Key Points
Bone marrow LGALS3 expression is associated with distinct clinical and biological features in patients with acute myeloid leukemia.
Higher bone marrow LGALS3 expression is an independent poor prognostic factor for overall survival and may serve as a potential therapeutic target.
Abstract
Alterations of galectin-3 expression are often seen in cancers and may contribute to tumorigenesis, cancer progression, and metastasis. The studies concerning clinical implications of galectin-3 expression in patients with acute myeloid leukemia (AML) are scarce. We investigated the expression of LGALS3, the gene encoding galectin-3, in the bone marrow (BM) mononuclear cells from an original cohort comprising 280 adults with primary non-acute promyelocytic leukemia. Higher LGALS3 expression was closely associated with older age, French-American-British M4/M5 subtypes, CD14 expression on leukemic cells, and PTPN11 mutation, but negatively correlated with CEBPA mutation and FLT3-ITD. Compared with patients with lower LGALS3 expression, those with higher expression had lower complete remission rates, higher primary refractory rates, and shorter overall survival. This result was validated in an independent validation cohort. A scoring system incorporating higher LGALS3 expression and 8 other risk factors, including age, white blood cell count, cytogenetics, and gene mutations, into survival analysis proved to be very useful to stratify patients with AML into different prognostic groups (P < .001). In conclusion, BM LGALS3 expression may serve as a new biomarker to predict clinical outcome in patients with AML, and galectin-3 may serve as a potential therapeutic target in those patients with higher expression of this protein.
Introduction
Recent research has demonstrated that change in glycan structures and the interaction of these structures with endogenous carbohydrate-binding proteins, known as lectins, play an important role in tumor progression.1,2 Galectins belong to a family of animal lectins characterized by their specific ability to recognize β-galactoside-containing carbohydrates and the evolutionary conserved amino acid sequences3 and are widely distributed from nematodes to mammals. All galectins contain highly conserved carbohydrate-recognition domains that are responsible for carbohydrate binding.4 At this time, 15 mammalian galectins have been identified.
Galectin-3, encoded by LGALS3 and located on chromosome 14 q21-q22 in human, is one of the galectin family members. It contains a single carbohydrate-recognition domain that is connected to an extended N-terminal proline- and glycine-rich domain. Galectin-3 is widely expressed in human tissues, including epithelial cells,5 myeloid cells, and immune cells.6,7 Increasing evidence demonstrates that the expression of galectin-3 is associated with cell adhesion, migration, proliferation, and apoptosis.8,9 Alterations of galectin-3 expression are often seen in cancers and may contribute to tumorigenesis, cancer progression, angiogenesis, and metastasis.1,10,11
Increased galectin-3 expression has been shown to be correlated with tumor progressions12 and poor prognosis in some cancers, such as hepatocellular carcinoma13 and primary cutaneous melanoma.14 For diffuse large B-cell lymphomas, galectin-3 is an antiapoptotic factor,15 and its overexpression is associated with a worse prognosis.16 In addition, it may promote drug resistance in chronic myeloid leukemia.17 In contrast, decreased galectin-3 expression is associated with tumor progressions18 and poor prognosis in other cancers, such as gastric cancer19 and clear cell renal carcinoma.20
To date, little is known about the clinical implication of galectin-3 expression in acute myeloid leukemia (AML). In this study, we investigated the bone marrow (BM) LGALS3 expression by real-time quantitative polymerase chain reaction (RQ-PCR) in a cohort of 280 adults with de novo non-acute promyelocytic leukemia (non-M3 AML) and correlated the results with clinical features and outcomes of the patients. To the best of our knowledge, this is the first report to address the prognostic implication of LGALS3 expression in patients with AML. We found that higher BM LGALS3 expression is an independent unfavorable prognostic factor for overall survival (OS) in these patients, and the finding was also validated in an independent validation cohort.
Materials and methods
Patients and samples
An original cohort comprised 280 adults 15 years of age or older with newly diagnosed de novo non-M3 AML at the National Taiwan University Hospital (NTUH) from July 1994 to December 2007 who had complete clinical data and enough cryopreserved BM samples for analysis. Thirty normal marrow donors were also enrolled for comparison. Diagnosis and classification of AML were made according to French-American-British (FAB) Cooperative Group Criteria. Patients with antecedent hematological diseases or therapy-related AML were excluded. The expression of BM LGALS3 was determined before treatment. Among the 280 patients, 211 (75.4%) received standard induction chemotherapy (idarubicin 12 mg/m2 per day on days 1 to 3 and cytarabine 100 mg/m2 per day on days 1 to 7) and then consolidation chemotherapy with 2 to 4 courses of high-dose cytarabine (2000 mg/m2 every 12 hours from days 1 to 4; total, 8 doses), with or without an anthracycline (idarubicin or mitoxantrone), after achieving complete remission (CR).21,22 Seventy patients received allogeneic hematopoietic stem cell transplantation (HSCT): 24 in CR1, 34 in CR2 or beyond, and 12 in relapsed/refractory status. The remaining 69 patients received palliative therapy with supportive care and/or low-dose chemotherapy because of underlying comorbidity or based on the decision of the patients. The validation cohort comprised 42 adults 15 years of age or older with newly diagnosed non-M3 AML at NTUH from Jan 2008 to June 2009. These patients were treated with the same regimens as those in the original cohort and were used to validate the prognostic effect of BM LGALS3 expression in AML. This study was approved by the Institutional Review Board of the NTUH and was done in accordance with the Declaration of Helsinki. All patients and normal donors signed the informed consents.
RQ-PCR
BM mononuclear cells from both cohorts and 30 healthy transplantation donors were isolated and cryopreserved until use. Total RNA was extracted and reverse transcribed. The gene expression level was quantified using TaqMan technology on the Applied Biosystems 7500/7500 Fast Real-Time PCR System, as previously described.23 Gene-specific primers and probe of LGALS3 were available as TaqMan Gene Expression Assay (Assay id, Hs00173587_m1; Applied Biosystems). Each sample was tested at least twice independently. The amount of the target gene was normalized to the housekeeping gene RPLP0. The copies of target gene were quantified only after successful amplification of the internal control, using the standard curves derived from cloned plasmids. All data were presented as log ratio of the target gene/RPLP0.
