Key Points
PML selectively activates NLRP3 inflammasome.
Targeting to PML could be used to attenuate NLRP3 inflammasome–associated pathogenesis.
Abstract
The functional activities of the tumor suppressor promyelocytic leukemia protein (PML) are mostly associated with its nuclear location. In the present study, we discovered an unexpected role of PML in NLRP3 inflammasome activation. In PML-deficient macrophages, the production of IL-1β was strongly impaired. The expression of pro–IL-1β, NLRP3, ASC, and procaspase-1 was not affected in Pml−/− macrophages. PML deficiency selectively reduced the processing of procaspase-1. We further showed that PML is required for the assembly of the NLRP3 inflammasome in reconstitution experiment. All PML isoforms were capable of stimulating NLRP3 inflammasome activation. In Pml−/− macrophages, the generation of reactive oxygen species and release of mitochondrial DNA were decreased. The involvement of PML in inflammasome activation constitutes an important activity of PML and reveals a new mechanism underlying the inflammasome activation. In addition, downregulation of PML by arsenic trioxide suppressed monosodium urate (MSU)-induced IL-1β production, suggesting that targeting to PML could be used to treat NLRP3 inflammasome–associated diseases.
Introduction
Promyelocytic leukemia protein (PML), first identified for its oncogenic translocation to fuse with the retinoic acid receptor α in acute promyelocytic leukemia, is a tumor suppressor with diverse activities in cell growth, cell death, stress responses, or senescence.1-4 Alternative splicing of the human PML gene produces seven PML isoforms (PML I-VII), all of which possess the same N-terminal region composed of RBCC domains connecting to different C-terminal sequences.5,6 With the exception of PML-VII, all other PML isoforms contain the nuclear localization sequence. PML-I, with a nuclear exclusion signal, is situated both in the nucleus and cytoplasm.6 Most of the physiologic functions of PML are attributed to PML nuclear bodies.1,2,4,7 A few activities have been identified with the cytoplasmic form of PML. For example, cytoplasmic PML participates in transforming growth factor–β signaling and inhibits p53 function.8,9 A cytoplasmic PML mutant enhances the suppressive activity of PML–retinoic alpha receptor-α on retinoic acid (RA)-mediated transcription.10 In addition, cytoplasmic PML inhibits herpes simplex virus–1 infection.11 More recently, PML was found to also localize at the endoplasmic reticulum (ER) and mitochondria-associated membranes (MAM),12 contributing to apoptosis induction. The function of cytoplasmic PML remains incompletely characterized.
The NACHT domain-, leucine-rich repeat (LRR)-, and pyrin domain–containing protein 3 (NLRP3, also known as NALP3, or cryopyrin) inflammasome is activated by a variety of agents including pathogens, microbial products, and environmental and endogenous danger signals.13-18 Inappropriate activation of NLRP3 inflammasome leads to a variety of autoinflammatory diseases as a result of overproduction of IL-1β.19-21 The assembly of the NLRP3 inflammasome is dependent on 2 sequential signals. The first inflammatory signal, through nuclear factor–kappa B (NF-κB) activation, promotes the expression of pro–IL-1β and NLRP3.22 The second activation signal stimulates the formation of the NLRP3 inflammasome, presumably by initial reversal of the self-inhibitory association of LRR, followed by the binding of NLRP3 to apoptosis-associated speck-like protein containing a CARD domain (ASC) and procaspase-1. The successful formation of inflammation generates caspase-1, which is essential for the cleavage of pro–IL-1β and pro–IL-18 to produce mature IL-1β and IL-18. Exactly how the assembly of the NLRP3 inflammasome is triggered remains largely elusive.13,14,16,18 The difficulties to characterize the biochemical processes underlying inflammasome formation could be partly attributed to the fact that cell lysis induces spontaneous inflammasome assembly in vitro as the consequence of cell membrane damage.23 Reactive oxygen species (ROS), membrane pore formation, intracellular K+ reduction, and lysosomal protease cathepsin B have been postulated as triggers of the NLRP3 inflammasome formation.13,14,16,18
Recent studies revealed that NLRP3 is colocalized with structures of the ER and MAMs after inflammasome stimulation.24,25 ROS generated by the mitochondrial respiratory chain promotes mitochondrial DNA release and NLRP3 inflammasome activation.24,25 ROS production in mitochondria is regulated by the transport of Ca2+ ions from MAMs to mitochondria.24 Interestingly, PML forms a complex with the inositol 1,4,5-triphosphate receptor (IP3R) and is required for IP3R-mediated Ca2+ release.12 In the present study, we found that PML is required for full NLRP3 inflammasome activation. IL-1β production in macrophages was profoundly compromised in the absence of PML. The expression of pro–IL-1β,and NLRP3 was independent of PML. Instead, PML is indispensable for NLRP3 inflammasome formation. We further demonstrated that targeting to PML effectively suppressed IL-1β production, suggesting a novel way to regulate NLRP3 inflammasome.
Methods
Reagents
Human PML-I cDNA was obtained from GeneCopoeia (Germantown, MD). Human PML-II, PML-III, PML-IV, and PML-V cDNAs were gifts by Dr. Kun-Sang Chang (M. D. Anderson Cancer Center, Houston, TX). Human PML-VI cDNA was a gift by Dr. Ron Evans (Salk Institute, San Diego, CA). pCMX-PML-VI∆SUMO, which carries 3 mutations (K65R, K160R, and K490R), was previously described.26 PML-I∆SUMO (PMLI-3KR)27 was a gift of Dr. Ruey-Hwa Chen (Academia Sinica, Taipei, Taiwan). PMLex1-4, the truncated form containing PML exons 1-4, was generated from PML-VI by polymerase chain reaction (PCR) and is analogous to PML-VII minus exon7a. PML-I∆NLS was generated by replacing both Lys487 and Lys490 of NLS with Ala using PCR. The primer used was: 5′ CCC AGT GCC CCA GGG CGG TCA TCG CGA TGG AGT CTG AGG 3′. AIM2 cDNA was obtained from Addgene (Cambridge, MA). 12-O-Tetradecanoylphorbol-13-acetate (TPA, also known as PMA), leptomycin B, Flag-M2 antibodies, and Flag-M2 beads were purchased from Sigma-Aldrich (St. Louis, MO). R837, LPS (lipopolysaccharide), Pam3CSK4, nigericin, alum crystals, dAdT, and monosodium urate (MSU) were obtained from InvivoGen (San Diego, CA). The antibody suppliers and the respective antibodies were: Abcam (Cambridge, MA) for PML (PML-97); Axxora (San Diego, CA) for ASC (AL177), human NLRP3 (clone nalpy3-b), and mouse NLRP3 (clone cryo-2); BD Biosciences (Franklin Lakes, NJ) for Ly-6G and CD11b; Cell Signaling Technology (Beverly, MA) for processed IL-1β p17 (Asp116) and AIM2; MBL (Nagoya, Japan) for ASC; R&D (Minneapolis, MN) for cathepsin B; Santa Cruz Biotechnology (Santa Cruz, CA) for human caspase-1 (A-19), mouse caspase-1 p10 (M-20), GAPDH (glyceraldehyde 3-phosphate dehydrogenase), Myc (clone 9E10), PML (PG-M3), PML (H-238), and RelA; and Upstate (Lake Placid, NY) for mouse PML (clone 36.1-104).
