 E-08 | ARAP3 | −1.61 | 6.63 E-08 | PDLIM7 | −2.77 | 5.34 E-10 | |
| EGR1 | −1.56 | 9.69 E-06 | SLAMF8 | −1.64 | 8.99 E-08 | EBPL | −2.77 | 1.21 E-12 | |
| CLDN1 | −1.57 | 3.34 E-06 | PLA2G4A | −1.64 | 1.76 E-07 | MTERFD1 | −2.81 | 2.61 E-10 | |
| TAGAP | −1.61 | 1.53 E-08 | KCNN4 | −1.66 | 1.74 E-07 | PPIAL4A | −2.84 | 1.61 E-04 | |
| PLA2G4A | −1.65 | 3.49 E-07 | EXT1 | −1.70 | 2.85 E-05 | STEAP3 | −2.92 | 1.91 E-10 | |
| FNIP2 | −1.69 | 8.25 E-08 | DMPK | −1.70 | 6.05 E-06 | EBAG9 | −2.94 | 1.34 E-11 | |
| KCNN4 | −1.69 | 3.23 E-07 | ADORA2B | −1.72 | 8.57 E-03 | FAM26F | −2.96 | 1.56 E-12 | |
| DAB2 | −1.69 | 2.95 E-06 | CCR2 | −1.74 | 3.98 E-02 | CHCHD10 | −3.00 | 1.81 E-09 | |
| CDR2 | −1.70 | 3.63 E-04 | SLC4A7 | −1.76 | 1.81 E-03 | TMEM41B | −3.02 | 9.03 E-12 | |
| SPRY2 | −1.71 | 1.45 E-04 | CLDN1 | −1.81 | 1.53 E-07 | EPCAM | −3.03 | 1.80 E-11 | |
| PTX3 | −1.73 | 7.12 E-09 | ENTPD1 | −1.86 | 3.14 E-09 | GRHPR | −3.08 | 9.66 E-11 | |
| TRIB2 | −1.81 | 4.97 E-10 | TRIB2 | −1.95 | 4.13 E-11 | IGSF6 | −3.16 | 5.83 E-10 | |
| MYC | −1.83 | 2.08 E-08 | ZC3H8 | −1.97 | 9.34 E-08 | CLK1 | −3.22 | 6.68 E-10 | |
| ADORA2B | −1.86 | 8.93 E-03 | PPAP2B | −2.04 | 1.73 E-10 | PATZ1 | −3.39 | 8.81 E-13 | |
| FLT1 | −1.88 | 1.69 E-07 | FOS | −2.20 | 1.00 E-08 | MTFMT | −3.41 | 1.21 E-12 | |
| PPAP2B | −2.08 | 4.25 E-10 | FLT1 | −2.32 | 5.60 E-09 | DNM1L | −3.43 | 1.42 E-13 | |
| CCL7 | −2.18 | 9.23 E-07 | IL1R2 | −2.43 | 1.00 E-08 | PNKD | −3.44 | 2.71 E-14 | |
| FOS | −2.26 | 1.62 E-08 | EHD1 | −2.43 | 2.21 E-12 | CCR2 | −3.61 | 7.47 E-05 | |
| CCL2 | −2.46 | 1.79 E-06 | PTX3 | −2.47 | 3.75 E-12 | TMEM167A | −3.77 | 2.79 E-13 | |
| EHD1 | −2.81 | 1.34 E-13 | CCL2 | −2.61 | 3.05 E-07 | G3BP2 | −3.83 | 2.71 E-14 | |
| DUSP6 | −3.80 | 1.34 E-13 | DUSP6 | −3.05 | 4.66 E-12 | MRPL33 | −3.88 | 2.49 E-13 | |
Validation of the transcription data and functional predictions
To verify the above predictions, we focused on genes whose expression showed a potent U0126 responsiveness (FOS, DUSP6, THBS1) (Table 1) or that have a large impact on immune responses (IDO1). First, the effect of the structurally unrelated MEK-ERK inhibitor PD98059 on the expression of U0126-responsive genes was determined. Like U0126, PD98059 reduced the level of ERK activation (Figure 2A), reduced the mRNA expression level of FOS and DUSP6 at all time points analyzed, and enhanced that of IDO1 and THBS1 (Figure 2B) in MDDCs. Second, the expression of U0126-dependent genes was monitored in MDDCs nucleofected with either a constitutively active (MEK-E) or a dominant negative (MEK-A) form of MEK. Overexpression of MEK-E resulted in enhanced phosphorylation of ERK in immature MDDCs and led to downregulation of U0126-upregulated genes (IDO1 and THBS1) and increase of U0126-downregulated genes like DUSP6 (Figure 2C). Altogether, these results validated the data from the global analysis of gene expression and confirmed the relevance of the MEK-ERK signaling pathway in the maintenance of the gene expression profile of immature MDDCs.