IHC for galectin-3 protein
We performed immunohistochemical staining (IHC) to assess the galectin-3 protein expression in the BM biopsy specimens from 30 selected patients with AML in the original cohort, with 15 having higher LGALS3 mRNA expression and another 15 having lower LGALS3 mRNA expression. The tissue section was prepared from formalin-fixed, paraffin-embedded tissue. After deparaffinization and rehydration, as previously described,23 the slides were boiled in 0.01 mol/L citrate buffer at pH 6.1 for 15 minutes to retrieve the antigens. The endogenous peroxidase activity was blocked by incubation with 3% H2O2 in methanol for 30 minutes. Then the sections were blocked with 10% normal horse serum (VECTASTAIN Elite ABC kit; Vector Laboratories). The primary antibody we used was mouse monoclonal antihuman galectin-3 (clone 9C4, Neo Markers; Laboratory Vision Corporation) at a 1:40 dilution. A negative, no-antibody control was included for each staining. The slides were incubated with a biotinylated secondary antibody (1:200 dilution; horse antimouse IgG; VECTASTAIN Elite ABC kit) for 60 minutes at room temperature, and AB reagent was applied based on the manufacturer's instructions (VECTASTAIN Elite ABC kit). The antigen detection was conducted by a color reaction with 3,3′-diaminobenzidine (DAB Peroxidase Substrate Kit; Vector Laboratories). Finally, the sections were counterstained with hematoxylin and mounted. Galectin-3 expression was assessed by staining intensity and frequency of positively stained cells. A score of 0 to 4 was calculated for each specimen, according to the addition of the score of staining intensity (0 = none, 1 = weak, 2 = strong) and the score of percentage of myeloid cells positively stained (0 = 0∼10%, 1 = 11∼50%, 2 = 51∼100%) by the pathologists, who were blind to the results of LGALS3 mRNA expression. Erythroid cells were excluded from calculation.
ELISA for protein determination
Enzyme-linked immunosorbent assay (ELISA) was performed to measure concentrations of the galectin-3 protein in BM plasma from 22 patients in the original cohort, using a previously described method.24 One-step sandwich enzyme immunoassay was done using commercially available kits with monoclonal antibody to galectin-3 protein (catalog number DGAL30; R&D Systems).
Immunophenotyping
A panel of monoclonal antibodies to myeloid associated antigens (CD13, CD33, CD11b, CD15, CD14, and CD41a), lymphoid-associated antigens (CD2, CD5, CD7, CD19, CD10, and CD20), and lineage-nonspecific antigens (HLA-DR, CD34, and CD56) was used to characterize the immunophenotypes of leukemia cells as previously described.25
Cytogenetic study
BM cells were harvested directly or after 1 to 3 days of unstimulated culture as described previously.26 The metaphase cells were banded by trypsin-Giemsa technique and karyotyped according to the International System for Human Cytogenetic Nomenclature.
Mutation analysis
Mutation analysis of 14 relevant molecular marker genes, including NPM1,27 CEBPA,28 FLT3-ITD,29 FLT3-TKD,30 RAS,31 KIT,32 MLL-PTD,33 PTPN11,34 WT1,22 AML1/RUNX1,21 ASXL1,35 IDH1,36 IDH2,37 and TET238 was performed as previously reported. Abnormal sequencing results were confirmed by at least 2 repeated analyses.
Statistical analysis
The discrete variables of patients with lower and higher LGALS3 expression were compared using the χ-square tests or Fisher’s exact test. We used the Mann-Whitney U-test to compare continuous variables and medians of distributions. Correlation between the LGALS3 mRNA expression and its protein expression was accessed by the Spearman’s rank correlation. OS was measured from the date of first diagnosis to death from any cause or the last follow-up, and relapse was defined as a reappearance of at least 5% leukemic blasts in a BM aspirate or new extramedullary leukemia in patients with a previously documented CR.39 Disease-free status indicated that the patient achieved CR and did not relapse by the end of this study, and disease-free survival was defined as the time from recruitment to the first of 3 events: treatment failure, leukemia relapse, or death from any cause.21,39 To exclude confounding influences of different treatment regimens, patients who received allogeneic HSCT were censored on the day of cell infusion.22 We adopted Kaplan-Meier estimation to plot survival curves and used log-rank tests to examine the difference between groups. Hazard ratio and 95% confidence interval were estimated by Cox proportional hazards regression models to determine independent risk factors associated with survival in multivariate analyses. Two-sided P values less than 0.05 were considered statistically significant. The whole patient population was included for analyses of the correlation between LGALS3 expression and clinical characteristics; however, only those receiving conventional standard chemotherapy, as mentioned above, were included in analyses of survivals. All statistical analyses were accomplished with the SPSS 17 (SPSS Inc.) and Statsdirect (2.7.8b, 2011).
Results
Correlation of BM LGALS3 expression with clinical features and laboratory data in the original cohort
LGALS3 expression in BM mononuclear cells of the patients with AML, as determined by RQ-PCR, varied greatly and overlapped with that of normal controls (supplemental Figure 1). The median value of BM LGALS3 expression in the total number of patients with AML was used as the cutoff point to define lower- and higher-expression groups. The comparison of clinical characteristics of patients with lower and higher BM LGALS3 expression is shown in Table 1. Patients with higher BM LGALS3 expression were older (median, 59 vs 45.5 years; P < .001) and had lower blast counts and serum lactate dehydrogenase levels than those with lower expression. Compared with patients with lower BM LGALS3 expression, those with higher expression more frequently had FAB M4 and M5 subtypes (both P = .003) and CD14 expression on leukemic cells but less frequently had FAB M1 subtype (P < .001) and CD7 expression on leukemic cells (supplemental Table 1). There was no difference in other variables including sex, hemoglobin levels, and platelet counts between the 2 groups.