PML knockout mice
PML knockout mice were originally generated by Wang et al.28 The frozen Pml−/− embryos (01XF8, 129/Sv-Pmltm1Ppp) were obtained from the NCI-Frederick MMHCC Repository, National Cancer Institute (Frederick, MD). The Pml−/− embryos were thawed and implanted into CD-1 foster mothers. Mice were maintained in the SPF mouse facility of the Institute of Molecular Biology, Academia Sinica. Mice used in this study were back-crossed with C57BL/6 mice for 8 generations or more. All mouse experiments were conducted with approval from the Institutional Animal Care and Utilization Committee, Academia Sinica.
Cell cultures
Murine bone marrow cells were flushed out from tibias and femurs by cold RMPI medium, and were cultured in complete DMEM medium with 10% FCS (Invitrogen/Life Technologies), 10 mM glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and 50 μM 2-ME (complete DMEM medium), with additional 20% L929 cell-conditioned medium to generate bone marrow–derived macrophages. The human monocyte cell line THP-1 was cultured in RPMI-1640 medium with the same supplements (complete RPMI medium). PML was knocked down in THP-1 cells using SMARTpool siRNA purchased from Dharmacon (Lafayette, CO), with sequences: 5′ GCA ACC AGT CGG TGC GTG A, 5′ GGA CAT GCA CGG TTT CCT G, 5′ GAG CTC AAG TGC GAC ATC A, and 5′ GGA AAG ATG CAG CTG TAT C.
Secretion and measurement of IL-1β
Mouse bone marrow–derived macrophages and peritoneal macrophages were plated in 12-well plates for overnight culture, replaced with fresh medium, and primed with LPS (0.5 μg/mL), Pam3CSK4 (1 μg/mL), or R837 (15 μg/mL) for 4 hours, followed by stimulation with adenosine triphosphate (ATP) (4 mM) for 40 minutes, or with MSU (150 μg/mL) or alum crystals (500 μg/mL) for 4 hours. THP-1 cells were primed with PMA (0.5 μg/mL) for 4 hours for differentiation into macrophages, replenished with fresh complete RPMI medium, and cultured overnight. THP-1 cells were activated with nigericin (10 μg/mL) and MSU in serum-free RPMI medium for 4 and 6 hours, respectively. At the end of activation, cells and supernatants were collected, and the levels of IL-1β or tumor necrosis factor-alpha (TNF-α) in supernatants were determined by enzyme-linked immunosorbent assay (ELISA) using Human IL-1β DuoSet, human TNF-α DuoSet, murine IL-1β DuoSet, and murine TNF-α DuoSet (R&D). Two volumes of cold acetone were added to the remaining cultured supernatants for precipitation at −20°C for 1 hour. Precipitates were spun down and resuspended in RIPA buffer (50 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 1mM EDTA, 1 mM Na3VO4, 10 mM NaF, protease inhibitor mixture [Roche]). Cell lysates and supernatant precipitates were resolved by 4% to 20% gradient SDS-PAGE, and generation of caspase-1 and IL-1β determined by immunoblots.
NLRP3 inflammasome reconstitution in 293T cells
293T cells were transfected with 200 ng pcDNA4-proIL-1β, 25 ng pcDNA4-NLRP3-Myc, 20 ng pcDNA-ASC-GFP, and 10 ng pcDNA4-procaspase-1-Myc, with an additional 100 ng of PML. Empty vector (pcDNA4) was included so that the quantity of DNA transfected was equal in the same experiment. Cells and culture supernatants were collected 48 hours after transfection and total cell lysates and supernatant precipitates prepared as mentioned previously. The IL-1β secreted was also measured by ELISA.
Preparation of nuclear and cytosolic extracts
Macrophages were washed and lysed with NP-40 buffer (0.25% NP40, 10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1 mM Na3VO4, 10 mM NaF, protease inhibitor mixture [Roche]) at 0°C for 4 minutes. The supernatants collected after centrifuge at 3000 rpm for 4 minutes on an Eppendorf microfuge were cytosolic extracts. The pellets were resuspended, washed 3 times at 4°C, and lysed in RIPA lysis buffer at 0°C for 30 minutes. Lysates were centrifuged at 13 200 rpm for 10 minutes, and supernatants were collected as nuclear extracts. The purity of cytosolic extracts and nuclei were assessed by the exclusive expression of glyceraldehyde GAPDH and Lamin B, respectively.
In situ proximity ligation assay
In situ proximity ligation assay was conducted as described by Söderberg et al.29 The Duolink Proximity Ligation in situ reagent kit (Olink, Uppsala, Sweden) was used. Seeded bone marrow–derived macrophages were untreated or treated with LPS or LPS plus ATP. The protocol was performed according to the instructions provided with the kit. Images were obtained on a Zeiss Axioplan 2 imaging microscope with a plan Apochromat 63×/1.4 Oil DIC objective at room temperature. Texas red–linked oilgonucleotide was detected using a 598 nm (Ex) and 613 nm (Em) filter set, whereas DAPI was detected using a 360 nm (Ex) and 460 nm (Em) filter set. The pinholes are: Ch3-1:106 (red), Ch2-2:82 (blue), and ChD-3:0. Images were acquired by LSM 510 META digital camera and processed with Zeiss LSM Image Browser version 4.2.0.121 software.
Mitochondrial DNA release
The release of mitochondrial DNA into cytosol was performed according to Nakahira et al.,25 with identical quantitative PCR primers for 18S and mouse cytochrome c. The PCR condition was 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
Results
Reduced caspase-1 generation and diminished IL-1β production in Pml−/− macrophages
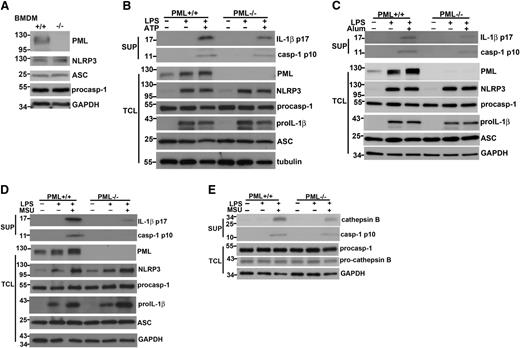
Macrophages from normal littermate control (Pml+/+) and Pml−/− mice were first examined on their capacities to produce IL-1β. IL-1β generation in macrophages required two signals: stimulation of macrophages with LPS alone did not generate IL-1β; only after subsequent treatment with ATP or MSU was IL-1β production detected (Figure 1A). Similarly, combinatory treatments of macrophages with either Pam3CSK4 plus ATP or R848 plus ATP led to IL-1β secretion (Figure 1B-C). The production of IL-1β was significantly impaired in PML-deficient macrophages at all stimulations examined (Figure 1A-C). The requirement of PML was IL-1β–specific because PML knockout did not alter TNF-α generation in macrophages activated by LPS+ATP or Pam3CSK4+ATP (Figure 1D).