Analysis of ERK-dependent gene expression in immature MDDCs. (A) Effect of U0126 (UO) or PD98059 (PD) treatment on ERK1/2 phosphorylation in immature DCs after the indicated time of treatment, as determined by western blot. The levels of total ERK1/2 and GAPDH protein were determined in parallel. The histogram illustrates the ratio p-ERK/ERK for each sample, as determined by densitometric analysis of the western blot. One of 3 representative experiments is presented. (B) Relative expression of the indicated genes in immature MDDCs treated with DMSO (-), U0126 (UO), or PD98059 (PD) after the indicated time of treatment. Results are expressed as relative expression, which indicates the expression of each gene in each sample relative to its expression in DMSO-treated immature DCs at every time point (*P < .05). Shown are the means and standard deviations (SDs) of three experiments for UO and two experiments for PD-treated samples. (C) Relative expression of the indicated genes in immature MDDCs nucleofected with either an empty vector (-) or expression vector for a constitutively active (E) or a dominant negative (A) forms of MEK, as determined by real time quantitative reverse transcriptase polymerase chain reaction 24 hours after nucleofection. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in immature DCs nucleofected with a constitutively active (E) form of MEK. The graph represents the mean and standard deviation of two different experiments. The top panels illustrate the level of ERK phosphorylation (left) in one representative experiment, and the ratio p-ERK/ERK after densitometric analysis of the western blot (right) in each sample.
Analysis of ERK-dependent gene expression in immature MDDCs. (A) Effect of U0126 (UO) or PD98059 (PD) treatment on ERK1/2 phosphorylation in immature DCs after the indicated time of treatment, as determined by western blot. The levels of total ERK1/2 and GAPDH protein were determined in parallel. The histogram illustrates the ratio p-ERK/ERK for each sample, as determined by densitometric analysis of the western blot. One of 3 representative experiments is presented. (B) Relative expression of the indicated genes in immature MDDCs treated with DMSO (-), U0126 (UO), or PD98059 (PD) after the indicated time of treatment. Results are expressed as relative expression, which indicates the expression of each gene in each sample relative to its expression in DMSO-treated immature DCs at every time point (*P < .05). Shown are the means and standard deviations (SDs) of three experiments for UO and two experiments for PD-treated samples. (C) Relative expression of the indicated genes in immature MDDCs nucleofected with either an empty vector (-) or expression vector for a constitutively active (E) or a dominant negative (A) forms of MEK, as determined by real time quantitative reverse transcriptase polymerase chain reaction 24 hours after nucleofection. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in immature DCs nucleofected with a constitutively active (E) form of MEK. The graph represents the mean and standard deviation of two different experiments. The top panels illustrate the level of ERK phosphorylation (left) in one representative experiment, and the ratio p-ERK/ERK after densitometric analysis of the western blot (right) in each sample.