Comparison of clinical manifestations and laboratory features between patients with AML with lower and higher LGALS3 expression
| Variables . | Total (n = 280) . | Lower LGALS3 expression (n = 140) . | Higher LGALS3 expression (n = 140) . | P value . |
|---|---|---|---|---|
| Sex* | .81 | |||
| Male | 157 | 80 (51) | 77 (49) | |
| Female | 123 | 60 (48.8) | 63 (51.2) | |
| Age (years)† | 50 (15-90) | 45.5 (15-85) | 59 (15-90) | <.001 |
| Laboratory data† | ||||
| WBC (/μL) | 23 855 (650-423 000) | 28 390 (1530-324 000) | 21 810 (650-423 000) | .051 |
| Hemoglobin (g/dL) | 8 (3.3-16.2) | 8 (3.9-13.1) | 8.1 (3.3-16.2) | .664 |
| Platelet (×1000 /μL) | 45 (3-802) | 43 (5-712) | 46 (3-802) | .746 |
| Blast (/μL) | 12 621 (0-369 070) | 17 165 (0-245 592) | 7002 (0-369 070) | .002 |
| LDH (U/L) | 882 (274-13 130) | 1005 (284-13 130) | 792 (274-8116) | .011 |
| FAB* | ||||
| M0 | 5 | 3 (60) | 2 (40) | >.999 |
| M1 | 66 | 51 (77.3) | 15 (22.7) | <.001 |
| M2 | 98 | 52 (53.1) | 46 (46.9) | .531 |
| M4 | 84 | 30 (35.7) | 54 (64.3) | .003 |
| M5 | 16 | 2 (12.5) | 14 (87.5) | .003 |
| M6 | 4 | 2 (50) | 2 (50) | >.999 |
| Undetermined | 7 | 0 (0) | 7 (100) | .014 |
| Induction response*,‡ | 211 | 120 | 91 | |
| CR | 155 | 99 (82.5) | 56 (61.5) | .001 |
| PR | 8 | 5 (4.2) | 3 (3.3) | >.999 |
| Refractory | 34 | 13 (10.8) | 21 (23.1) | .023 |
| Induction death | 14 | 3 (2.5) | 11 (12.1) | .01 |
| Allo-HSCT | 70 | 45 (37.5) | 25 (27.5) | .141 |
| CR1 | 24 | 15 | 9 | >.999 |
| ≥CR2 | 34 | 25 | 9 | .14 |
| Primary refractory | 12 | 5 | 7 | .1 |
| Variables . | Total (n = 280) . | Lower LGALS3 expression (n = 140) . | Higher LGALS3 expression (n = 140) . | P value . |
|---|---|---|---|---|
| Sex* | .81 | |||
| Male | 157 | 80 (51) | 77 (49) | |
| Female | 123 | 60 (48.8) | 63 (51.2) | |
| Age (years)† | 50 (15-90) | 45.5 (15-85) | 59 (15-90) | <.001 |
| Laboratory data† | ||||
| WBC (/μL) | 23 855 (650-423 000) | 28 390 (1530-324 000) | 21 810 (650-423 000) | .051 |
| Hemoglobin (g/dL) | 8 (3.3-16.2) | 8 (3.9-13.1) | 8.1 (3.3-16.2) | .664 |
| Platelet (×1000 /μL) | 45 (3-802) | 43 (5-712) | 46 (3-802) | .746 |
| Blast (/μL) | 12 621 (0-369 070) | 17 165 (0-245 592) | 7002 (0-369 070) | .002 |
| LDH (U/L) | 882 (274-13 130) | 1005 (284-13 130) | 792 (274-8116) | .011 |
| FAB* | ||||
| M0 | 5 | 3 (60) | 2 (40) | >.999 |
| M1 | 66 | 51 (77.3) | 15 (22.7) | <.001 |
| M2 | 98 | 52 (53.1) | 46 (46.9) | .531 |
| M4 | 84 | 30 (35.7) | 54 (64.3) | .003 |
| M5 | 16 | 2 (12.5) | 14 (87.5) | .003 |
| M6 | 4 | 2 (50) | 2 (50) | >.999 |
| Undetermined | 7 | 0 (0) | 7 (100) | .014 |
| Induction response*,‡ | 211 | 120 | 91 | |
| CR | 155 | 99 (82.5) | 56 (61.5) | .001 |
| PR | 8 | 5 (4.2) | 3 (3.3) | >.999 |
| Refractory | 34 | 13 (10.8) | 21 (23.1) | .023 |
| Induction death | 14 | 3 (2.5) | 11 (12.1) | .01 |
| Allo-HSCT | 70 | 45 (37.5) | 25 (27.5) | .141 |
| CR1 | 24 | 15 | 9 | >.999 |
| ≥CR2 | 34 | 25 | 9 | .14 |
| Primary refractory | 12 | 5 | 7 | .1 |
The median value of LGALS3 expression in total population was used as the cutoff point to define lower- and higher-expression groups.
PR, partial remission.
Number (%) of patients.
Median (range).
Only the 211 patients who received conventional intensive induction chemotherapy, and then consolidation chemotherapy if CR was achieved, were included in the analysis.
Correlation between LGALS3 RNA expression and protein expression in the original cohort
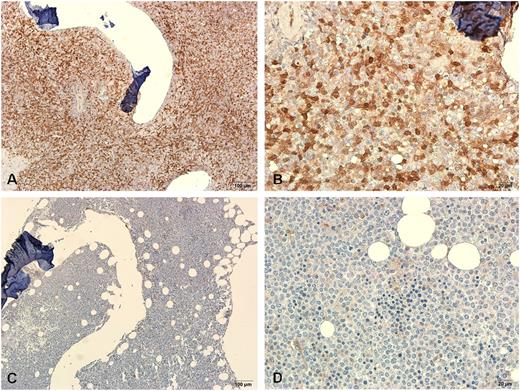
Galectin-3 protein expression measured by scoring IHC of BM biopsy specimens correlated well with LGALS3 mRNA expression in the 30 patients studied (P = .043 by Spearman’s rank correlation). Representative IHC of a sample with a high score and another with a low score was demonstrated in Figure 1. Further, the scatter plot of the galectin-3 protein levels in BM plasma measured by ELISA vs log-transformed mRNA expression of LGALS3 for all 22 patients tested showed close association of the 2 (supplemental Figure 2; P = .003 by Spearman’s rank correlation).
Representative immunohistochemical stainings of galectin-3 protein in BM biopsy specimens from two patients. One specimen from a patient with higher BM LGALS3 mRNA expression showed strong staining (A-B) and another one from a patient with lower BM LGALS3 mRNA expression showed weak staining (C-D). Magnification ×100 and ×400, respectively.
Representative immunohistochemical stainings of galectin-3 protein in BM biopsy specimens from two patients. One specimen from a patient with higher BM LGALS3 mRNA expression showed strong staining (A-B) and another one from a patient with lower BM LGALS3 mRNA expression showed weak staining (C-D). Magnification ×100 and ×400, respectively.
Correlation of BM LGALS3 expression with karyotype and molecular gene mutations in the original cohort
Chromosome data were available in 267 patients at diagnosis. The comparison of karyotypes of patients with lower and higher BM LGALS3 expression is shown in supplemental Table 2. There was no association of BM LGALS3 expression with any chromosome change. Regarding the molecular gene mutations (Table 2), higher BM LGALS3 expression was closely associated with PTPN11 mutation (10% vs 1.4%; P = .003) and showed a trend to be associated with TET2 mutation (17.9% vs 9.3%; P = .054) but was inversely correlated with FLT3-ITD (19.3% vs 31.4%; P = .028) and CEBPA mutations (8.6% vs 19.3%; P = .015).