Deficiency in PML impairs IL-1β secretion in macrophages. (A-C) Reduced IL-1β secretion in PML-null macrophages. Bone marrow–derived macrophages (BMDMs) from normal littermate control (Pml+/+) and Pml−/− mice were primed with LPS (0.5 μg/mL) (A), Pam3CSK4 (Pam3, 1 μg/mL) (B), or R837 (15 μg/mL) (C) for 4 hours, followed by incubation with ATP (4 mM) for 40 minutes (A-C), or MSU (150 μg/mL) for 4 hours (A). The secreted IL-1β in the supernatants was determined by ELISA. (D) Normal TNF-α secretion in PML-deficient macrophages. Bone marrow–derived macrophages from wild-type and Pml−/− mice were stimulated with LPS or Pam3CSK4 for 6 hours, followed by ATP or nigericin (Nig, 10 μM), and secreted TNF-α was measured by ELISA. **P < .01; ***P < .001 for paired t-test. Data represent mean values and SD of a specific experiment with triplicate samples. All experiments were repeated 4 times with similar results.
Deficiency in PML impairs IL-1β secretion in macrophages. (A-C) Reduced IL-1β secretion in PML-null macrophages. Bone marrow–derived macrophages (BMDMs) from normal littermate control (Pml+/+) and Pml−/− mice were primed with LPS (0.5 μg/mL) (A), Pam3CSK4 (Pam3, 1 μg/mL) (B), or R837 (15 μg/mL) (C) for 4 hours, followed by incubation with ATP (4 mM) for 40 minutes (A-C), or MSU (150 μg/mL) for 4 hours (A). The secreted IL-1β in the supernatants was determined by ELISA. (D) Normal TNF-α secretion in PML-deficient macrophages. Bone marrow–derived macrophages from wild-type and Pml−/− mice were stimulated with LPS or Pam3CSK4 for 6 hours, followed by ATP or nigericin (Nig, 10 μM), and secreted TNF-α was measured by ELISA. **P < .01; ***P < .001 for paired t-test. Data represent mean values and SD of a specific experiment with triplicate samples. All experiments were repeated 4 times with similar results.
PML deficiency did not affect the capacity of macrophage to take up particulate substrates such as MSU (data not shown), in agreement with a recent report showing the normal phagocytic function in Pml−/− macrophages.30 The attenuated IL-1β production in Pml−/− macrophages was not caused by impaired expression of NLRP3 inflammasome components. The expression of NLRP3, ASC, and procaspase-1 was normal in bone marrow–derived macrophages (Figure 2A). Analysis of cell lysates and culture supernatants revealed 2 sequential biochemical events matching with the two-signal requirement: the activation by LPS led to the induction of pro–IL-1β and NLRP3 (Figure 2B-D), and the addition of ATP or MSU resulted in the generation of p10 active caspase-1 and p17 mature IL-1β in culture supernatants (Figure 2B-D). No defect was found for the first stage of activation in Pml−/− macrophages. The activation of Il1b expression in macrophages was not affected by PML deficiency (supplemental Figure 1A). Both the induction of pro–IL-1β protein and the upregulation of NLRP3 protein were normal in PML-null macrophages stimulated by LPS (Figure 2B-D). NF-κB activation was in fact moderately increased in Pml−/− macrophages relative to Pml+/+ macrophages (supplemental Figure 2), consistent with a previous study reporting that PML inhibits NF-κB activation.31 LPS stimulation also led to a significant increase in PML protein (Figure 2B-D) as well as in Pml transcript (supplemental Figure 1B). By contrast, a clear defect was found in the second stage of activation: the cleavage of procaspase-1 to active caspase-1 (p10) was decreased and the processing of pro–IL-1β to mature IL-1β (p17) was diminished in Pml−/− macrophages (Figure 2B-D). Because the expression of NLRP3, ASC, and procaspase-1 in activated PML-null macrophages was comparable with that in control macrophages (Figure 2B-D), the decreased procaspase-1 processing in Pml−/− macrophages suggests a likely defect at the activation of the NLRP-3 inflammasome. In addition to caspase-1, generation of active cathepsin B was attenuated in PML-deficient macrophages, whereas the levels of procathepsin B were unchanged (Figure 2E).
Reduced caspase-1 generation and IL-1β processing in Pml−/− macrophages. (A) PML deficiency did not affect the expression of NLRP3, ASC, and procaspase-1. The expression of PML, NLRP3, ASC, procaspase-1, and GAPDH proteins was determined in BMDMs from Pml+/+ and Pml−/− mice. Numbers to the left indicate molecular weight in kDa. (B-D) PML deficiency decreased active caspase-1 generation and IL-1β processing. Pml+/+ and Pml−/− macrophages were primed with LPS for 4 hours, followed by stimulation with ATP for 40 minutes (B), alum crystal for 4 hours (C), or MSU for 4 hours (D). Culture supernatants and BMDMs were collected before and after LPS activation, or after stimulation with ATP, alum crystal, or MSU. Total cell lysates (TCL) were generated from BMDMs at each stage. Proteins in supernatants (SUP) were precipitated by cold (–20°C) acetone. Both supernatant precipitates and TCL were resolved by SDS-PAGE. The contents of mature IL-1β (p17) and active caspase-1 (p10) in supernatants, and PML, NLRP3, procaspase-1, pro-IL-1β, ASC, and GAPDH in total cell lysates, were assessed by Western blotting. (E) Reduced cathepsin B processing in PML-null macrophages. Pml+/+ and Pml−/− macrophages were primed with LPS, followed by MSU stimulation as in (B). The levels of cathepsin B and active caspase-1 p10 in supernatants, and the contents of procaspase-1 and pro-cathepsin B in total cell lysates were determined. (A-E) Results shown were representative of at least 3 independent experiments.