Contribution of MEK/ERK to maintenance of the immature state of MDDCs
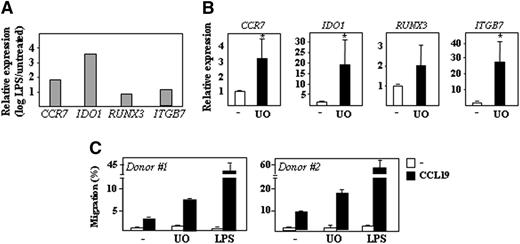
LPS triggers a dramatic modulation of the gene expression profile of MDDCs.20 To illustrate the role of the MEK-ERK signaling pathway in the maintenance of the immature MDDC transcriptional profile, U0126-dependent gene expression changes were compared with those that take place upon LPS-induced MDDC maturation (Figure 3A). U0126 modified the expression of genes whose levels are highly dependent on the state of maturation of MDDC, including CCR7, IDO1, RUNX3, ITGB7, CXCL16, SCARB1, OLR1, CCR2, and CXCR421,22 (data not shown). In agreement with the profiling results, inhibition of ERK1/2 activation resulted in significantly higher expression of CCR7, IDO1, RUNX3, and ITGB7 (Figure 3B). At the functional level, and as predicted from their higher expression of CCR7, U0126-treated immature MDDCs exhibited a higher chemotactic response toward CCL19 than DMSO-treated MDDCs (Figure 3C). Therefore, inhibition of the phosphorylation state of ERK upregulates genes/functions that constitute a hallmark for MDDC maturation, thus linking ERK phosphorylation to the maintenance of the MDDC immature state.
Effect of U0126 on the expression of DC maturation-associated genes. (A) Relative expression of the indicated genes in LPS-treated mature MDDCs. Results are expressed as relative expression (log scale), which indicates the expression of each gene in mature MDDCs relative to its expression in immature MDDCs. (B) Relative expression of the indicated genes in immature DCs from four different donors treated with DMSO (-) or U0126 (UO) for 24 hours. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in DMSO-treated MDDCs (*P < .05). (C) CCL19-directed migration of immature DCs exposed to either DMSO (-), U0126 (UO), or DMSO + LPS (LPS) for 24 hours, as determined in Transwell migration assays on immature DCs from two independent donors. Results are expressed as percentage of migrated cells relative to the input in the presence and absence of CCL19.
Effect of U0126 on the expression of DC maturation-associated genes. (A) Relative expression of the indicated genes in LPS-treated mature MDDCs. Results are expressed as relative expression (log scale), which indicates the expression of each gene in mature MDDCs relative to its expression in immature MDDCs. (B) Relative expression of the indicated genes in immature DCs from four different donors treated with DMSO (-) or U0126 (UO) for 24 hours. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in DMSO-treated MDDCs (*P < .05). (C) CCL19-directed migration of immature DCs exposed to either DMSO (-), U0126 (UO), or DMSO + LPS (LPS) for 24 hours, as determined in Transwell migration assays on immature DCs from two independent donors. Results are expressed as percentage of migrated cells relative to the input in the presence and absence of CCL19.
Immature MDDCs display a potent ability for capturing and internalizing extracellular material.2 The prediction that U0126-regulated genes contribute to lipoprotein binding prompted us to evaluate whether inhibition of ERK phosphorylation influences such an effector function. U0126-treated cells displayed a significantly enhanced expression of genes involved in lipoprotein binding (CD36, OLR1, SCARB1, and CXCL16) (Figure 4A) as well as a significantly elevated lipoprotein binding ability (Figure 4B). Therefore, the MEK-ERK signaling pathway regulates LDL binding by MDDCs.
Effect of U0126 on the expression of CCL2 and genes involved in LDL binding in immature MDDCs. (A) Relative expression of the indicated genes in immature DCs treated with DMSO (-) or U0126 (UO) for 24 hours. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in DMSO-treated MDDCs (*P < .05). Shown are the means and SDs of three independent experiments. (B) LDL binding to immature DCs exposed to either DMSO or U0126 for 24 hours and exposed to LDL for the indicated periods of time, as determined by flow cytometry. The lower panel illustrates the percentage of LDL-positive cells in each sample (*P < .05). The means and SDs of five independent experiments are presented. (C) Relative expression of the CCL2 gene in immature DCs treated with DMSO (-), U0126 (UO), or PD98059 (PD) after the indicated time of treatment. The means and SDs of three independent experiments for UO and two experiments for PD-treated MDDCs are presented. Results are expressed as relative expression, which indicates CCL2 expression in each sample relative to its expression in DMSO-treated MDDCs (*P < .05). (D) Expression of CCL2 (MCP1) in immature DCs treated with either DMSO (-), U0126 (UO), or PD98059 (PD) after the indicated time of treatment, as determined by enzyme-linked immunosorbent assay. Shown are the means and SDs of four independent experiments (*P < .05).