Comparison of genetic alterations between patients with AML who have lower and higher LGALS3 expression
| Variant . | Number (%) of patients with the gene mutation . | P value . | ||
|---|---|---|---|---|
| Whole cohort (n = 280) . | Lower LGALS3 expression (n = 140) . | Higher LGALS3 expression (n = 140) . | ||
| CEBPA | 39 (13.9) | 27 (19.3) | 12 (8.6) | .015 |
| CEBPAsingle-mut | 11 (3.9) | 8 (5.7) | 3 (2.1) | .125 |
| CEBPAdouble-mut | 28 (10) | 19 (13.6) | 9 (6.4) | .045 |
| FLT3-ITD | 71 (25.4) | 44 (31.4) | 27 (19.3) | .028 |
| NPM1 | 74 (26.4) | 35 (25) | 39 (27.9) | .684 |
| FLT3-TKD | 21 (7.5) | 11 (7.9) | 10 (7.1) | >.999 |
| PTPN11 | 16 (5.7) | 2 (1.4) | 14 (10) | .003 |
| RAS | 42 (15) | 17 (12.1) | 25 (17.9) | .241 |
| MLL-PTD | 15 (5.4) | 6 (4.3) | 9 (6.4) | .597 |
| KIT | 9 (3.2) | 5 (3.6) | 4 (2.9) | .75 |
| WT1 | 21 (7.5) | 12 (8.6) | 9 (6.4) | .651 |
| AML1/RUNX1 | 37 (13.2) | 13 (9.3) | 24 (17.1) | .077 |
| ASXL1 | 29 (10.4) | 16 (11.4) | 13 (9.3) | .695 |
| IDH1 | 17 (6.1) | 12 (8.6) | 5 (3.6) | .131 |
| IDH2 | 39 (13.9) | 17 (12.1) | 22 (15.7) | .49 |
| TET2 | 38 (13.6) | 13 (9.3) | 25(17.9) | .054 |
| Variant . | Number (%) of patients with the gene mutation . | P value . | ||
|---|---|---|---|---|
| Whole cohort (n = 280) . | Lower LGALS3 expression (n = 140) . | Higher LGALS3 expression (n = 140) . | ||
| CEBPA | 39 (13.9) | 27 (19.3) | 12 (8.6) | .015 |
| CEBPAsingle-mut | 11 (3.9) | 8 (5.7) | 3 (2.1) | .125 |
| CEBPAdouble-mut | 28 (10) | 19 (13.6) | 9 (6.4) | .045 |
| FLT3-ITD | 71 (25.4) | 44 (31.4) | 27 (19.3) | .028 |
| NPM1 | 74 (26.4) | 35 (25) | 39 (27.9) | .684 |
| FLT3-TKD | 21 (7.5) | 11 (7.9) | 10 (7.1) | >.999 |
| PTPN11 | 16 (5.7) | 2 (1.4) | 14 (10) | .003 |
| RAS | 42 (15) | 17 (12.1) | 25 (17.9) | .241 |
| MLL-PTD | 15 (5.4) | 6 (4.3) | 9 (6.4) | .597 |
| KIT | 9 (3.2) | 5 (3.6) | 4 (2.9) | .75 |
| WT1 | 21 (7.5) | 12 (8.6) | 9 (6.4) | .651 |
| AML1/RUNX1 | 37 (13.2) | 13 (9.3) | 24 (17.1) | .077 |
| ASXL1 | 29 (10.4) | 16 (11.4) | 13 (9.3) | .695 |
| IDH1 | 17 (6.1) | 12 (8.6) | 5 (3.6) | .131 |
| IDH2 | 39 (13.9) | 17 (12.1) | 22 (15.7) | .49 |
| TET2 | 38 (13.6) | 13 (9.3) | 25(17.9) | .054 |
Effect of BM LGALS3 expression on response to therapy and clinical outcome in the original cohort
Of the 211 patients with AML undergoing conventional intensive induction chemotherapy, 155 (73.5%) patients achieved a CR (Table 1). Patients with higher BM LGALS3 expression, compared with those with lower expression, had lower CR rates (61.5% vs 82.5%; P = .001) and higher primary refractory rates (23.1% vs 10.8%; P = .023). However, the relapse rate was similar between the 2 groups once CR was achieved (59% vs 44.6%; P = .096). After a median follow-up time of 69.5 months (range, 1.25 to 182 months), patients with higher BM LGALS3 expression had a significant shorter OS than those with lower expression (median, 16.3 vs 39.8 months; P = .02) (Table 3; Figure 2A). There was no significant difference of disease-free survival between these 2 groups (P = .811). We further separated the total patients into 4 quartiles on the basis of the expression of BM LGALS3 levels. The Kaplan-Meier curve for OS showed that patients in the highest quartile of LGALS3 expression had the worst prognosis, whereas those in the lowest quartile had the best prognosis (Figure 2B; P = .004). These findings made the prognostic relevance of BM LGALS3 expression more credible. In the subgroup analysis of the 109 patients with normal karyotype (cytogenetically normal; CN-AML), the difference in OS between patients with higher and lower BM LGALS3 expression became even more obvious (median, 17 vs 95 months; P = .008; Figure 2C).
Univariate analysis of the effect of clinical parameters, BM LGALS3 expression, and molecular alterations on overall survival in patients with AML
| Variable . | No. of patients . | Overall survival (months) . | |
|---|---|---|---|
| Median* . | P value . | ||
| Age, years | .001 | ||
| ≤50 | 133 | 39.8 | |
| >50 | 78 | 12.3 ± 3 | |
| Sex | .677 | ||
| Male | 110 | 25 ± 7.7 | |
| Female | 101 | 18 ± 5.3 | |
| WBC | .046 | ||
| ≤50000/μL | 149 | 26 ± 16.6 | |
| >50000/μL | 62 | 15.5 ± 5.2 | |
| Karyotype† | .013 | ||
| Favorable | 33 | NR | |
| Intermediate | 152 | 22 ± 9.5 | |
| Unfavorable | 18 | 9.5 ± 1.5 | |
| LGALS3 | .02 | ||
| Lower expression | 120 | 39.8 ± 14.4 | |
| Higher expression | 91 | 16.3 ± 3.4 | |
| CEBPA | .008 | ||
| Double mutation | 28 | NR | |
| Others | 183 | 19 ± 3 | |
| NPM1+/ FLT3-ITD- | .038 | ||
| Yes | 26 | 95 | |
| Others | 185 | 22 ± 3.4 | |
| AML1/RUNX1 | .001 | ||
| Mutated | 21 | 8 ± 1 | |
| Wild | 190 | 25.5 ± 14 | |
| WT1 | .024 | ||
| Mutated | 21 | 11 ± 1.4 | |
| Wild | 190 | 25 ± 15.4 | |
| MLL-PTD | .004 | ||
| Yes | 10 | 10.5 ± 6.1 | |
| No | 201 | 25 ± 7.6 | |
| RAS | .89 | ||
| Mutated | 33 | 17 ± 7.3 | |
| Wild | 178 | 23.5 ± 7 | |
| PTPN11 | .21 | ||
| Mutated | 7 | 17.5 ± 13.6 | |
| Wild | 204 | 23.5 ± 6.8 | |
| FLT3-TKD | .542 | ||
| Yes | 17 | 39.8 | |
| No | 194 | 23 ± 3.4 | |
| KIT | .525 | ||
| Mutated | 8 | 12 ± 1.9 | |
| Wild | 203 | 25 ± 7.3 | |
| ASXL1 | .697 | ||
| Mutated | 14 | 26 ± 5.7 | |
| Wild | 197 | 23 ± 7.6 | |
| IDH1 | .798 | ||
| Mutated | 15 | 19 ± 1.8 | |
| Wild | 196 | 25 ± 7.4 | |
| IDH2 | .116 | ||
| Mutated | 27 | NR | |
| Wild | 184 | 22 ± 3.4 | |
| TET2 | .547 | ||
| Mutated | 22 | 13 ± 10.9 | |
| Wild | 189 | 23.5 ± 7 | |
| Variable . | No. of patients . | Overall survival (months) . | |
|---|---|---|---|
| Median* . | P value . | ||
| Age, years | .001 | ||
| ≤50 | 133 | 39.8 | |
| >50 | 78 | 12.3 ± 3 | |
| Sex | .677 | ||
| Male | 110 | 25 ± 7.7 | |
| Female | 101 | 18 ± 5.3 | |
| WBC | .046 | ||
| ≤50000/μL | 149 | 26 ± 16.6 | |
| >50000/μL | 62 | 15.5 ± 5.2 | |
| Karyotype† | .013 | ||
| Favorable | 33 | NR | |
| Intermediate | 152 | 22 ± 9.5 | |
| Unfavorable | 18 | 9.5 ± 1.5 | |
| LGALS3 | .02 | ||
| Lower expression | 120 | 39.8 ± 14.4 | |
| Higher expression | 91 | 16.3 ± 3.4 | |
| CEBPA | .008 | ||
| Double mutation | 28 | NR | |
| Others | 183 | 19 ± 3 | |
| NPM1+/ FLT3-ITD- | .038 | ||
| Yes | 26 | 95 | |
| Others | 185 | 22 ± 3.4 | |
| AML1/RUNX1 | .001 | ||
| Mutated | 21 | 8 ± 1 | |
| Wild | 190 | 25.5 ± 14 | |
| WT1 | .024 | ||
| Mutated | 21 | 11 ± 1.4 | |
| Wild | 190 | 25 ± 15.4 | |
| MLL-PTD | .004 | ||
| Yes | 10 | 10.5 ± 6.1 | |
| No | 201 | 25 ± 7.6 | |
| RAS | .89 | ||
| Mutated | 33 | 17 ± 7.3 | |
| Wild | 178 | 23.5 ± 7 | |
| PTPN11 | .21 | ||
| Mutated | 7 | 17.5 ± 13.6 | |
| Wild | 204 | 23.5 ± 6.8 | |
| FLT3-TKD | .542 | ||
| Yes | 17 | 39.8 | |
| No | 194 | 23 ± 3.4 | |
| KIT | .525 | ||
| Mutated | 8 | 12 ± 1.9 | |
| Wild | 203 | 25 ± 7.3 | |
| ASXL1 | .697 | ||
| Mutated | 14 | 26 ± 5.7 | |
| Wild | 197 | 23 ± 7.6 | |
| IDH1 | .798 | ||
| Mutated | 15 | 19 ± 1.8 | |
| Wild | 196 | 25 ± 7.4 | |
| IDH2 | .116 | ||
| Mutated | 27 | NR | |
| Wild | 184 | 22 ± 3.4 | |
| TET2 | .547 | ||
| Mutated | 22 | 13 ± 10.9 | |
| Wild | 189 | 23.5 ± 7 | |
Two hundred eleven patients with non-M3 AML who received conventional intensive induction chemotherapy, and then consolidation chemotherapy if CR was achieved, were included in the analysis.
NR, not reached.
Median ± standard deviation.
Eight patients without chromosome data at diagnosis were excluded for analysis. Favorable, t(8;21), inv(16); unfavorable, −7, del(7q), −5, del(5q), 3q abnormality, complex abnormalities; intermediate, normal karyotype and other abnormalities.
Kaplan–Meier survival curves for overall survival stratified by BM LGALS3 mRNA expression in the original cohort. In 211 non-M3 AML patients who received conventional intensive chemotherapy, patients with higher LGALS3 expression had shorter OS (A). The survival curves stratified by LGALS3 mRNA expression into four quartiles showing patients in the highest (4th) quartile of LGALS3 expression had the worst prognosis, whereas those in the lowest (1st) quartile had the best prognosis (B). In the subgroup of 109 patients with CN-AML, patients with higher LGALS3 expression had shorter OS (C). The median value of BM LGALS3 expression in the original cohort of 280 patients was used as the cutoff point to define lower- and higher-expression groups (A and C).
Kaplan–Meier survival curves for overall survival stratified by BM LGALS3 mRNA expression in the original cohort. In 211 non-M3 AML patients who received conventional intensive chemotherapy, patients with higher LGALS3 expression had shorter OS (A). The survival curves stratified by LGALS3 mRNA expression into four quartiles showing patients in the highest (4th) quartile of LGALS3 expression had the worst prognosis, whereas those in the lowest (1st) quartile had the best prognosis (B). In the subgroup of 109 patients with CN-AML, patients with higher LGALS3 expression had shorter OS (C). The median value of BM LGALS3 expression in the original cohort of 280 patients was used as the cutoff point to define lower- and higher-expression groups (A and C).
In multivariate analysis (Table 4) including variables significantly associated with clinical outcome in univariate analysis (Table 3), the independent poor risk factors for OS were age older than 50 years, white blood cell (WBC) counts more than 50 000/μL, unfavorable karyotype, higher BM LGALS3 expression, AML1/RUNX1 mutation, WT1 mutation, and MLL-PTD. On the contrary, CEBPAdouble-mut and NPM1+/FLT3-ITD− were independent favorable prognostic factors. Among patients with CN-AML, higher BM LGALS3 expression was still an independent poor prognostic factor for OS (hazard ratio, 2.737; 95% confidence interval, 1.434 to 5.225; P = .002; Table 4).
Multivariate analysis (Cox regression) on the overall survival in patients with AML
| Variables . | Hazard ratio . | 95% confidence interval . | P value . |
|---|---|---|---|
| Non-M3 AML group | |||
| Age* | 2.623 | 1.693-4.062 | <.001 |
| WBC† | 2.414 | 1.517-3.841 | <.001 |
| Karyotype‡ | 3.317 | 1.578-6.972 | .002 |
| LGALS3§ | 1.736 | 1.133-2.662 | .011 |
| NPM1/FLT3-ITD|| | 0.337 | 0.157-0.723 | .005 |
| CEBPA¶ | 0.359 | 0.163-0.788 | .011 |
| AML1/RUNX1 | 2.017 | 1.073-3.789 | .029 |
| MLL-PTD | 3.155 | 1.207-8.245 | .019 |
| WT1 | 2.704 | 1.385-5.279 | .004 |
| Normal karyotype group | |||
| Age* | 3.389 | 1.757-6.536 | <.001 |
| WBC† | 3.01 | 1.56-5.808 | .001 |
| LGALS3§ | 2.737 | 1.434-5.225 | .002 |
| NPM1/FLT3-ITD|| | 0.337 | 0.141-0.806 | .015 |
| CEBPA¶ | 0.257 | 0.096-0.687 | .007 |
| WT1 | 3.208 | 1.228-8.378 | .017 |
| Variables . | Hazard ratio . | 95% confidence interval . | P value . |
|---|---|---|---|
| Non-M3 AML group | |||
| Age* | 2.623 | 1.693-4.062 | <.001 |
| WBC† | 2.414 | 1.517-3.841 | <.001 |
| Karyotype‡ | 3.317 | 1.578-6.972 | .002 |
| LGALS3§ | 1.736 | 1.133-2.662 | .011 |
| NPM1/FLT3-ITD|| | 0.337 | 0.157-0.723 | .005 |
| CEBPA¶ | 0.359 | 0.163-0.788 | .011 |
| AML1/RUNX1 | 2.017 | 1.073-3.789 | .029 |
| MLL-PTD | 3.155 | 1.207-8.245 | .019 |
| WT1 | 2.704 | 1.385-5.279 | .004 |
| Normal karyotype group | |||
| Age* | 3.389 | 1.757-6.536 | <.001 |
| WBC† | 3.01 | 1.56-5.808 | .001 |
| LGALS3§ | 2.737 | 1.434-5.225 | .002 |
| NPM1/FLT3-ITD|| | 0.337 | 0.141-0.806 | .015 |
| CEBPA¶ | 0.257 | 0.096-0.687 | .007 |
| WT1 | 3.208 | 1.228-8.378 | .017 |
Only variables with P value less than .05 in univariate analysis were incorporated into the multivariate Cox proportional hazard regression analysis.
Age older than 50 y relative to age 50 y or younger.
WBC greater than 50 000/μL vs less than or equal to 50 000/μL.
Unfavorable cytogenetics versus others. Eight patients without chromosome data were not included in the analysis.
Higher LGALS3 expression vs lower LGALS3 expression.
NPM1+/FLT3-ITD− vs other subtypes.
CEBPAdouble-mut vs other subtypes.
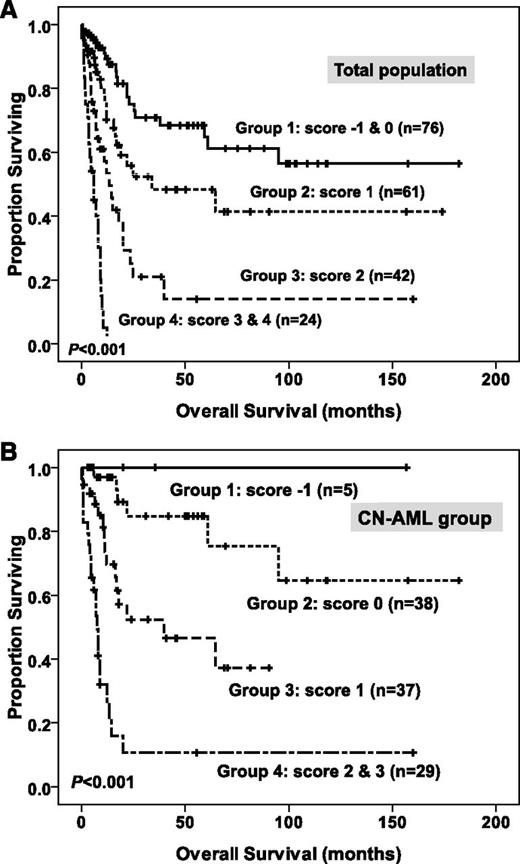
To better stratify these patients into different risk groups, a scoring system incorporating 9 prognostic markers, including age, WBC counts and karyotype at diagnosis, BM LGALS3 expression, NPM1/FLT3-ITD, MLL-PTD, CEBPAdouble-mut, and mutations of AML1/RUNX1 and WT1 into survival analysis was formulated on the basis of the results of our Cox proportional hazards model. A score of +1 was assigned for each parameter associated with an adverse prognosis (older age, high WBC counts, higher BM LGALS3 expression, MLL-PTD, mutation of AML1/RUNX1, and WT1), whereas a score of −1 was assigned for each factor associated with a favorable outcome (CEBPAdouble-mut and NPM1+/FLT3-ITD−). The karyotypes were stratified into 3 groups (unfavorable, +1; intermediate, 0; and favorable, −1). Each patient had a final score by the algebraic summation of these scores. The 8 patients without chromosome data were not included in the analysis. This scoring system divided the patients with AML into 4 groups with different clinical outcomes (P < .001 for OS; Figure 3A). Further, patients with CN-AML could also be stratified into 4 groups with significant different prognosis, using 6 parameters, as previously reported (age, WBC counts, BM LGALS3 expression, NPM1/FLT3-ITD, CEBPAdouble-mut, and WT1 mutation),22,40 based on the scoring system mentioned earlier (P < .001 for OS; Figure 3B).
Kaplan-Meier survival curves for overall survival in patients with non-M3 AML (A) and in patients with CN-AML (B) in the original cohort, according to the scoring system (both P < .001). In the non-M3 AML group, patients were grouped by the scoring system based on BM LGALS3 expression and 8 other prognostic markers (age, karyotype, WBC counts, CEBPAdouble-mut, NPM1/FLT3-ITD, MLL-PTD, and mutations of WT1 and AML1/RUNX1). A score of -1 was assigned for each parameter associated with a favorable outcome (CEBPAdouble-mut and NPM1+/FLT3-ITD-), whereas a score of +1 for each factor was associated with an adverse outcome (older age, high WBC counts at diagnosis, higher BM LGALS3 expression, MLL-PTD, mutations of WT1 and AML1/RUNX1). The karyotypes were stratified into 3 groups (unfavorable, −1; intermediate, 0; and favorable, +1). Each patient had a final score by the algebraic summation of these scores. Eight patients without chromosome data were not included in the analysis. Furthermore, using 6 parameters (age, WBC counts, BM LGALS3 expression, NPM1/FLT3-ITD, CEBPAdouble-mut, and WT1 mutation) based on the scoring system mentioned earlier, patients in the CN-AML group could also easily be stratified into 4 groups with significant different prognosis (B).
Kaplan-Meier survival curves for overall survival in patients with non-M3 AML (A) and in patients with CN-AML (B) in the original cohort, according to the scoring system (both P < .001). In the non-M3 AML group, patients were grouped by the scoring system based on BM LGALS3 expression and 8 other prognostic markers (age, karyotype, WBC counts, CEBPAdouble-mut, NPM1/FLT3-ITD, MLL-PTD, and mutations of WT1 and AML1/RUNX1). A score of -1 was assigned for each parameter associated with a favorable outcome (CEBPAdouble-mut and NPM1+/FLT3-ITD-), whereas a score of +1 for each factor was associated with an adverse outcome (older age, high WBC counts at diagnosis, higher BM LGALS3 expression, MLL-PTD, mutations of WT1 and AML1/RUNX1). The karyotypes were stratified into 3 groups (unfavorable, −1; intermediate, 0; and favorable, +1). Each patient had a final score by the algebraic summation of these scores. Eight patients without chromosome data were not included in the analysis. Furthermore, using 6 parameters (age, WBC counts, BM LGALS3 expression, NPM1/FLT3-ITD, CEBPAdouble-mut, and WT1 mutation) based on the scoring system mentioned earlier, patients in the CN-AML group could also easily be stratified into 4 groups with significant different prognosis (B).
Intriguingly, the unfavorable prognostic effect of higher BM LGALS3 expression on OS was lost among the 70 patients receiving allogeneic HSCT. Furthermore, in patients with higher BM LGALS3 expression, those who received allogeneic HSCT (n = 25) had a trend of better OS than those who did not (median, 48.9 vs 13.5 months; P = .051). It implies that HSCT may ameliorate the poor survival effect of higher BM LGALS3 expression. However, further investigations with more patients recruited are needed to verify this point.
The prognostic effect of BM LGALS3 expression in the validation cohort
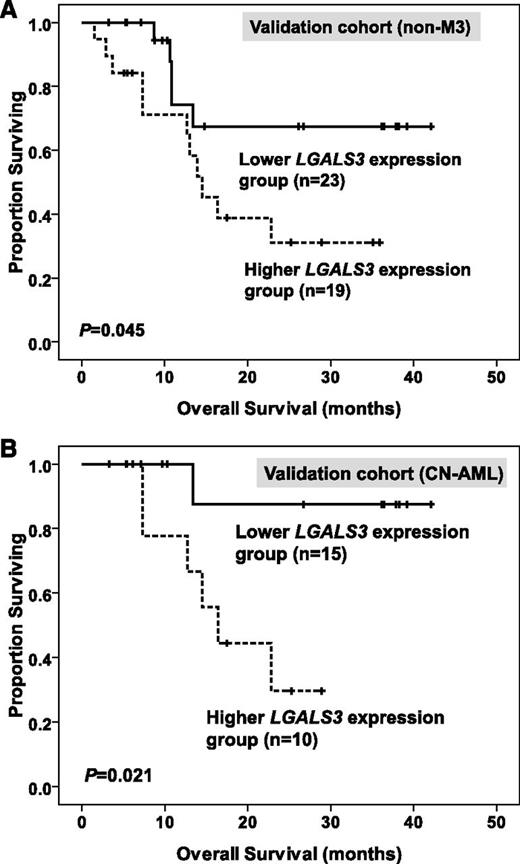
The median value of BM LGALS3 expression in the original cohort was used as a cutoff point to define lower- and higher-expression groups in the validation cohort. After a median follow-up time of 35.8 months (range, 3.3 to 42.1 months), we found that higher BM LGALS3 expression was still a significant unfavorable prognostic factor for OS in total patients and, in the CN-AML group, of the validation cohort (median, 14.5 months vs not reached [P = .045; Figure 4A] and median, 16.4 months vs not reached [P = .021; Figure 4B], respectively).
Kaplan-Meier survival curves for overall survival in 42 patients with non-M3 AML (A) and 25 patients with CN-AML (B) in the validation cohort. The median value of BM LGALS3 expression in the original cohort of 280 patients was used as the cutoff point to define lower- and higher-expression groups.
Kaplan-Meier survival curves for overall survival in 42 patients with non-M3 AML (A) and 25 patients with CN-AML (B) in the validation cohort. The median value of BM LGALS3 expression in the original cohort of 280 patients was used as the cutoff point to define lower- and higher-expression groups.
Discussion
A variety of studies demonstrated a role of galectin-3 expression in tumor progression,12,18 including hematological malignancies.15,17,41 However, little is known about the prognostic implication of its expression in AML. To the best of our knowledge, this is the first report to demonstrate that higher expression of BM LGALS3, the gene encoding human galectin-3, is an independent poor prognostic factor for OS in patients with AML, irrespective of age, WBC counts, karyotype, and other genetic markers. The poor prognostic implication of higher BM LGALS3 expression on OS could also be demonstrated in patients with CN-AML.
Previous studies concerning the relationship between the expression of galectin-3 and prognosis in solid cancers showed somewhat conflicting results.13,14,19,20 Regarding the hematological malignancies, higher expression of LGALS3 in peripheral blood mononuclear cells is observed in patients with indolent chronic lymphocytic leukemia compared with those with progressive disease.41 In contrast, endothelial expression of galectin-3 is identified as a bad prognostic factor in primary central nervous system lymphoma,42 and increased serum galectin-3 expression may indicate a worse prognosis in diffuse large B-cell lymphomas.16 It has been shown that galectin-3 localized in the cytosol protects the cell from apoptosis, but that in the nucleus, it has the opposite effect.43 The variable effects of galectin-3 on cancers are probably related to the cell types involved and the subcellular localization of the protein.
In this study, we demonstrated that BM LGALS3 expression higher than the median level of total patients with AML was associated with a poorer OS in patients with AML, especially in the CN-AML group. The prognostic effect of higher BM LGALS3 expression was validated in the independent validation cohort, although the patient numbers were limited in that cohort. It seems that galectin-3 is located in both cytosol and nucleus of leukemic cells, as shown by the IHC (Figure 1). The underlying mechanisms associating higher LGALS3 expression with poor prognosis in AML remain to be determined. It has been shown that galectin-3 inhibits cell apoptosis induced by chemotherapeutic drugs.44 Cheng et al45 also found that increased galectin-3 expression in K562 cells stabilized antiapoptotic Bcl-2 family proteins and facilitated leukemia cell survival from apoptotic stimuli. Yamamoto-Sugitani et al17 demonstrated that galectin-3 induced by leukemia microenvironment supported molecular signaling pathways and promoted drug resistance in chronic myeloid leukemia. Although the above studies only focus on solid cancers and chronic myeloid leukemia cells, their results are consistent with our findings that patients with AML who have higher LGALS3 expression were more refractory to chemotherapy and had lower CR rates. Additional research will be needed to elucidate the effects of galectin-3 expression in AML cells.
The finding that higher BM LGALS3 expression was closely associated with PTPN11 mutation in this study was interesting. PTPN11 encodes SHP-2, a cytoplasmic protein tyrosine phosphatase containing 2 tandemly arranged Src homology 2 (SH2) domains at the N terminus.46 SHP-2 is a positive modulator of rat sarcoma (RAS) signaling.46 Somatic gain of function mutations of PTPN11 can be detected in patients with juvenile myelomonocytic leukemia, myelodysplastic syndrome, and AML, especially the FAB M4/M5 subtypes.34,47 It has been shown that galectin-3-mediated cell transformation is partly attributed to its interaction with oncogenic RAS.48 Galectin-3 may more actively interact with RAS signaling molecules in patients with PTPN11 mutations and lead to AML. Further studies are needed to clarify this point.
Recently, many gene mutations were detected in AML, and some were found to be independent prognostic factors.21,22,40 To better stratify patients with AML into different risk groups, a survival scoring system incorporating BM LGALS3 expression and 8 other prognostic factors (age, WBC counts, karyotype, NPM1/FLT3-ITD, MLL-PTD, CEBPAdouble-mut, mutations of AML1/RUNX1, and WT1) into survival analysis was formulated. This scoring system was more powerful than a single marker to separate patients with non-M3 AML into different prognostic groups. Moreover, using 6 parameters (age, WBC counts, BM LGALS3 expression, NPM1/FLT3-ITD, CEBPAdouble-mut, and WT1 mutation), the CN-AML group could also easily be stratified into 4 groups with significant different prognosis. Further trials with independent cohort will be needed to validate this molecularly based risk assessment and stratification of patients with AML.
The limitation in our study is that BM biopsies were not done in all patients; therefore, galectin-3 protein expression could not be measured by IHC in every case. However, we distinctly showed that there was a significant correlation between galectin-3 protein and mRNA expression in leukemic cells from the 30 selected patients with AML. Interestingly, the concentration of galectin-3 protein in BM plasma also correlated well with LGALS3 mRNA expression in leukemic cells from the 22 selected patients. To correlate the galectin-3 protein expression levels with clinical outcome, we analyzed the patients who had protein analyses and had received standard chemotherapy, including 25 patients with IHC and 16 with ELISA studies, respectively. We found the patients with higher IHC scoring or higher galectin-3 values in BM plasma measured by ELISA seemed to have a reduced OS compared with those with lower IHC scoring or lower plasma values (IHC: median survival, 13.5 vs 22 months [P = .437]; ELISA: 16.3 months vs not reached [P = .248], respectively), although the difference did not reach statistical significance. Further studies with large cohorts are necessary to clarify this point. GCS-100, a novel galectin-3 antagonist, has been shown to have an antimyeloma effect.49 Galectin-3-targeted conjugates are also suggested to be a promising treatment policy in prostate carcinoma.50 Galectin-3-targeted agents in combination with chemotherapy may be an attractive strategy for the treatment of patients with AML patients who have higher expression of this protein.
In conclusion, this study demonstrated that higher BM LGALS3 expression was closely associated with older age, FAB M4/M5 subtypes, and PTPN11 mutation, but negatively correlated with FLT3-ITD and CEBPA mutations. Furthermore, higher BM LGALS3 expression was an independent poor prognostic factor for OS in patients with non-M3 AML and CN-AML. The unfavorable prognostic effect of higher BM LGALS3 expression was also validated in an independent validation cohort. Incorporation of higher BM LGALS3 expression with 8 other prognostic factors into survival analyses can better stratify patients into different risk groups. Higher BM LGALS3 expression may serve as a new biomarker for foreseeing the clinical outcome of patients with AML, and galectin-3-targeted therapy may represent a potential new approach for patients with AML who have higher expression of this protein.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was partially sponsored by grants from the National Science Council (NSC 97-2314-B002-015-MY3, NSC-97-2628-B-002-002-MY3, NSC 100-2325-B002-032 and NSC 100-2628 -B-002-003-MY3), the Department of Health (DOH99-TD-C- 111-001), and the Department of Medical Research, National Taiwan University Hospital (NTUH 99P14 and 100P07).
Authorship
Contribution: C.-L.C. designed the study, performed gene expression study, analyzed and interpreted data, did statistical analysis, and wrote the manuscript. H.-A.H. designed and planned the study, contributed data management and statistical analysis, and wrote the manuscript. M.-C.L. instructed and helped to perform gene expression study and interpreted data. C.-Y.L. was responsible for statistical analysis. J.-Y.J., C.-W.L., and Y.-J.L. performed immunohistochemical staining and interpreted results. H.-Y.C. and F.-T.L. were responsible for data management and interpretation. W.-C.C., C.-Y.C., J.-L.T., M.Y., S.-Y.H., B.-S.K., S.-J.W., and W.T. participated in data collection and provision of patients. H.-F.T. designed and planned the study, wrote the manuscript, and coordinated the study during the entire period.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hwei-Fang Tien, Department of Internal Medicine, National Taiwan University Hospital, No. 7, Chung-Shan South Rd, Taipei, 100 Taiwan; e-mail: hftien@ntu.edu.tw.
References
Author notes
C.-L.C., H.-A.H., and M.-C.L. contributed equally to this study.