Reduced caspase-1 generation and IL-1β processing in Pml−/− macrophages. (A) PML deficiency did not affect the expression of NLRP3, ASC, and procaspase-1. The expression of PML, NLRP3, ASC, procaspase-1, and GAPDH proteins was determined in BMDMs from Pml+/+ and Pml−/− mice. Numbers to the left indicate molecular weight in kDa. (B-D) PML deficiency decreased active caspase-1 generation and IL-1β processing. Pml+/+ and Pml−/− macrophages were primed with LPS for 4 hours, followed by stimulation with ATP for 40 minutes (B), alum crystal for 4 hours (C), or MSU for 4 hours (D). Culture supernatants and BMDMs were collected before and after LPS activation, or after stimulation with ATP, alum crystal, or MSU. Total cell lysates (TCL) were generated from BMDMs at each stage. Proteins in supernatants (SUP) were precipitated by cold (–20°C) acetone. Both supernatant precipitates and TCL were resolved by SDS-PAGE. The contents of mature IL-1β (p17) and active caspase-1 (p10) in supernatants, and PML, NLRP3, procaspase-1, pro-IL-1β, ASC, and GAPDH in total cell lysates, were assessed by Western blotting. (E) Reduced cathepsin B processing in PML-null macrophages. Pml+/+ and Pml−/− macrophages were primed with LPS, followed by MSU stimulation as in (B). The levels of cathepsin B and active caspase-1 p10 in supernatants, and the contents of procaspase-1 and pro-cathepsin B in total cell lysates were determined. (A-E) Results shown were representative of at least 3 independent experiments.
We observed a similar defect in the human macrophage cell line THP-1. In PMA-primed THP-1 cells, stimulation with nigericin led to the generation of active caspase-1 and mature p17 IL-1β (supplemental Figure 3). Knockdown of PML by siRNA, not affecting the levels of NLRP3, procaspase-1, and ASC (supplemental Figure 3), reduced the production of p10 caspase-1 and p17 IL-1β in THP-1 cells, further supporting a role of PML in caspase-1 activation and pro–IL-1β processing.
PML increases IL-1β production in reconstituted NLRP3 inflammasome
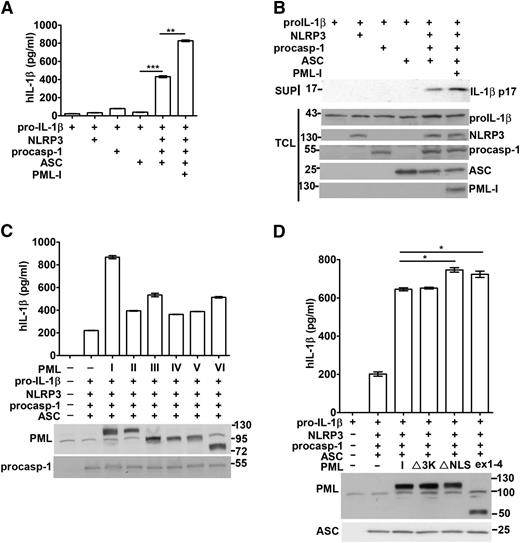
We further evaluated the involvement of PML in the reconstituted NLRP3 inflammasome in 293T cells. ASC and NLRP3 are absent in 293T cells, and functional NLRP3 inflammasome could be reconstituted through transfection of plasmids encoding pro–IL-1β, NLRP3, procaspase-1, and ASC. The expression of each transfected gene in 293T cells was confirmed by Western blotting (Figure 3B). Cotransfection of pro–IL-1β, procaspase-1, NLRP3, and ASC resulted in the secretion of IL-1β into culture supernatants (Figure 3A-B). The strict requirement for the full reconstitution of NLRP3 inflammasome was shown by the little IL-1β that was produced in the absence of any components (Figure 3A; supplemental Figure 4).
PML enhances the reconstitution of the NLRP3 inflammasome. (A-B) PML-I increased the activation of the NLRP3 inflammasome reconstituted in 293T cells. 293T cells were transfected with 200 ng pro–IL-1β, 25 ng NLRP3, 10 ng procaspase-1, and 20 ng ASC, in addition to 100 ng of PML-I. Forty-eight hours after transfection, culture supernatants were harvested and TCL were isolated. The IL-1β secreted was measured by ELISA (A). The protein levels of pro–IL-1β, NLRP3, procaspase-1, ASC, and PML-I in TCL and p17 IL-1β in the supernatants were determined (B). (C) The function of NLRP3 inflammasome reconstituted in 293T cells was enhanced by PML-II, PML-III, PML-IV, PML-V, or PML-VI. PML-II, PML-III, PML-IV, PML-V, or PML-VI was transfected with NLRP3 inflammasome components as in (A). The transfected PML and procaspase-1 were confirmed by immunoblots and the levels of IL-1β quantified. (D) SUMO mutant, NLS mutant, or truncated PML was as effective as PML-I in promoting NLRP3 inflammasome activation. PML-I, PML-I∆SUMO (∆3K), PML-I∆NLS (K487A, K490A), or PMLex1-4 was introduced into 293T cells with NLRP3 inflammasome components as in (A). The levels of IL-1β were measured. Empty vector (pcDNA4) was included (A,C,D) so that the amount of DNA transfected in each set was equal. All experiments were repeated 3 times and produced similar results. *P < .05; **P < .01; ***P < .001, for paired t-test.
PML enhances the reconstitution of the NLRP3 inflammasome. (A-B) PML-I increased the activation of the NLRP3 inflammasome reconstituted in 293T cells. 293T cells were transfected with 200 ng pro–IL-1β, 25 ng NLRP3, 10 ng procaspase-1, and 20 ng ASC, in addition to 100 ng of PML-I. Forty-eight hours after transfection, culture supernatants were harvested and TCL were isolated. The IL-1β secreted was measured by ELISA (A). The protein levels of pro–IL-1β, NLRP3, procaspase-1, ASC, and PML-I in TCL and p17 IL-1β in the supernatants were determined (B). (C) The function of NLRP3 inflammasome reconstituted in 293T cells was enhanced by PML-II, PML-III, PML-IV, PML-V, or PML-VI. PML-II, PML-III, PML-IV, PML-V, or PML-VI was transfected with NLRP3 inflammasome components as in (A). The transfected PML and procaspase-1 were confirmed by immunoblots and the levels of IL-1β quantified. (D) SUMO mutant, NLS mutant, or truncated PML was as effective as PML-I in promoting NLRP3 inflammasome activation. PML-I, PML-I∆SUMO (∆3K), PML-I∆NLS (K487A, K490A), or PMLex1-4 was introduced into 293T cells with NLRP3 inflammasome components as in (A). The levels of IL-1β were measured. Empty vector (pcDNA4) was included (A,C,D) so that the amount of DNA transfected in each set was equal. All experiments were repeated 3 times and produced similar results. *P < .05; **P < .01; ***P < .001, for paired t-test.