Effect of U0126 on the expression of CCL2 and genes involved in LDL binding in immature MDDCs. (A) Relative expression of the indicated genes in immature DCs treated with DMSO (-) or U0126 (UO) for 24 hours. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in DMSO-treated MDDCs (*P < .05). Shown are the means and SDs of three independent experiments. (B) LDL binding to immature DCs exposed to either DMSO or U0126 for 24 hours and exposed to LDL for the indicated periods of time, as determined by flow cytometry. The lower panel illustrates the percentage of LDL-positive cells in each sample (*P < .05). The means and SDs of five independent experiments are presented. (C) Relative expression of the CCL2 gene in immature DCs treated with DMSO (-), U0126 (UO), or PD98059 (PD) after the indicated time of treatment. The means and SDs of three independent experiments for UO and two experiments for PD-treated MDDCs are presented. Results are expressed as relative expression, which indicates CCL2 expression in each sample relative to its expression in DMSO-treated MDDCs (*P < .05). (D) Expression of CCL2 (MCP1) in immature DCs treated with either DMSO (-), U0126 (UO), or PD98059 (PD) after the indicated time of treatment, as determined by enzyme-linked immunosorbent assay. Shown are the means and SDs of four independent experiments (*P < .05).
Contribution of MEK/ERK to effector functions of MDDCs in inflammatory responses
The CCL2 chemokine directs monocyte/macrophage recruitment into tissues under resting and inflamed conditions.23 Since U0126-regulated genes appeared to be significantly associated with the “Inflammatory Response” GO function, the influence of MEK-ERK activation on CCL2 expression was evaluated. U0126 or PD98059 significantly reduced the levels of CCL2 mRNA in immature MDDCs at all time points (Figure 4C), and both inhibitors significantly reduced CCL2 production at early time points (Figure 4D). Therefore, the MEK-ERK pathway regulates the release of CCL2, a chemokine constitutively produced by immature MDDCs.
Contribution of MEK/ERK to the gene expression profile of peripheral blood myeloid DCs
To determine whether the results observed on MDDCs could be extrapolated to ex vivo isolated cells, peripheral blood CD1c+ myeloid DCs were exposed to U0126 for 20 hours before analyzing the expression of ERK-dependent genes. U0126 downregulated the expression of the well-established ERK-target gene DUSP6 and, as in MDDCs, enhanced the expression of the maturation-associated ITGB7 gene SCARB1, which codes for the LDL-binding scavenger SRB1, CD36, and the AhR-target genes CYP1B1 and AHRR (Figure 5). In addition, U0126 treatment downregulated CCL2 and SERPINB2 gene expression (Figure 5). Therefore, inhibition of MEK/ERK activation in peripheral blood myeloid DCs results in gene expression changes that resemble those observed in U0126-treated MDDCs, because it modulates the expression of genes associated with DC maturation (ITGB7) or involved in the capture of extracellular material (SCARB1, CD36) and myeloid cell recruitment (CCL2).
Effect of U0126 on gene expression in peripheral blood myeloid DC. Relative expression of the indicated genes in CD1c+ myeloid DCs treated with either DMSO (-) or U0126 (UO) for 20 hours. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in DMSO-treated cells (*P < .05). Shown are the means and SDs of three independent experiments.
Effect of U0126 on gene expression in peripheral blood myeloid DC. Relative expression of the indicated genes in CD1c+ myeloid DCs treated with either DMSO (-) or U0126 (UO) for 20 hours. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in DMSO-treated cells (*P < .05). Shown are the means and SDs of three independent experiments.