Next, the effect of PML on caspase-1 activation and IL-1β generation in reconstituted inflammasome was assessed. PML-I cotransfection led to a clear increase in IL-1β production in the reconstituted NLRP3 inflammasome (Figure 3A). PML-I expression did not alter the apparent levels of pro–IL-1β, NLRP3, and procaspase-1 (Figure 3B), suggesting an effect of PML-I on the activation of the reconstituted NLRP3 inflammasome. We further tested other PML isoforms for their capacity to promote IL-1β generation. Overexpression of PML-II, PML-III, PML-IV, PML-V, and PML-VI also increased the production of IL-1β in 293T cells with reconstituted NLRP3 inflammasome (Figure 3C). In addition, PML-I∆3K (PML-I 3KR27 ), the mutant that cannot be sumoylated, exerted an additive effect on the generation of IL-1β indistinguishable from wild-type PML-I in the same system (Figure 3D, ∆3K). A similar result in NLRP3 inflammasome activation was found with PML-VI∆3K when compared with PML-VI (supplemental Figure 5). Therefore, PML increased IL-1β generation in reconstituted NLRP3 inflammasome independent of PML sumoylation.
We also elucidated whether PML participated in the function of the NLRP3 inflammasome in vivo. To assay for in vivo activation of the NLRP3 inflammasome, phosphate-buffered saline alone or MSU crystals were intraperitoneally administered to Pml+/+ and Pml−/− mice, and the influx of neutrophils into the peritoneal cavity as a consequence of IL-1β generation was determined.32,33 As shown in supplemental Figure 6A-B, peritoneal infiltration of polymorphonuclear neutrophils was significantly reduced in Pml−/− mice compared with Pml+/+ mice. Therefore, PML is also required for NLRP3 inflammasome activation in vivo.
PML does not regulate the activation of AIM2 inflammasome
We further examined whether PML was able to promote the activation of other inflammasome. AIM2 inflammasome is activated by the binding of cytosolic dsDNA.14-18 Transfection of macrophages with dAdT resulted in production of IL-1β, which was not affected by PML deficiency (Figure 4A). In addition, expression of PML-I did not add to the activation of the reconstituted AIM2 inflammasome, in contrast to the enhancing effect of PML-I in the reconstituted NLRP3 inflammsome (Figure 4B). Therefore, PML does not regulate the activation of AIM2 inflammasome, suggesting that PML selectively targets to NLRP3 inflammasome but not to a common mechanism downstream of different inflammasomes.
Activation of AIM2 inflammasome is not regulated by PML. (A) Normal AIM2-mediated IL-1β production in PML-deficient macrophages. Pml+/+ and Pml−/− BMDMs were primed with LPS for 3 hours, followed by nigericin treatment for 4 hours or dAdT for 6 hours, and the amount of IL-1β secreted was quantitated. dAdT (1.5 μg) was transfected using Lipofectamine 2000. (B) PML-I did not increase the activation of the reconstituted AIM2 inflammasome. 293T cells were transfected with the indicated components of AIM2/NLRP3 inflammasome, with or without PML-I. Empty vector was included to ensure the same amount of DNA was transfected in each set. The production of IL-1β was determined 48 hours later. Experiments were independently repeated 3 times. **P < .01; ***P < .001, for paired t test.
Activation of AIM2 inflammasome is not regulated by PML. (A) Normal AIM2-mediated IL-1β production in PML-deficient macrophages. Pml+/+ and Pml−/− BMDMs were primed with LPS for 3 hours, followed by nigericin treatment for 4 hours or dAdT for 6 hours, and the amount of IL-1β secreted was quantitated. dAdT (1.5 μg) was transfected using Lipofectamine 2000. (B) PML-I did not increase the activation of the reconstituted AIM2 inflammasome. 293T cells were transfected with the indicated components of AIM2/NLRP3 inflammasome, with or without PML-I. Empty vector was included to ensure the same amount of DNA was transfected in each set. The production of IL-1β was determined 48 hours later. Experiments were independently repeated 3 times. **P < .01; ***P < .001, for paired t test.
Cytoplasmic PML may contribute to NLRP3 inflammasome activation
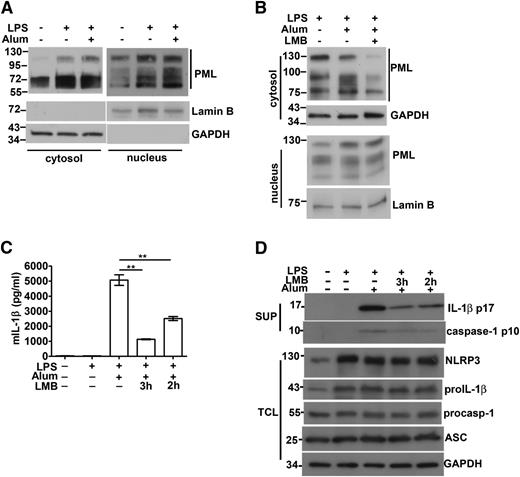
Even though PML is located mainly in the nucleus, previous reports have demonstrated the presence of PML in the cytosol.6,8-12 We investigated whether cytosolic PML also contributes to the activation of the NLRP3 inflammasome. We produced PML-I mutant with mutation at NLS (PML-I∆NLS). The predominant cytoplasmic localization of PML-I∆NLS, relative to PML-I, was confirmed by immunofluorescence (supplemental Figure 7). In reconstituted NLRP3 inflammasome, PML-I∆NLS led to a moderate increase in enhancing IL-1β production relative to the wild-type PML-I (Figure 3D). We also generated a truncated PML analogous to PML-VII, the cytosolic form of PML. PMLex1-4 was preferentially expressed in cytoplasm (supplemental Figure 7) and exhibited an enhancing effect, also like PML-I∆NLS, in the activation of reconstituted NLRP3 inflammasome (Figure 3D). Results from PML-I∆NLS and PMLex1-4 suggest that cytosolic PML also facilitates NLRP3 inflammasome activation. We detected an elevated level of cytoplasmic PML in LPS-primed macrophages (Figure 5A), likely because of an overall increase in PML proteins in macrophages stimulated with LPS (Figure 2B-D). The enhanced cytoplasmic presence was observed with PML of different molecular weights, suggesting the increase was not limited to a specific PML isoform in the activated macrophages (Figure 5A). Cotreatment of macrophages with LPS and alum crystal did not further increase the level of cytoplasmic PML (Figure 5A). LPS-induced cytosolic appearance of PML was attenuated by leptomycin B (Figure 5B), suggesting that some of the cytosolic PML is exported from the nucleus. It may be noted that cytoplasmic PML represents a smaller fraction of total cellular PML than nuclear PML. Quantitation analysis by densitometry indicates that 15% to 20% of total PML was situated in cytoplasm in macrophages.
Association of cytosolic PML with caspase-1 activation and IL-1β production. (A) The level of cytosolic PML in macrophages was increased by LPS stimulation. BMDM cells were primed with LPS for 4 hours, followed by treatment with alum crystal (400 μg/mL) for 1 hour. The amounts of PML in cytosolic extracts (40 μg) and nuclear extracts (25 μg) were determined. The exposure time during development was 4 seconds for cytosolic extracts and 1 second for nuclear extracts. Lamin B and GAPDH were markers for nuclear and cytoplasmic extracts, respectively. (B) Leptomycin B (LMB) decreased the cytosolic presence of PML. LMB (10 ng/mL) was included in the LPS priming stage, and the contents of cytoplasmic and nuclear PML in BMDM determined as in (A). (C-D) LMB treatment suppressed caspase-1 activation and IL-1β production. BMDMs were treated with LPS for 1 to 2 hours, followed by addition of LMB for 2 to 3 hours (4 hours total LPS priming time). Next, alum crystals were added and incubated for an additional 4 hours. Culture supernatants were isolated and cell lysates were prepared as indicated. The levels of IL-1β in supernatants were measured by ELISA (C) and immunoblotting (D). The contents of NLRP3, pro–IL-1β, procaspase-1, ASC, and caspase-1 in cell lysates or supernatants were assessed by Western blotting (D). Representative results of 3 independent experiments are shown. **P < .01 for paired t-test.
Association of cytosolic PML with caspase-1 activation and IL-1β production. (A) The level of cytosolic PML in macrophages was increased by LPS stimulation. BMDM cells were primed with LPS for 4 hours, followed by treatment with alum crystal (400 μg/mL) for 1 hour. The amounts of PML in cytosolic extracts (40 μg) and nuclear extracts (25 μg) were determined. The exposure time during development was 4 seconds for cytosolic extracts and 1 second for nuclear extracts. Lamin B and GAPDH were markers for nuclear and cytoplasmic extracts, respectively. (B) Leptomycin B (LMB) decreased the cytosolic presence of PML. LMB (10 ng/mL) was included in the LPS priming stage, and the contents of cytoplasmic and nuclear PML in BMDM determined as in (A). (C-D) LMB treatment suppressed caspase-1 activation and IL-1β production. BMDMs were treated with LPS for 1 to 2 hours, followed by addition of LMB for 2 to 3 hours (4 hours total LPS priming time). Next, alum crystals were added and incubated for an additional 4 hours. Culture supernatants were isolated and cell lysates were prepared as indicated. The levels of IL-1β in supernatants were measured by ELISA (C) and immunoblotting (D). The contents of NLRP3, pro–IL-1β, procaspase-1, ASC, and caspase-1 in cell lysates or supernatants were assessed by Western blotting (D). Representative results of 3 independent experiments are shown. **P < .01 for paired t-test.
We also examined whether the reduction of cytosolic PML by leptomycin B affected IL-1β production in macrophages. The addition of leptomycin B for 2 hours before alum crystal stimulation suppressed IL-1β secretion in LPS-primed macrophages, whereas longer incubation with leptomycin B (3 hours) led to further inhibition of IL-1β generation (Figure 5C). The presence of leptomycin B did not affect the levels of NLRP3, pro-IL-1β, procaspase-1, and ASC in macrophages, but profoundly suppressed the generation of p10 caspase-1 and mature IL-1β (Figure 5D). Reduction in cytoplasmic PML decreased caspase-1–dependent IL-1β production, suggesting that cytosolic PML may contribute to NLRP3 inflammasome activation.
We next investigated whether PML is associated with NLRP3. Immunoprecipitation analysis revealed a PML-I-NLRP3 interaction when NLRP3-Myc and PML-I-FLAG were overexpressed in 293T cells (supplemental Figure 8A). We subsequently identified that the LRR domain of NLRP3 interacted with PML-I (supplemental Figure 8B). However, upon examining the binding of other PML isoforms to NLRP3, we did not detect interaction between NLRP3 and PML-II, PML-III, PML-IV, PML-V, and PML-VI (supplemental Figure 8C; data not shown for PML-II, PML-III, PML-IV, and PML-V). Therefore, association of NLRP3 with PML is not essential for PML to enhance NLRP3 inflammasome activation.
PML in NLRP3 inflammasome formation in situ
We also measured the assembly of NLRP3 inflammasome in situ. Proximity ligation assay28 was used to detect the close association between NLRP3 and ASC upon inflammasome formation. In proximity ligation assay, the in situ interaction between 2 proteins brings their respective antibody to close proximity, allowing the oligonucleotides linked to the antibodies to prime for circular DNA strands amplification. Consistent with the requirement of dual stimulation by LPS and ATP for NLRP3 inflammasome activation in normal macrophages, no association between NLRP3 and ASC could be detected in untreated macrophages or in macrophages stimulated with LPS alone (Figure 6A). NLRP3 became closely proximal to ASC after the addition of ATP to LPS-treated macrophages. ATP-induced NLRP3-ASC interaction was diminished in LPS-primed, PML-deficient macrophages (Figure 6A-B), consistent with a requirement of PML for NLRP3 inflammasome formation. Similarly, LPS plus ATP-induced NLRP3-caspase-1 close association was largely eliminated in Pml−/− macrophages (Figure 6C-D). The proximity ligation assay was also used to determine the in situ interaction between PML and caspase-1 during inflammasome formation. Similar to the NLRP3-caspase-1 interaction, PML was spatially associated with caspase-1 in macrophages treated with LPS and ATP, but not with LPS alone (Figure 6E-F). NLRP3 was also brought to close proximity of PML when macrophages were activated by LPS and ATP (Figure 6E,G).
In situ association of NLRP3 and ASC in macrophages is PML-dependent as measured by proximity ligation assay. (A-B) In situ association of NLRP3 and ASC in macrophages activated by LPS and ATP. BMDMs were plated overnight and sequentially stimulated with LPS and ATP as indicated. Macrophages were stained overnight at 4°C with anti-NLRP3 (Cryo-2, mouse MAb) and anti-ASC (AL177, rabbit pAb), followed by oligonucleotide-linked anti-mouse antibodies and anti-rabbit antibodies. After the addition of the template oligonucleotides, the primers in close proximity were allowed to anneal and circularize. The products generated by rolling-circle amplification were detected by hybridization with a Texas red–conjugated probe and were examined under a confocal laser scanning microscope (A). Amplicons were counted in at least 40 cells and divided by the number of cells to obtain spots/ν (nucleus) (B). (C-D) Close proximity between NLRP3 and caspase-1 in activated macrophages. The association of NLRP3 with caspase-1 in macrophages was analyzed (C) and quantitated (D) as in (A) and (B). (E-G) In situ association of PML with NLRP3 or caspase-1 in macrophages activated by LPS and ATP. BMDMs were treated with LPS and ATP, stained with anti-PML (H-238, rabbit pAb) and anti-NLRP3, or with anti-PML and anti-caspase-1, and the association of PML to caspase-1 or NLRP3 was measured (E) and quantified (F-G). The bar represents 10 μm. *P < .05; **P < .01 for paired t-test.