The p38MAPK inhibitor SB203580 activates ERK and modulates the expression of ERK-dependent genes in immature MDDCs
Since SB203580 enhances ERK phosphorylation in various cellular systems,24,25 including MDDCs,5 we determined gene expression changes in SB203580-treated MDDCs (supplemental Figure 1A). SB203580 inhibited p38 phosphorylation at early time points, in agreement with previous reports,26 and enhanced ERK phosphorylation throughout the whole period of analysis (supplemental Figure 1B). In fact, SB203580 modified the expression of more than 2000 genes in MDDCs (supplemental Figure 1C), including well-known p38MAPK targets27,28 and numerous U0126-independent genes (527 at 4 hours, 1052 at 10 hours, and 1357 at 24 hours) (supplemental Figure 1D). However, a significant number of genes were oppositely regulated by SB203580 and U0126 (supplemental Figure 1D-E,G), a finding further confirmed at the protein level for RUNX3 and CCL2 (supplemental Figure 1F,H). Since the alternative p38MAPK inhibitor BIRB0796 did not promote ERK phosphorylation (supplemental Figure 1B) and had no effect on the expression of several SB203580-responsive genes (supplemental Figure 1E,G), these results suggested that some SB203580-induced gene expression changes in immature MDDCs may depend, at least partly, on the ability of this inhibitor to enhance ERK phosphorylation.
Role of the AhR transcription factor in U0126-dependent gene expression changes
The search for overrepresented cognate sequences in the proximal regulatory regions of U0126-regulated genes using the JASPAR database on Pscan sofware29 revealed a statistically significant enrichment of AhR-binding sequences (supplemental Table 4). The relevance of this prediction was also supported by the high percentage of known AhR-target genes30,31 within the set of U0126-upregulated genes in MDDCs (Table 1 and Figure 6A) and ex vivo isolated myeloid DCs (Figure 5). To assess whether AhR mediates the transcriptional effects of U0126, the influence of pharmacologic modulators of AhR on U0126-regulated gene expression was assayed. Like U0126, the AhR ligand TCDD led to significant enhancement of AhR-target genes in immature MDDCs (Figure 6A). Furthermore, the U0126-dependent upregulation of these genes was inhibited by the AhR antagonist alpha naphthoflavone (α-NF; Figure 6B), an effect confirmed at the protein level for THBS1 (Figure 6C). The expression of U0126-dependent genes that are related to T-cell stimulatory activity of MDDCs (RUNX3, IDO1) or that are altered upon MDDC maturation (ITGB7) was also modulated by TCDD (Figure 6D), although only ITGB7 expression was significantly inhibited by α-NF (Figure 6E). Moreover, the U0126-dependent upregulation of CYP1B1 and AHRR was also inhibited by α-NF in monocyte-derived macrophages (supplemental Figure 2A) but was not seen in peripheral blood myeloid DCs (supplemental Figure 2B). Altogether, these results strongly suggested the involvement of AhR in the transcriptional effects of U0126 in immature MDDC and macrophages.
Contribution of AhR to the ERK-dependent gene expression profile in immature DCs. (A) Relative expression of the indicated genes in immature MDDCs from five independent donors after treatment with either DMSO (-), U0126 (UO; 2.5 μM), or TCDD (10 nM) for 24 hours. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in DMSO-treated cells (*P < .05). (B) Relative expression of the indicated genes in immature MDDCs treated for 24 hours with U0126 (UO; 2.5 μM), α-NF (1 μM), or both. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in U0126-treated immature MDDCs (*P < .05). The means and SDs of four independent experiments are presented. (C) Expression of THBS1 in immature MDDCs treated for 24 hours with DMSO (-), U0126 (UO; 2.5 μM), or α-NF (1 μM). The levels of AhR, phosphorylated ERK, and GAPDH protein were determined in parallel. (D) Relative expression of CCR7, RUNX3, IDO1, and ITGB7 genes in immature MDDCs treated for 24 hours with either DMSO (-) or TCDD (10 nM). Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in DMSO-treated immature MDDCs (*P < .05). (E) Relative expression of CCR7, RUNX3, IDO1, and ITGB7 genes in immature MDDCs treated for 24 hours with either U0126 (UO; 2.5 μM), α-NF (1 μM), or both. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in U0126-treated immature MDDCs (*P < .05). In (D) and (E), the means and SDs of four independent experiments are presented.
Contribution of AhR to the ERK-dependent gene expression profile in immature DCs. (A) Relative expression of the indicated genes in immature MDDCs from five independent donors after treatment with either DMSO (-), U0126 (UO; 2.5 μM), or TCDD (10 nM) for 24 hours. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in DMSO-treated cells (*P < .05). (B) Relative expression of the indicated genes in immature MDDCs treated for 24 hours with U0126 (UO; 2.5 μM), α-NF (1 μM), or both. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in U0126-treated immature MDDCs (*P < .05). The means and SDs of four independent experiments are presented. (C) Expression of THBS1 in immature MDDCs treated for 24 hours with DMSO (-), U0126 (UO; 2.5 μM), or α-NF (1 μM). The levels of AhR, phosphorylated ERK, and GAPDH protein were determined in parallel. (D) Relative expression of CCR7, RUNX3, IDO1, and ITGB7 genes in immature MDDCs treated for 24 hours with either DMSO (-) or TCDD (10 nM). Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in DMSO-treated immature MDDCs (*P < .05). (E) Relative expression of CCR7, RUNX3, IDO1, and ITGB7 genes in immature MDDCs treated for 24 hours with either U0126 (UO; 2.5 μM), α-NF (1 μM), or both. Results are expressed as relative expression, which indicates the expression of each gene relative to its expression in U0126-treated immature MDDCs (*P < .05). In (D) and (E), the means and SDs of four independent experiments are presented.
To establish a link between MEK-ERK inhibition and AhR activity, the influence of U0126 on the in vivo DNA-binding and the transcriptional activity of AhR was assessed. DNA-binding of AhR in vivo was evaluated by chromatin immunoprecipitation on control and U0126-treated cells, using the occupancy of the AhR cognate sequences within the CYP1B1 gene promoter region as readout. The −790/–890 CYP1B1 gene promoter region was readily amplified in anti-AhR–precipitated DNA from U0126-treated MDDCs, whereas much lower levels of amplification were detected in anti-AhR–precipitated DNA from control MDDCs (Figure 7A) or after immunoprecipitation with an isotype-matched antibody (immunoglobulin G, Figure 7A). Therefore, U0126 enhances the in vivo occupancy of AhR-binding elements in MDDCs.
Effect of U0126 on the transcriptional activity of AhR. (A) In vivo occupancy of the CYP1B1 AhR-binding element by AhR. Shown are two independent chromatin immunoprecipitations on DMSO- and U0126-treated immature MDDCs using a monoclonal antibody specific for AhR (αAhR) or a nonspecific isotype-matched antibody (IgG). Immunoprecipitated chromatin was analyzed by PCR (30 amplification cycles) using a pair of CYP1B1 promoter-specific primers that amplify the −790/–890 promoter region. Input DNA lanes represent the PCR analysis performed on DNA isolated from 1% of the starting sonicated lysate. In the right panel, a dashed vertical line has been inserted to indicate repositioned gel lanes. (B-D) HepG2, MDDCs, or human monocytes were transfected with either the AhR-dependent XRE-TATA-luc construct (B-D) or a TATA-luc construct (B). Fourteen to 24 hours after transfection, cells were treated with DMSO (-), U0126, or TCDD, and 12 or 18 hours later, cells were lysed and the level of firefly luciferase was determined. For each individual reporter construct, relative transcriptional activity represents the luciferase activity yielded by each expression vector relative to the activity measured in DMSO-treated cells. In all cases, firefly luciferase levels were normalized according to the transfection efficiency, as determined through the use of a cotransfected Renilla luciferase–coding construct. Data represent the means and SDs of eight (HepG2; B), five (MDDCs; C) or three (monocytes; D) independent experiments.
Effect of U0126 on the transcriptional activity of AhR. (A) In vivo occupancy of the CYP1B1 AhR-binding element by AhR. Shown are two independent chromatin immunoprecipitations on DMSO- and U0126-treated immature MDDCs using a monoclonal antibody specific for AhR (αAhR) or a nonspecific isotype-matched antibody (IgG). Immunoprecipitated chromatin was analyzed by PCR (30 amplification cycles) using a pair of CYP1B1 promoter-specific primers that amplify the −790/–890 promoter region. Input DNA lanes represent the PCR analysis performed on DNA isolated from 1% of the starting sonicated lysate. In the right panel, a dashed vertical line has been inserted to indicate repositioned gel lanes. (B-D) HepG2, MDDCs, or human monocytes were transfected with either the AhR-dependent XRE-TATA-luc construct (B-D) or a TATA-luc construct (B). Fourteen to 24 hours after transfection, cells were treated with DMSO (-), U0126, or TCDD, and 12 or 18 hours later, cells were lysed and the level of firefly luciferase was determined. For each individual reporter construct, relative transcriptional activity represents the luciferase activity yielded by each expression vector relative to the activity measured in DMSO-treated cells. In all cases, firefly luciferase levels were normalized according to the transfection efficiency, as determined through the use of a cotransfected Renilla luciferase–coding construct. Data represent the means and SDs of eight (HepG2; B), five (MDDCs; C) or three (monocytes; D) independent experiments.
Evaluation of the AhR-dependent transcriptional activity was accomplished through the use of the AhR-responsive reporter construct XRE-TATA-luc, which encodes the firefly luciferase gene under the control of tandem repeats of the XRE transcriptional response element. In HepG2 human hepatoma cells, the XRE-TATA-luc construct was responsive to U0126 at two different time points, whereas the weak activity of the control TATA-luc reporter construct was not affected (Figure 7B). More importantly, the activity of the AhR-responsive luciferase construct was significantly enhanced by U0126 in both MDDCs (Figure 7C) and human monocytes (Figure 7C). Therefore, inhibition of the MEK-ERK signaling pathway by U0126 in MDDCs results in enhanced AhR transcriptional activity and higher occupancy of AhR-binding elements in vivo, thus establishing AhR as a factor contributing to the maintenance of the immature state of MDDCs.
Discussion
Previous studies have demonstrated the opposite influence of ERK and p38MAPK activation on DC maturation,4 emphasizing that MEK-ERK signaling pathway blockade accelerates DC maturation and increases proinflammatory cytokine production.5,7,32 We now report that the MEK-ERK signaling helps in the maintenance of the immature state of MDDCs and that AhR mediates some of the ERK-dependent transcriptional effects.
The role of ERK in maintaining immature DC homeostasis agrees well with its positive effect on the expression of CCL2, a chemokine whose deficiency impairs IL-10 production by macrophages and DCs,33 as well as with the upregulation of maturation-associated functions/markers upon MEK-ERK inhibition. Of note, U0126 upregulates CCR7 expression and CCL19-directed migration, both of which are maturation-associated properties.1 The U0126-augmented CCL19-dependent migratory response of MDDCs implies a role for MEK-ERK in DC retention within peripheral tissues and suggests that ERK inhibition would endow DCs with a higher capacity for homing to draining lymph nodes. Since U0126 also enhances CXCL16, a chemokine that mediates activated T-lymphocyte migration,34 MEK-ERK inhibition in DCs would enhance the chances for DC–T-cell encounters within the lymph node. Conversely, inhibition of MEK-ERK increases the expression of genes implicated in LDL binding.35 The capture of ac-LDL reflects antigen uptake/endocytic ability,36 a defining property of immature DCs. However, an increased ability to capture antigens via receptor-mediated endocytosis has also been demonstrated in mature DCs37 and has been proposed to confer mature DCs with the ability to capture and present specific antigens and pathogens previously recognized by the immune system. Therefore, the elevated antigen-capture ability and the increased CCL19-directed migration would predict that ERK-defective DCs exhibit an increased ability to promote effector T-cell responses within the lymph node.
The analysis of the proximal regulatory regions of MEK-ERK–dependent genes revealed a significant enrichment in DNA elements recognized by NFκB, a master regulator of the DC maturation process,38 Egr1, Klf4, and AhR. The latter is a ligand-activated transcription factor involved in the metabolic response to environmental contaminants such as dioxins (e.g., 2,3,7,8-tetrachlorodibenzo-p-dioxin [TCDD]) and whose deficiency deregulates cell proliferation, differentiation, and immune homeostasis.39-41 Our results demonstrate that MEK inhibition increases AhR transcriptional activity in MDDCs because (1) the AhR partial antagonist α-NF42 impairs U0126-induced gene upregulation; (2) total AhR protein levels are reduced after U0126 treatment (Figure 6C), in agreement with the fast 26S proteasome-dependent degradation of activated AhR;43 (3) U0126 enhances the activity of an AhR-dependent reporter construct in MDDCs; and (4) U0126 treatment leads to in vivo occupancy of AhR-binding DNA elements. Therefore, AhR mediates, at least in part, the changes in gene expression that take place in MDDCs upon exposure to MEK-ERK inhibitors.
Whereas ERK inhibition upregulated AhR-dependent transcription in in vitro generated monocyte-derived cells (MDDCs and macrophages), α-NF did not inhibit the U0126-induced upregulation of AhR-target genes in peripheral blood CD1c+ DCs. The cell-type specificity of the ERK–AhR link may reflect the distinct developmental origin (monocyte vs common DC progenitor) of MDDCs and CD1c+ DCs44 that display opposite immunoregulatory functions in response to E. coli.45 Additionally, MDDCs and CD1c+ DCs also differ in their cytokine requirements (GM-CSF + IL-4 vs Flt3).44 Thus, although the transcriptional profile of in vitro–generated MDDCs is more related to that of CD1c+ blood DCs than to other primary DC populations,46 the gene expression signature of MDDCs is also considerably skewed by IL-4 and, interestingly, it reveals a significantly higher expression of the prototypical AhR-target gene CYP1B1 than other in vitro–generated DC populations.46 Therefore, it is conceivable that differentiation-driving cytokines might determine the level of expression or activation of AhR in CD1c+ DCs and MDDCs, thus explaining the cell-context dependency of the ERK–AhR link that we have observed.
Our results are compatible with previous reports on the contribution of AhR to immune and inflammatory responses, which are dependent on the activating ligand and the cellular context. AhR exhibits anti-inflammatory and immunosuppressive roles in several settings;47 AhR agonists have immunosuppressive effects48 and repress NFκB-responsive genes, and AhR deficiency enhances proinflammatory cytokine production by murine macrophages.47 However, AhR ligands can also transactivate NFκB-responsive genes49 and promote Th17 polarization.47 These dual effects of AhR have also been described for DCs, in which AhR agonists increase MHC-II and CD80 and CD86 levels; impair the LPS-induced secretion of IL-6, tumor necrosis factor alpha, IL-10, and IL-12;47 and negatively regulate DC immunogenicity.50 Thus, our findings demonstrate that AhR mediates part of the MEK-ERK-dependent maintenance of the immature state of MDDCs. Since MEK-ERK inhibitors are currently tested in cancer clinical trials, our observations may contribute to the development of novel therapeutic strategies that consider the modulation of intratumoral DC maturation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Ministerio de Ciencia e Innovación (SAF2011-23801), Genoma España (Molecular and Cellular Mechanisms in Chronic Inflammatory and Autoimmune Diseases project), Instituto de Salud Carlos III (Spanish Network for the Research in Infectious Diseases REIPI RD06/0008, and AIDS Research Network RIS RD06/0006/1016), and Programa de Actividades de I + D de la Comunidad de Madrid (RAPHYME).
Authorship
Contribution: N.A.-M., S.C., C.N., O.M.P., B.A., and A.D.-S. performed research; F.S.-C. and A.D. performed the statistical analysis of microarray data; P.M.F.-S., J.L.R.-F., O.M.P., V.A., A.C. and S.S.-R. provided crucial reagents; N.A.-M., S.C., and A.L.C. designed research and contributed to the preparation of the manuscript; and A.L.C. conceived the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Angel L. Corbí, Centro de Investigaciones Biológicas, CSIC, Ramiro de Maeztu, 9, Madrid 28040 Spain; e-mail: acorbi@cib.csic.es
References
Author notes
N.A.-M., S.C., and C. N. contributed equally to this study.