In situ association of NLRP3 and ASC in macrophages is PML-dependent as measured by proximity ligation assay. (A-B) In situ association of NLRP3 and ASC in macrophages activated by LPS and ATP. BMDMs were plated overnight and sequentially stimulated with LPS and ATP as indicated. Macrophages were stained overnight at 4°C with anti-NLRP3 (Cryo-2, mouse MAb) and anti-ASC (AL177, rabbit pAb), followed by oligonucleotide-linked anti-mouse antibodies and anti-rabbit antibodies. After the addition of the template oligonucleotides, the primers in close proximity were allowed to anneal and circularize. The products generated by rolling-circle amplification were detected by hybridization with a Texas red–conjugated probe and were examined under a confocal laser scanning microscope (A). Amplicons were counted in at least 40 cells and divided by the number of cells to obtain spots/ν (nucleus) (B). (C-D) Close proximity between NLRP3 and caspase-1 in activated macrophages. The association of NLRP3 with caspase-1 in macrophages was analyzed (C) and quantitated (D) as in (A) and (B). (E-G) In situ association of PML with NLRP3 or caspase-1 in macrophages activated by LPS and ATP. BMDMs were treated with LPS and ATP, stained with anti-PML (H-238, rabbit pAb) and anti-NLRP3, or with anti-PML and anti-caspase-1, and the association of PML to caspase-1 or NLRP3 was measured (E) and quantified (F-G). The bar represents 10 μm. *P < .05; **P < .01 for paired t-test.
Reduced ROS generation and mitochondrial DNA release in PML-null macrophages
PML has been shown to mediate the release of Ca2+ from the ER,12 and uptake of Ca2+ into mitochondria results in ROS production, which is linked to NLRP3 inflammasome activation.24,25 We used okadaic acid to block PML-dependent and PP2a-mediated Ca2+ release from the ER.12 Treatment with okadaic acid effectively suppressed NLRP3 inflammasome activation in wild-type macrophages, as shown by diminished IL-1β production (supplemental Figure 9A). By contrast, TNF-α generation was not affected by okadaic acid (supplemental Figure 9B). We also investigated whether a deficiency in PML affected the generation of ROS in mitochondria. The induction of the population marked by Mitotracker green–positive and Mitotracker deep red–low, representing mostly ROS-producing mitochondria,24 was attenuated in PML-knockout macrophages stimulated with LPS+ATP (supplemental Figure 9C). Another mediator associated with NLRP3 inflammasome activation is cytosolic mitochondrial DNA.25 Mitochondrial DNA release was reduced in Pml−/− macrophages treated with LPS and ATP, relative to Pml+/+ macrophages (supplemental Figure 9D). Together, these data suggest a possibility that PML deficiency is coupled to decreased mitochondrial ROS production and attenuated mitochondrial DNA release, leading to impaired NLRP3 inflammasome activation.
Downregulation of PML by arsenic oxide suppresses IL-1β production
PML and PML-RARα fusion proteins are effectively degraded by arsenic trioxide.34 We examined whether arsenic trioxide treatment could be used to suppress inflammasome activation. Co-incubation of cells with arsenic trioxide for 1 hour was sufficient to eliminate most of PML in THP-1 cells (Figure 7A). NLRP3, constitutively expressed in THP-1 cells, and the upregulated pro–IL-1β were not affected by arsenic trioxide (Figure 7A, TCL). MSU-induced mature IL-1β production was profoundly suppressed in THP-1 cells pretreated with arsenic trioxide (Figure 7A, SUP). PML was also effectively downregulated by arsenic trioxide in bone marrow–derived macrophages (Figure 7B). Activation of caspase-1 and generation of IL-1β stimulated by LPS/ATP were largely inhibited in arsenic trioxide–treated macrophages (Figure 7B-C). In contrast, the induction of NLRP3 and pro–IL-1β was not affected by arsenic trioxide treatment in macrophages (Figure 7B). Because a high concentration of arsenic trioxide is known to trigger apoptosis in many cell types,35 we also monitored cell viability and found no apparent cell death at the dose and the duration of the experiment (supplemental Figure 10A,C). In addition, the production of TNF-α was not affected by the treatment with arsenic trioxide (Figure 7D; supplemental Figure 10B). Therefore, targeted degradation of PML effectively prevents the activation of NLRP3 inflammasome.
Arsenic trioxide suppresses MSU-induced IL-1β production by PML downregulation. (A) Arsenic trioxide promoted PML degradation and reduced IL-1β production in THP-1 cells. PMA-primed THP-1 cells were treated with or without arsenic trioxide (50 μM) for 1 hour, washed, and then stimulated with MSU for 6 hours. Culture supernatants and cell lysates were isolated and the contents of IL-1β and inflammasome components assessed by immunoblots. (B-C) Downregulation of PML by arsenic trioxide suppressed IL-1β generation in bone marrow–derived macrophages. BMDMs were stimulated with LPS for 4 hours, followed by arsenic trioxide (250 μM) for 1 hour, as indicated. Macrophages were washed and stimulated with ATP for an additional 30 minutes. IL-1β in the supernatants was determined by immunoblots (B) and ELISA (C). NLRP3 inflammasome components in TCLs were determined by immunoblots. (D) Arsenic trioxide did not affect TNF-α production. BMDMs were stimulated as in (B-C) and TNF-α in the supernatants was quantitated by ELISA. ***P < .001 for paired t-test.
Arsenic trioxide suppresses MSU-induced IL-1β production by PML downregulation. (A) Arsenic trioxide promoted PML degradation and reduced IL-1β production in THP-1 cells. PMA-primed THP-1 cells were treated with or without arsenic trioxide (50 μM) for 1 hour, washed, and then stimulated with MSU for 6 hours. Culture supernatants and cell lysates were isolated and the contents of IL-1β and inflammasome components assessed by immunoblots. (B-C) Downregulation of PML by arsenic trioxide suppressed IL-1β generation in bone marrow–derived macrophages. BMDMs were stimulated with LPS for 4 hours, followed by arsenic trioxide (250 μM) for 1 hour, as indicated. Macrophages were washed and stimulated with ATP for an additional 30 minutes. IL-1β in the supernatants was determined by immunoblots (B) and ELISA (C). NLRP3 inflammasome components in TCLs were determined by immunoblots. (D) Arsenic trioxide did not affect TNF-α production. BMDMs were stimulated as in (B-C) and TNF-α in the supernatants was quantitated by ELISA. ***P < .001 for paired t-test.
Discussion
In the present study, we illustrate that NLRP3 inflammasome–mediated IL-1β production is severely compromised in the absence of PML. Most of the physiological activities of PML have been associated with PML nuclear body formation.1,2,4 PML binds directly to many transcription factors and modulates their transcription activities.7,36 PML also regulates gene expression by sequestering a transcription repressor into the PML nuclear body.26,37 The PML nuclear body is also the major site where posttranslational modification and degradation of PML-interacting proteins takes place.4,38 We examined the possibility that PML participates in IL-1β expression through transcription regulation and found that the levels of the known NLRP3 components—NLRP3, procaspase-1, ASC, and pro–IL-1β—were not affected by PML deficiency. By contrast, the second stage of NLRP3 inflammasome activation, characterized by the formation of the NLRP3 inflammasome, processing of caspase-1, and cleavage of pro-IL-1β, was severely impaired in PML-deficient macrophages. This was also confirmed by the fact that reconstitution of NLRP3 inflammasome was significantly increased by further coexpression of PML. In addition, the in situ interaction between NLRP3 and ASC was diminished in Pml−/− macrophages. Together, defects in caspase-1 activation and IL-1β production in PML-null macrophages could be attributed to a defective assembly of the NLRP3 inflammasome. Consistent with an involvement of PML in NLRP3 inflammasome activation, PML is expressed at high levels in macrophages.39 Notably, PML deficiency or PML overexpression did not affect the activation of AIM2 inflammasome (Figure 4), suggesting that PML targets to NLRP3 inflammasome but not to a common mechanism downstream of different inflammasomes.
We found that all PML isoforms exhibited similar activity in promoting NLRP3 inflammasome assembly (Figure 5C). A PML-VII analog PMLex1-4 was as effective as PML-I in enhancing NLRP3 inflammasome formation (Figure 5D), suggesting the N-terminal region of PML encoded by exons 1-4, shared by all PML isoforms, is sufficient to facilitate NLRP3 inflammasome activation. This is in contrast to the reported association of distinct nuclear functions with different PML isoforms.2,5,6,26 For example, PML-IV, but not other PML isoforms, triggers premature senescence.40 PML-I and PML-VI, but not PML-IV, promote the transcription activation of nuclear factor of activated T cells.36 The isoform-independent enhancement of IL-1β production by PML agrees with a nucleus-independent activity of PML. In addition to a few cytoplasmic functions of PML previously documented,8-12 our results suggest that PML may promote the formation of the NLRP3 inflammasome.
PML forms complexes with the inositol 1,4,5-triphosphate receptor (IP3R), Akt, and protein phosphatase 2a (PP2a) at the ER for IP3R-mediated Ca2+ release.12 The voltage-dependent anion channel is involved in the transport of Ca2+ from MAMs to mitochondria, contributing to ROS generation as well as inflammasome activation.24 In addition, NLRP3 is redistributed to the ER and mitochondria organelle clusters after inflammasome activation.24 Together, these findings suggest a possibility that PML enhances Ca2+ release from the ER into MAMs, and the subsequent voltage-dependent anion channel–mediated uptake of Ca2+ promotes ROS production and NLRP3 inflammasome activation. In the present study, we found that inhibition of PP2a by okadaic acid decreased IL-1β production and PML-deficiency impaired mitochondrial ROS production and reduced mitochondrial DNA release (supplemental Figure 9). PML may thus contribute to NLRP3 inflammasome activation at stages before ROS production and mitochondrial DNA release.12,24,25 Further studies will be required to establish the biochemical coupling from PML, Ca2+ release, and ROS production to NLRP3 inflammasome activation.
The exact biochemical processes involved in NLRP3 inflammasome activation remain incompletely understood. K+ efflux, ROS, and cathepsin B are a few non–mutually exclusive mediators involved in inflammasome activation.13,14,16,18 Notably, PML deficiency also impaired release of cathepsin B (Figure 2E), revealing an essential role of PML in both ROS production and cathepsin B activation. This is in contrast to disruption of autophage proteins that affects only ROS generation but not cathepsin B release.25 The observed enhancing function of PML in NLRP3 inflammasome is likely the result of a combination of both ROS-dependent and ROS-independent pathways. It may be noted that PML is involved in the posttranslational modification, sequestration, or degradation of the nuclear body–associated proteins.4,38 Even though we have shown that sumoylation mutant of PML-I and PML-VI promoted NLRP3 inflammation (Figure 3D; supplemental Figure 5), we cannot exclude the possibility that some PML partners modified at the nuclear body enter cytoplasm and participate in the NLRP3 inflammasome formation. We are making progress to delineate the complete mechanisms of how PML promotes NLRP3 inflammamsome assembly.
NLRP3 inflammasome activation is implicated in diseases including familial cold autoinflammatory syndrome, gout, silicosis, asbestosis, and type 2 diabetes mellitus.19-21 IL-1β and IL-18 also regulate a wide variety of lymphocyte functions, including T-cell costimulation, production of Th2 response, and generation of Th17 responses.41,42 Consequently, excess IL-1β contributes to the development of allergic and inflammatory diseases. Antiinflammatory compounds, such as agents to block IL-1β activity or suppress caspase-1, have been developed to counteract inflammasome- and IL-1β–associated diseases.43 Our results demonstrate that PML is involved in NLRP3 inflammasome activation, and downregulation of PML helps reduce inflammasome-mediated pathogenesis. PML is likely another viable target for therapy against NLRP3 inflammasome–mediated diseases. This is supported by the fact that arsenic trioxide, the well-known PML antagonist, suppressed MSU-induced IL-1 production (Figure 7). Arsenic oxide has been used successfully to reduce IL-18 levels.44 Even though arsenic oxide exhibits multiple actions,35 some of the effects are likely caused by PML degradation. These results suggest the potential application to control inflammasome-associated diseases by downregulation of PML expression. Further investigation to develop reagent-specific targeting to PML may help delineate such possibility.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Yu-Chen Lin, Drs Kun-Sang Chang, Ruey-Hwa Chen, Ron Evans, and I-Chen Ho for critical reagents; Sue-Ping Lee and the staff of the Confocal Core of the Institute of Molecular Biology, Academia Sinica, for confocal microscopy; and Dr Heiko Kuhn for editing the manuscript.
This work was supported by grant NSC98-2321-B001-015 from the National Science Council, an Academia Sinica Investigator Award from Academia Sinica, and grant NHRI-EX100-10012SI from the National Health Research Institute, Taiwan, R.O.C.
Authorship
Contribution: Y.H.L., Y.W.H., Y.H.W., C.S.T., and S.T.M. performed experiments. Y.C.L., W.C.K., Y.T.C., and H.M.S. contributed to generation of constructs. S.T.J. generated Pml−/− mice from frozen embryo and M.Z.L. conceived of, designed, and supervised the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ming-Zong Lai, Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan, R.O.C.; e-mail: mblai@imb.sinica.edu.tw.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal