Abstract
Recent studies of the spectrum of somatic genetic alterations in acute myeloid leukemia (AML) have identified frequent somatic mutations in genes that encode proteins important in the epigenetic regulation of gene transcription. This includes proteins involved in the modification of DNA cytosine residues and enzymes which catalyze posttranslational modifications of histones. Here we describe the clinical, biological, and therapeutic relevance of mutations in epigenetic regulators in AML. In particular, we focus on the role of loss-of-function mutations in TET2, gain-of-function mutations in IDH1 and IDH2, and loss-of-function mutations in ASXL1 and mutations of unclear impact in DNMT3A in AML pathogenesis and therapy. Multiple studies have consistently identified that mutations in these genes have prognostic relevance, particularly in intermediate-risk AML patients, arguing for inclusion of mutational testing of these genetic abnormalities in routine clinical practice. Moreover, biochemical, biological, and epigenomic analyses of the effects of these mutations have informed the development of novel therapies which target pathways deregulated by these mutations. Our understanding of the effects of these mutations on hematopoiesis and potential for therapeutic targeting of specific AML subsets is also reviewed here.
Introduction
Acute myeloid leukemia (AML) is a clonal disorder of hematopoiesis characterized by the accumulation of immature myeloid cells accompanied by impaired normal hematopoiesis. Given the need to improve outcome in AML, multiple studies aimed at genetic characterization of AML have been performed in the hopes of furthering our understanding of AML pathogenesis and identifying new therapeutic approaches. To this end, a number of targeted sequencing, exome sequencing, and whole-genome sequencing studies have been performed in AML patients in the last 5 to 10 years. From these studies, a number of recurrently mutated genes have been identified and, interestingly, many of the newly identified recurrently mutated genes encode proteins that normally function in the epigenetic regulation of transcription. Although the definition of the term epigenetics has been a matter of debate, in this review, we refer to epigenetics as any process that directly affects modifications of DNA cytosine residues or posttranslational modifications of histones. This includes frequent mutations in the genes DNMT3A (DNA nucleotide methyltransferase 3A), TET2 (ten-eleven translocation 2), IDH1/2 (isocitrate dehydrogenase 1/2), and ASXL1 (the addition of sex combs like 1) in AML patients (mutations and translocations in MLL1 will be discussed in another accompanying review in this series) (Table 1). Although prior DNA methylation profiling studies in AML have identified the presence of a profoundly abnormal epigenome in this disease,1,2 the identification of mutations in epigenetic regulatory genes now provides a link between the altered epigenome in AML and somatic genetic alterations in this disorder.
Mutations in recurrently mutated epigenetic modifiers in adults with AML and their clinical relevance and associations
| Gene . | Mutation effect on gene . | Mutational frequency in AML, % . | Clinical associations . | |
|---|---|---|---|---|
| 16-60 y . | >60 y . | |||
| ASXL16,19,46,47,65 | Affected by loss-of-function mutations and copy-number loss | 3-6.8 | 16.2-25 | Mutations repeatedly identified as enriched in elderly AML patients, AML with an antecedent MDS, and AML patients with RUNX1 mutations. Associated with adverse OS in patients 16-60 y and >60 y of age. In some studies, the adverse effect is only in the subset of CN-AML or intermediate-risk AML. |
| DNMT3A7,14,16,19,66 | Appear to be affected by loss-of-function mutations, effect of the recurrent Arg882 mutation not clear | 17.8-23 | 16-22 | DNMT3A mutations confer adverse risk to intermediate-risk AML patients in multiple series. In most series, this adverse risk is restricted to those patients with FLT3-ITD mutant intermediate-risk AML patients. DNMT3A-mutant patients appear to have improved outcome with high-dose induction daunorubicin therapy (90 mg/m2) as opposed to conventional dose (45 mg/m2). |
| IDH1/219,22,35,36,67-71 | Gain of function | IDH1: 7-10.9 | IDH1: 9.6-14 | Multiple conflicting reports on the prognostic impact of IDH1/2 mutations in AML. IDH1/2 mutations consistently reported to be significantly associated with NPM1 mutations and identified favorable effect of IDH2R140Q mutations on OS and especially favorable outcome of patients with both IDH1 or mutually exclusive with TET2 mutations. Studies with uniform treatments IDH2 and NPM1 mutations. |
| IDH2: 8-12.1 | IDH2: 8-19 | |||
| TET24,19,21,24,72 | Affected by loss-of-function mutations and copy-number loss | 7-10 | 19-24.5 | Repeatedly associated with adverse OS in the subset of patients with intermediate-risk AML. In several studies, prognostic effect of TET2 mutations is independent of FLT3-ITD in intermediate-risk AML. |
| Gene . | Mutation effect on gene . | Mutational frequency in AML, % . | Clinical associations . | |
|---|---|---|---|---|
| 16-60 y . | >60 y . | |||
| ASXL16,19,46,47,65 | Affected by loss-of-function mutations and copy-number loss | 3-6.8 | 16.2-25 | Mutations repeatedly identified as enriched in elderly AML patients, AML with an antecedent MDS, and AML patients with RUNX1 mutations. Associated with adverse OS in patients 16-60 y and >60 y of age. In some studies, the adverse effect is only in the subset of CN-AML or intermediate-risk AML. |
| DNMT3A7,14,16,19,66 | Appear to be affected by loss-of-function mutations, effect of the recurrent Arg882 mutation not clear | 17.8-23 | 16-22 | DNMT3A mutations confer adverse risk to intermediate-risk AML patients in multiple series. In most series, this adverse risk is restricted to those patients with FLT3-ITD mutant intermediate-risk AML patients. DNMT3A-mutant patients appear to have improved outcome with high-dose induction daunorubicin therapy (90 mg/m2) as opposed to conventional dose (45 mg/m2). |
| IDH1/219,22,35,36,67-71 | Gain of function | IDH1: 7-10.9 | IDH1: 9.6-14 | Multiple conflicting reports on the prognostic impact of IDH1/2 mutations in AML. IDH1/2 mutations consistently reported to be significantly associated with NPM1 mutations and identified favorable effect of IDH2R140Q mutations on OS and especially favorable outcome of patients with both IDH1 or mutually exclusive with TET2 mutations. Studies with uniform treatments IDH2 and NPM1 mutations. |
| IDH2: 8-12.1 | IDH2: 8-19 | |||
| TET24,19,21,24,72 | Affected by loss-of-function mutations and copy-number loss | 7-10 | 19-24.5 | Repeatedly associated with adverse OS in the subset of patients with intermediate-risk AML. In several studies, prognostic effect of TET2 mutations is independent of FLT3-ITD in intermediate-risk AML. |
CN-AML, cytogenetically normal AML; IDH, isocitrate dehydrogenase; MDS, myelodysplastic syndrome; OS, overall survival.
From the studies of these genes in different AML cohorts, several common themes have already emerged. First, mutations in recently identified epigenetic modifiers are enriched in adults with AML (ages 16 years old and above) compared with pediatric AML patients.3-5 Indeed, the frequency of mutations in epigenetic modifiers correlates with increasing age of AML patients.6-8 Second, from studies evaluating the impact of these mutations in the hematopoietic system of genetically engineered mice, it appears that mutations in several of these genes affect hematopoietic self-renewal and/or differentiation without being sufficient for leukemogenesis on their own.9-12 Consistent with this notion, somatic mutations in TET2 and DNMT3A have been described in the hematopoietic system of elderly individuals before the occurrence of clinically apparent myeloid malignancy.8,13 Lastly, a number of studies integrating mutational analysis with clinical outcome in the setting of prospective clinical trials in AML have identified mutations as markers of prognostic risk stratification in AML. In addition to the prognostic importance of mutations in epigenetic modifiers, there may be important therapeutic implications of specific mutations in epigenetic pathways. In this review, we describe the clinical, biological, and therapeutic implications of mutations in genes encoding epigenetic modifiers which have been identified to be mutated in >5% of patients with AML.
Mutations in genes which impact DNA cytosine modifications: DNMT3A and TET2 mutations
DNMT3A mutations
The DNA (cytosine-5)-methyltransferase 3A (DNMT3A) is a member of the DNA methyltransferase family which includes DNMT1, DNMT3A, DNMT3B, and DNMT3L (Figure 1). Mutations in DNMT3A were initially described by 3 independent groups in 4% to 22% of adult AML patients.14-16 Since that time, recurrence studies in additional AML cohorts have reported that DNMT3A is one of the most frequently mutated genes in AML, occurring in up to 36% of cytogenetically normal AML (CN-AML) patients in the largest series reported to date.17 Moreover, mutations in DNMT3A have been associated with adverse overall survival (OS) (Table 1 and reviewed recently by Thiede18 ). Despite the high frequency of mutations in DNMT3A in AML and their consistent association with adverse prognosis, the biochemical effect of DNMT3A mutations on DNA cytosine methylation and transcription has not been definitively delineated. Mutations in DNMT3A can result in nonsense, frameshift, and missense alterations throughout the open-reading frame.14-16 Of note, a recurrent heterozygous mutation at residue Arginine 882 accounts for 40% to 60% of DNMT3A mutations.14,16 Limited data suggest that R882 mutations result in a loss of methyltransferase activity in in vitro assays.15 However, in AML cells, R882 mutations always occur with retention of the wild-type allele, suggesting the R882 mutant can serve either as a dominant-negative regulator of wild-type DNMT3A or may result in acquisition of an undefined, neomorphic enzymatic activity. In one study, global levels of DNA methylation did not differ between DNMT3A wild-type and mutant patients as assessed using liquid chromatography–tandem mass spectrometry.14 Moreover, methylation studies using the HELP (HpaII tiny fragment enrichment by ligation-mediated polymerase chain reaction) assay have not thus far resolved a methylation-specific signature characteristic of DNMT3A mutant AML samples compared with DNMT3A wild-type samples.7
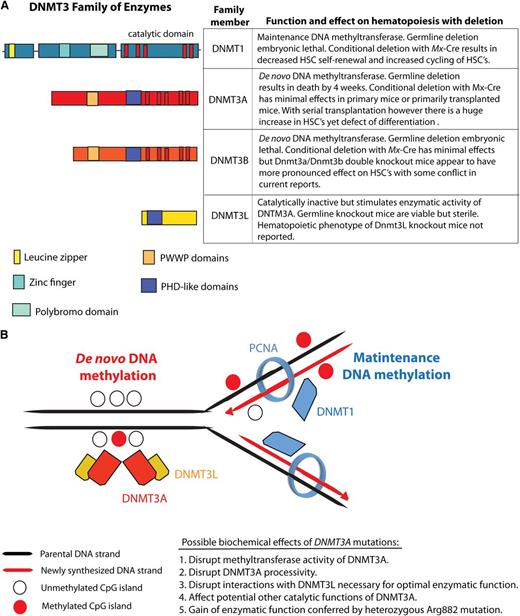
The DNMT family of enzymes and their known effects on hematopoiesis and DNA methylation. The DNA methyltransferases DNMT1, DNMT3A, and DNMT3B catalyze methylation of CpG dinucleotides in genomic DNA. (A) The conserved domains, function, and biological effect of each DNMT member. (B) Currently, it is understood that DNMT3A and DNMT3B are essential for the establishment of DNA cytosine methylation as they catalyze the addition of methyl groups onto the C5 position of DNA cytosine residues without regard for the methylation status of DNA. In contrast, DNMT1 appears to be essential for maintenance of DNA methylation after DNA replication as DNMT1 (1) binds PCNA and (2) has preferential enzymatic activity for hemimethylated DNA over unmethylated DNA. DNMT3L, in contrast, lacks catalytic activity but appears to physically interact with DNMT3A and stimulate its enzymatic activity. PCNA, proliferating cell nuclear antigen; PHD, plant homeodomain; PWWP, proline tryptophan tryptophan proline.
The DNMT family of enzymes and their known effects on hematopoiesis and DNA methylation. The DNA methyltransferases DNMT1, DNMT3A, and DNMT3B catalyze methylation of CpG dinucleotides in genomic DNA. (A) The conserved domains, function, and biological effect of each DNMT member. (B) Currently, it is understood that DNMT3A and DNMT3B are essential for the establishment of DNA cytosine methylation as they catalyze the addition of methyl groups onto the C5 position of DNA cytosine residues without regard for the methylation status of DNA. In contrast, DNMT1 appears to be essential for maintenance of DNA methylation after DNA replication as DNMT1 (1) binds PCNA and (2) has preferential enzymatic activity for hemimethylated DNA over unmethylated DNA. DNMT3L, in contrast, lacks catalytic activity but appears to physically interact with DNMT3A and stimulate its enzymatic activity. PCNA, proliferating cell nuclear antigen; PHD, plant homeodomain; PWWP, proline tryptophan tryptophan proline.
Currently, no specific therapies targeted toward DNMT3A have been developed to date. Recently, work from our group examining the associations of genetic mutations and response to high-dose (90 mg/m2) vs standard dose (45 mg/m2) daunorubicin-induction chemotherapy in AML patients from 16 to 60 years of age suggested that DNMT3A patients specifically benefit from high-dose daunorubicin induction therapy.19 In this study, only patients with mutations in DNMT3A, NPM1, or fusions in MLL1 appeared to benefit from this therapy indicating at least 1 therapeutic implication for the treatment of DNMT3A-mutant AML patients. Although this will need to be validated in other cohorts and investigated in functional systems, these clinical data suggest a mechanistic link between DNMT3A, NPM1, or fusions in MLL1 and relative resistance to anthracyclines.
TET2 mutations
TET2, located on 4q24, is a member of the TET family of proteins (TET1-3). TET2 was named based on the identification that the founding TET member, TET1 (located on chromosome 10q22) as a translocation member with MLL1 on chromosome 11q23 in a rare t(10;11)(q22;q23).20 TET2 is mutated in 8% to 23% of adult patients with AML.19,21-23 Mutations in TET2, however, are enriched in cytogenetically defined intermediate-risk AML or CN-AML where the frequency of TET2 mutations is 18% to 23%.4,24 In parallel with studies on the frequency and clinical importance of TET2 mutations in AML, biochemical studies revealed that the TET family of proteins have a dioxygenase enzymatic activity, dependent on Fe(II) and α-ketoglutarate (α-KG), located in a conserved C-terminal catalytic domain which catalyzes the conversion of the methyl group at the 5-position of cytosine of DNA (5-methylcytosine [5mC] to 5-hydroxymethylcytosine [5hmC])25 (Figure 2). 5hmC, an oxidized version of 5mC, represents a new base in genomic DNA which may have a specific effect on transcription, recruit and/or inhibit specific DNA binding protein, and/or may represent an intermediate in the process of DNA demethylation. Although genome-wide mapping studies of 5hmC in the hematopoietic system have not yet been reported, genome-wide mapping of 5hmC in embryonic stem cells has identified that 5hmC is most often present at transcription start sites and within gene bodies.26,27 5hmC is also more commonly enriched at gene exons compared with introns. Shortly after the discovery of the enzymatic function of the TET family, 2 studies noted that patients with myeloid malignancies with mutations in TET2 had decreased global levels of 5hmC.28,29 However, the effects of TET2 mutations on the global and site-specific levels of 5mC were conflicting in these studies. Recently developed technologies to map 5hmC occupancy on a genome-wide scale, and to resolve 5hmC from 5mC at a single nucleotide base resolution level will hopefully help to determine the effect of TET2 mutations on 5mC, 5hmC, and the relationship of these modified bases to gene transcription.
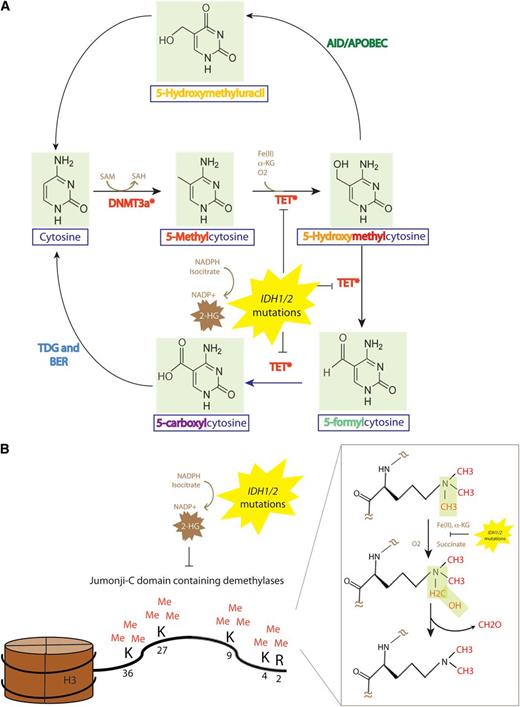
The DNA methylation and demethylation pathway, and effect of TET2 and IDH1/2 mutations on epigenetic DNA and histone modifications. (A) The DNMT family of DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B) each may place a methyl group on the C5 position of DNA cytosine residues in a reaction which requires SAM as a cofactor. (B) Members of the TET family of enzymes (TET1, TET2, TET3) may then oxidize 5-mC to 5hmC an enzymatic reaction which requires Fe(II) and α-KG as substrates. The TET family may also then iteratively oxidize 5hmC further to 5-formylcytosine followed by 5-caC. 5-caC can be directly recognized by the enzyme TDG followed by excision with the BER pathway (an enzymatic activity that is unable to excise 5hmC or 5-mC) to generate unmethylated cytosine. The AID-APOBEC DNA repair pathway can also convert 5hmC to 5-hydroxymethyluracil which activates the BER using TDG or the SMUG1 to generate unmethylated cytosines. TET-mediated enzymatic processes are dependent on α-KG. The presence of an IDH1/2 mutation results in the production of 2-HG, which is structurally very similar to α-KG and can compete with α-KG to inhibit α-KG–dependent enzymatic processes. This includes inhibition of the α-KG–dependent family of JMJC containing histone demethylases. JMJC histone demethylases are responsible for demethylation of histone 3H residues at amino acid residues 2, 4, 9, 27, and 36 and histone H4 amino acid residue 3. 5-cac, 5-carboxylcytosine; AID, activation-induced cytidine deaminase; APOBEC, apolipoprotein B mRNA editing enzyme catalytic; BER, base-excision repair; SMUG1, single-strand-selective monofunctional uracil DNA glycosylase; TDG, thymine DNA glycosylase.
The DNA methylation and demethylation pathway, and effect of TET2 and IDH1/2 mutations on epigenetic DNA and histone modifications. (A) The DNMT family of DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B) each may place a methyl group on the C5 position of DNA cytosine residues in a reaction which requires SAM as a cofactor. (B) Members of the TET family of enzymes (TET1, TET2, TET3) may then oxidize 5-mC to 5hmC an enzymatic reaction which requires Fe(II) and α-KG as substrates. The TET family may also then iteratively oxidize 5hmC further to 5-formylcytosine followed by 5-caC. 5-caC can be directly recognized by the enzyme TDG followed by excision with the BER pathway (an enzymatic activity that is unable to excise 5hmC or 5-mC) to generate unmethylated cytosine. The AID-APOBEC DNA repair pathway can also convert 5hmC to 5-hydroxymethyluracil which activates the BER using TDG or the SMUG1 to generate unmethylated cytosines. TET-mediated enzymatic processes are dependent on α-KG. The presence of an IDH1/2 mutation results in the production of 2-HG, which is structurally very similar to α-KG and can compete with α-KG to inhibit α-KG–dependent enzymatic processes. This includes inhibition of the α-KG–dependent family of JMJC containing histone demethylases. JMJC histone demethylases are responsible for demethylation of histone 3H residues at amino acid residues 2, 4, 9, 27, and 36 and histone H4 amino acid residue 3. 5-cac, 5-carboxylcytosine; AID, activation-induced cytidine deaminase; APOBEC, apolipoprotein B mRNA editing enzyme catalytic; BER, base-excision repair; SMUG1, single-strand-selective monofunctional uracil DNA glycosylase; TDG, thymine DNA glycosylase.
In addition, biological studies examining the role of TET2 in hematopoiesis through genetically engineered mouse models have been very enlightening. Four mouse models with genetic deletion of Tet2 have shown that deletion of Tet2 results in progressive expansion of the hematopoietic stem progenitor compartment, increased hematopoietic stem cell (HSC) self-renewal, and progressive development of a proliferative myeloid malignancy most similar to human chronic myelomonocytic leukemia.11,12,30 By itself, loss of Tet2 does not appear to result in AML in mice. Further work to understand (1) the combined phenotype of mice with Tet2 loss and the presence of co-occurring genetic alterations and (2) the genetic targets of Tet2 loss which result in increased HSC self-renewal alone and in combination with other genetic events may improve our understanding of the pathogenesis and therapy of TET2 mutant AML.
The adverse prognostic effect of TET2 mutations in CN-AML patients with and without the FLT3-ITD mutations provides a rationale for considering the use of allogeneic transplantation in patients with FLT3 wild-type, TET2 mutant CN-AML in first complete remission. In addition, recent preclinical studies suggest inhibition of the EGLN family of prolyl hydroxylases may represent a novel therapeutic approach to the treatment of TET2 mutant patients31 (discussed in the next section, on IDH1/2 mutations). Given the relationship between TET2 and 5mC, several groups have attempted to understand whether patients with myeloid malignancies and TET2 mutations are more sensitive to DNA methyltransferase inhibitor therapy.32-34 However, these studies have had conflicting results and modest numbers of patients with limited genetic characterization beyond TET2 mutations, limiting conclusions from these studies.
Mutations in genes that affect both DNA cytosine modifications and histone posttranslational modifications: IDH1/2 mutations
Mutations in IDH1 and IDH2 are present in 5% to 30% of adults with AML overall. Mutations in IDH1/2 are enriched in CN-AML, where the mutational frequencies are 10% to 16% for IDH1 and 10% to 19% for IDH2, respectively.19,35,36 Unlike mutations in TET2 and DNMT3A, which are associated with adverse outcome in CN-AML, the prognostic relevance of mutations in IDH1 and IDH2 has yielded disparate results among studies despite being examined in multiple large cohorts of AML patients (reviewed recently by Abdel-Wahab37 ). Despite the relative ease of identifying mutations in IDH1 (which occur at Arg132) and IDH2 (which can occur at Arg140 or Arg172), the results from the correlative studies have been complicated by the fact that a number of studies (1) examined the clinical relevance of only 1 or 2 mutations of the 3 possible point mutations in IDH1 and IDH2, (2) have included a limited number of additional genetic alterations beyond IDH1/2 in the analysis, and/or (3) have studied patients receiving different therapies including dose-intense chemotherapy, transplantation, and less aggressive antileukemic therapies. Multiple clinical correlative studies of IDH1/2 mutations, however, have identified a statistically significant association between IDH1/2 mutations and NPM1 mutations and mutual exclusivity of IDH1/2 mutations and TET2 mutations.19,23 In a phase 3 trial of patients from the Eastern Cooperative Oncology Group (ECOG) E1900 cohort, IDH2 mutations at the Arg140 residue were specifically associated with a favorable outcome in AML, both in the entire cohort without regard to the cytogenetic subset as well as in cytogenetically defined intermediate-risk AML patients.19 Moreover, the subset of patients with both NPM1 mutations and IDH1 or IDH2 mutations had a particularly favorable outcome, superior to that of the cytogenetically favorable subset of AML patients (3-year OS of intermediate-risk AML patients with NPM1 and IDH1/IDH2 mutations was 89%).
IDH1/2 normally participates in the citric-acid cycle to convert isocitrate to α-KG while reducing NAD phosphate–positive (NADP+) to reduced NADP (NADPH). Mutations in IDH1 (at Arg132) or in IDH2 (at Arg140 or Arg172) confer a new enzymatic function to these enzymes, with acquisition of the ability to convert α-KG to 2-hydroxyglutarate (2-HG) via the conversion of NADPH to NADP+38,39 (Figure 2). This results in increased production of 2-HG than would occur in usual physiologic conditions. One of the downstream sequelae of this increased production of 2-HG is that enzymes that normally depend on α-KG as a substrate are functionally impaired in the presence of IDH1/2 mutations due to competitive inhibition by 2-HG. The spectrum of α-KG–dependent enzymes impaired by the presence of IDH1/2 mutations and/or 2-HG include the TET family of enzymes and the Jumonji-C domain-containing (JMJC) family of histone lysine demethylases (Figure 2).29,40
Recently, Sasaki et al41 have reported the phenotype of a conditional Idh1(R132H) knockin (KI) mouse model. The authors found that mice expressing the R132H mutation produced 10-fold higher serum 2-HG than wild-type littermates. Despite this elevation in serum 2-HG, the KI mice had a normal lifespan and were viable and fertile. At the same time, conditional expression of Idh1R132H using either system resulted in an age-dependent splenomegaly due to extramedullary hematopoiesis and a coincident development of a hypocellular bone marrow compared with age-matched wild-type counterparts. Despite the decreased cellularity of the marrow in the KI mice, there was an expansion in the most primitive stem/progenitor compartment in these mice due to an expansion of the multipotent progenitor population specifically. This suggests that mutant Idh1 actually results in an age-dependent blockade of hematopoietic differentiation. Similar to the situation with Dnmt3a and Tet2 null mice,9 however, Idh1 R132H expression in the hematopoietic system does not appear sufficient on its own to result in AML in vivo.
In addition to the above effects of mutant forms of IDH1/2 on hematopoiesis, 2-HG exposure results in impaired myeloid differentiation and cytokine-independent growth in TF-1 cells which normally depend on granulocyte macrophage–colony-stimulating factor for growth.31 Interestingly, using a cell-permeable form of the R-enantiomer and S-enantiomer of 2-HG (R-2HG and S-2HG, respectively), Losman et al31 have found that 2-HG is responsible for the growth-transforming effects of 2-HG while S-2HG is not transforming. Moreover, further analysis of this phenomenon suggests that the transforming effects of R-2HG may specifically be related to the fact that R-2HG acts as a cofactor to promote the hydroxylase activity of the EglN prolyl-4-hydroxylases, while S-2HG does not. In accord with this, the authors demonstrated that knockdown of EglN1 prevented transformation of TF-1 cells in the presence of 2-HG or mutant IDH1 expression. Interestingly, TF-1 cells transformed by TET2 loss were similarly inhibited by knockdown of EglN1. These findings suggest that therapeutic targeting of EglN prolyl-4-hydroxylase activity might be effective in the treatment of IDH1-mutant and TET2-mutant myeloid leukemias.
In addition to the potential therapeutic use of EglN inhibition for IDH1/2-mutant disease, small molecule selective inhibitors of mutant IDH1 and IDH2 have recently been developed. Data presented at the 2012 American Society of Hematology meeting reported that in vitro treatment with IDH inhibitors reverses hypermethylation of DNA as well as histones, and in vivo use lowers 2-HG by >90% in xenograft models.42 In addition to mutant-specific small-molecule inhibitors of IDH1 and IDH2, there may be therapeutic potential in inhibiting production of glutamine, the essential source of α-KG, by small-molecule inhibiting of the enzyme glutaminase.43 Further detailed description of preclinical studies in IDH1/2-mutant in vivo models is eagerly anticipated.
Mutations in genes which affect histone posttranslational modifications: ASXL1 and other Polycomb-group genes
In addition to mutations in genes that affect DNA cytosine modifications, genes which affect histone posttranslational modifications have also been found to be repeatedly mutated in AML. The earliest and most studied of such genes is MLL1, which is affected by translocations as well as in-frame duplications in AML.44 More recently, however, mutations in the Polycomb group of proteins have been found in patients with all myeloid malignancies. Of these, the most extensively studied in AML is ASXL1. ASXL1 is affected by somatic deletions as well as point mutations in patients with all myeloid malignancies. Mutations in ASXL1 were initially identified by Gelsi-Boyer et al45 in patients with myelodysplastic syndrome (MDS) and chronic myelomonocytic leukemia. More recently, a series of studies have extensively characterized the prognostic relevance of ASXL1 mutations in AML (Table 2). The findings from these studies have been remarkably consistent. First, while mutations in ASXL1 are present in 6% to 30% of AML patients, there is a positive correlation of ASXL1 mutations with advancing age, and ASXL1 mutations have been found in 16.2% to 25% of AML patients >60 years of age6,46 compared with 3.6% to 8% of adult AML patients <60 years of age.6,19 Studies by Metzeler et al6 and Schnittger et al46 indicate that ASXL1 mutations are 4 to 5 times more likely in AML patients >60 years of age compared with those from 16 to 60 years of age. Second, there appears to be an enrichment of ASXL1 mutations in AML patients with a history of preceding MDS.47 Most importantly, ASXL1 mutations have been associated with adverse outcome in all studies to date in AML, MDS, and myeloproliferative neoplasm.
Clinical correlative studies of ASXL1 mutations in patients with AML
| Study . | No. of patients . | Mutational frequency, % . | Clinical findings . | Patient population/comments on study design . |
|---|---|---|---|---|
| 65 | 63 | 19 | ASXL1 mutations exclusive with NPM1 mutations (validated in at least 1 larger study). | No specific subtype of AML studied. No effects on clinical outcome studied. |
| 47 | 67 | 25 | None. | No specific subtype of AML studied. No effects on clinical outcome studied. |
| 73 | 501 | 10.8 | Mutation associated with older patients. Mutant patients had lower complete remission rate and shorter OS but ASXL1 mutations not an independent predictor of adverse outcome in MVA. | All patients with de novo AML from Taiwan. ASXL1 mutations significantly associated with RUNX1 mutations. |
| 6 | 423 | 8.9 | ASXL1 mutations much more common in AML patients >60 y of age compared with younger patients. ASXL1 mutations associated with adverse OS only in ELN-favorable patients. ASXL1 mutations exclusive with NPM1 mutations. | All patients with CN-AML and all received cytarabine/daunorubicin-based first-line therapy on a CALGB trial. |
| 46 | 740 | 17.2 | Mutation associated with older patients. ASXL1 mutations were an adverse predictor of worsened OS. | All patients with cytogenetically defined intermediate-risk AML. ASXL1 mutations significantly associated with RUNX1 mutations. |
| 19 | 502 | 3 | ASXL1 mutations were an adverse predictor of worsened OS in overall cohort as well as intermediate-risk AML patients. | All patients aged 16-60 y of age from the ECOG E1900 randomized phase 3 trial of standard dose (45 mg/mg2) or high-dose daunorubicin (90 mg/mg2) plus cytarabine induction therapy. |
| Study . | No. of patients . | Mutational frequency, % . | Clinical findings . | Patient population/comments on study design . |
|---|---|---|---|---|
| 65 | 63 | 19 | ASXL1 mutations exclusive with NPM1 mutations (validated in at least 1 larger study). | No specific subtype of AML studied. No effects on clinical outcome studied. |
| 47 | 67 | 25 | None. | No specific subtype of AML studied. No effects on clinical outcome studied. |
| 73 | 501 | 10.8 | Mutation associated with older patients. Mutant patients had lower complete remission rate and shorter OS but ASXL1 mutations not an independent predictor of adverse outcome in MVA. | All patients with de novo AML from Taiwan. ASXL1 mutations significantly associated with RUNX1 mutations. |
| 6 | 423 | 8.9 | ASXL1 mutations much more common in AML patients >60 y of age compared with younger patients. ASXL1 mutations associated with adverse OS only in ELN-favorable patients. ASXL1 mutations exclusive with NPM1 mutations. | All patients with CN-AML and all received cytarabine/daunorubicin-based first-line therapy on a CALGB trial. |
| 46 | 740 | 17.2 | Mutation associated with older patients. ASXL1 mutations were an adverse predictor of worsened OS. | All patients with cytogenetically defined intermediate-risk AML. ASXL1 mutations significantly associated with RUNX1 mutations. |
| 19 | 502 | 3 | ASXL1 mutations were an adverse predictor of worsened OS in overall cohort as well as intermediate-risk AML patients. | All patients aged 16-60 y of age from the ECOG E1900 randomized phase 3 trial of standard dose (45 mg/mg2) or high-dose daunorubicin (90 mg/mg2) plus cytarabine induction therapy. |
CALGB, Cancer and Leukemia Group B; ELN, European Leukemia Net; MVA, multivariate analysis.
Despite the adverse prognostic relevance of ASXL1 mutations in AML patients, the biological function of ASXL1 has been less clearly delineated. ASXL1 has been reported to be involved in a large number of physical interactions where the biological and biochemical relevance of the majority of these interactions to hematopoiesis is unclear. ASXL1 was shown to interact with HP1α,48 LSD1,48 SRC1,49 and RARα49 in in vitro assays in nonhematopoietic contexts. More recently, ASXL1 has been shown to interact with the nuclear deubiquitinase BAP1 in a biochemical complex which serves to remove a ubiquitin from histone H2A at lysine 11950 (H2AK119). While this biochemical activity has been demonstrated in cell-free biochemical assays,50 the in vivo relevance of this interaction and regulation of H2AK119Ub levels is less clear, possibly related to redundancy in ASXL proteins and their interaction with BAP1.51,52 In addition to these interactions, we recently identified that mutations in ASXL1 result in loss of ASXL1 protein expression.51 Given that Asx in Drosophila appears to regulate Polycomb- as well as Trithorax-group gene function, we analyzed the effect of ASXL1 mutations and loss on the chromatin modifications placed by these enzymatic complexes. This analysis revealed a striking global decrease of histone 3H lysine 27 methylation, a histone mark associated with repression of transcription placed by the Polycomb-repressive complex 2 (PRC2). Concordant with the effect of ASXL1 on H3K27me3 levels, ASXL1 was found to physically associate with the core PRC2 complex members, and loss of ASXL1 diminished recruitment of PRC2 member PRC2 target loci, suggesting that ASXL1 may function in recruitment and/or stabilization of the PRC2 complex to specific loci in the mammalian genome.51
There have thus far been 2 studies evaluating the in vivo function of Asxl1 in hematopoiesis. First, soon after the discovery of ASXL1 mutations, Fisher et al10 reported the phenotype of mice with a germline gene-trap allele of Asxl1. Asxl1 attenuation resulted in significant perinatal lethality, although surviving mice had defects in B lymphopoiesis and developmental defects. In order to understand the biological effect of postnatal Asxl1 loss in an established MDS/myeloproliferative neoplasm model, we knocked down Asxl1 expression using small-hairpin RNA in vivo in a mouse model of hematopoietic NrasG12D overexpression.51 This resulted in accelerated disease latency and increased disease burden in vivo, underscoring the biological importance of Asxl1 alterations in myeloid transformation.
The therapeutic potential for epigenetic-targeted therapies in AML
The high frequency of somatic alterations in epigenetic modifiers in AML patients, combined with the established clinical importance of DNA methyltransferase inhibitors in the clinical management of patients with myeloid malignancies, has led to a great interest in the development of novel epigenetic therapies (Table 3). Proteins involved in epigenetic regulation often depend on critical protein-protein interactions for function as well as on cofactors for enzymatic activity. For instance, all known lysine and arginine histone methyltransferases require the cofactor S-Adenosyl methionine (SAM) for the enzymatic activity of transferring a methyl donor group. Thus, one challenge to developing targeted epigenetic therapies has been in developing therapies to specifically inhibit 1 epigenetic-regulatory protein or enzyme without affecting other epigenetic regulators. In addition, disruption of protein-protein binding interactions has been a recurrent challenge in drug development. Despite these technical challenges, a number of compounds have been developed which appear to target specific epigenetic-regulatory proteins or processes. These include inhibitors of bromodomains (recently reviewed in Dawson et al53 ), mutant IDH1 and IDH2 proteins,42 and selective competitive antagonists of the enzymes EZH2,54,55 LSD1,56-59 and DOT1L60,61 (Table 1). A number of these therapies have been tested specifically in preclinical contexts of MLL-translocated AML and studies in broader biological and genetic contexts are needed to understand their efficacy in a wider array of AML subtypes. Moreover, studies of these compounds and the impact of inhibiting epigenetic complexes on epigenomic patterning and on normal HSCs, differentiation, and homeostasis are in their earliest stages. For instance, at least 3 studies have identified that inhibition of PRC2 activity by genetic inactivation of EZH2 has potential therapeutic efficacy in the context of MLL-AF9–driven leukemias.62-64 Whether EZH2 inactivation may have therapeuticefficacy in other subtypes of AML or in other myeloid malignancies has not been studied. Moreover, while the rationale for EZH2 inhibition in lymphoma54,55 and solid tumors is becoming readily apparent, the long-term effects of EZH2 pharmacologic inhibition on normal hematopoiesis are not yet known. Nonetheless, the development of these novel therapeutics has already resulted in a number of phase 1 studies in patients with hematologic malignancies, including phase 1 trials of the DOT1L inhibitor EPZ-5676 and of the bromodomain inhibitor OTX015. These early-phase studies will hopefully yield critical information for further therapeutic studies of these agents. In parallel, the development of these epigenetic-targeted agents provide exciting reagents to probe epigenetic complexes and their roles in normal and malignant hematopoiesis. One important question to answer will be to identify the effect of epigenetic targeted therapies on differentiation of normal and leukemic hematopoietic cells as opposed to merely inducing leukemic cell death. Moreover, the unexpected adverse effects and possible mechanisms of resistance to these promising therapies are currently unknown.
Novel epigenetic-targeted pharmacologic agents in preclinical development in the treatment of myeloid malignancies
| Target . | Rationale . | Stage of clinical development . |
|---|---|---|
| BET family of bromodomain containing proteins (BRD2, BRD3, BRD4, and BRDt) | Inhibition of BET bromodomains has been repeatedly shown to have therapeutic efficacy in preclinical models of AML (both MLL translocated and MLL wild-type AML).74,75 A single underlying basis for this efficacy is not entirely clear but it is possibly related to downregulation of MYC transcription following BET inhibition and/or to inhibition of transcriptional elongation. | Phase 1 study of the BRD2/3/4 inhibitor OTX015 in patients with hematologic malignancies is ongoing. |
| DOT1L | Most MLL translocations result in recruitment of the histone 3H lysine 79 methyltransferase DOT1L (reviewed in Krivtsov et al44 ) and recruitment of DOT1L has been shown to be critical for the transforming properties of MLL fusion proteins.76 | Phase 1 and expanded cohort study of EPZ-5676 in advanced hematologic malignancies, including acute leukemia with MLL rearrangements. |
| IDH1/IDH2 | Recurrent gain-of-function mutations in IDH1 and IDH2 result in the production of 2-HG38,39 and both the presence of the mutant forms of IDH1/229,31,41 as well as the presence of 2-HG alone31 has been shown to be transforming for myeloid hematopoietic cells. Thus, mutant-selective inhibitors of IDH1 and IDH2 are in preclinical development.42 | Preclinical studies on the therapeutic potential of mutant IDH1/2 pharmacologic inhibition are ongoing.42 |
| EZH2 | Despite the fact that somatic deletions and mutations of EZH2 are present in patients with myeloid malignancies,77 genetic deletion of EZH2 has been shown to impair the oncogenicity of MLL-AF9 AML cells.62-64 | Preclinical studies on the therapeutic potential of EZH2 pharmacologic inhibition54,55 in the context of MLL wild-type and translocated AML are ongoing. |
| LSD1 | Downregulating LSD1 by RNA interference or pharmacologic inhibition with tranylcypromine analogs selectively targeted MLL translocated leukemic cells by inducing their differentiation in one study.58 In another study, combining LSD1 inhibition with ATRA resulted in terminal differentiation and abrogation of leukemogenesis of AML cell lines and primary AML samples (regardless of presence of MLL translocation).56 | Preclinical studies of tranylcypromine and compounds derived from it are ongoing. In addition, development of more potent and selective LSD1 inhibitors are in development.57,59,78 |
| UTX/JMJD3 | The recurrent presence of mutations which result in loss of H3K27me3 in myeloid malignancies (including ASXL1,51 EZH2,77 SUZ12,79 and EED79 deletions and somatic mutations) suggest a rationale for inhibiting demethylation of H3K27 in these disorders. | Preclinical studies on the therapeutic potential of UTX/JMJD3 pharmacologic inhibition80 in human malignancies are ongoing. |
| Target . | Rationale . | Stage of clinical development . |
|---|---|---|
| BET family of bromodomain containing proteins (BRD2, BRD3, BRD4, and BRDt) | Inhibition of BET bromodomains has been repeatedly shown to have therapeutic efficacy in preclinical models of AML (both MLL translocated and MLL wild-type AML).74,75 A single underlying basis for this efficacy is not entirely clear but it is possibly related to downregulation of MYC transcription following BET inhibition and/or to inhibition of transcriptional elongation. | Phase 1 study of the BRD2/3/4 inhibitor OTX015 in patients with hematologic malignancies is ongoing. |
| DOT1L | Most MLL translocations result in recruitment of the histone 3H lysine 79 methyltransferase DOT1L (reviewed in Krivtsov et al44 ) and recruitment of DOT1L has been shown to be critical for the transforming properties of MLL fusion proteins.76 | Phase 1 and expanded cohort study of EPZ-5676 in advanced hematologic malignancies, including acute leukemia with MLL rearrangements. |
| IDH1/IDH2 | Recurrent gain-of-function mutations in IDH1 and IDH2 result in the production of 2-HG38,39 and both the presence of the mutant forms of IDH1/229,31,41 as well as the presence of 2-HG alone31 has been shown to be transforming for myeloid hematopoietic cells. Thus, mutant-selective inhibitors of IDH1 and IDH2 are in preclinical development.42 | Preclinical studies on the therapeutic potential of mutant IDH1/2 pharmacologic inhibition are ongoing.42 |
| EZH2 | Despite the fact that somatic deletions and mutations of EZH2 are present in patients with myeloid malignancies,77 genetic deletion of EZH2 has been shown to impair the oncogenicity of MLL-AF9 AML cells.62-64 | Preclinical studies on the therapeutic potential of EZH2 pharmacologic inhibition54,55 in the context of MLL wild-type and translocated AML are ongoing. |
| LSD1 | Downregulating LSD1 by RNA interference or pharmacologic inhibition with tranylcypromine analogs selectively targeted MLL translocated leukemic cells by inducing their differentiation in one study.58 In another study, combining LSD1 inhibition with ATRA resulted in terminal differentiation and abrogation of leukemogenesis of AML cell lines and primary AML samples (regardless of presence of MLL translocation).56 | Preclinical studies of tranylcypromine and compounds derived from it are ongoing. In addition, development of more potent and selective LSD1 inhibitors are in development.57,59,78 |
| UTX/JMJD3 | The recurrent presence of mutations which result in loss of H3K27me3 in myeloid malignancies (including ASXL1,51 EZH2,77 SUZ12,79 and EED79 deletions and somatic mutations) suggest a rationale for inhibiting demethylation of H3K27 in these disorders. | Preclinical studies on the therapeutic potential of UTX/JMJD3 pharmacologic inhibition80 in human malignancies are ongoing. |
Preclinial and clinical trials involving novel DNA methyltransferase inhibitors and HDAC inhibitors are not mentioned. BET, bromodomain and extra-terminal; HDAC, histone deacetylase.
Conclusion
The discovery of mutations in specific epigenetic modifiers in patients with AML has furthered our understanding of the pathophysiology of this disorder. Genomic analysis of these genes in clinically annotated cohorts of AML patients strongly suggests that incorporation of mutational testing for DNMT3A, TET2, IDH1/2, and ASXL1 may improve our ability to risk-stratify patients with AML and potentially provide additional biomarkers for minimal residual disease detection. The identification of 5 of 10 recurrent molecular genetic alterations in AML patients with prognostic importance presents a challenge for implementing testing of these genetic alterations into clinical practice. Current genetic testing of AML clinical patient samples relies on characterization of metaphase karyotype, fluorescence in situ hybridization, restriction enzyme digestion of polymerase chain reaction products, and capillary sequencing. However, these currently clinically used technologies will be inadequate for comprehensive genetic characterization of AML patients in the future; conventional Sanger sequencing will be overly costly and unwieldy for these purposes as well. Second-generation sequencing technologies (eg, Illumina and SOLiD) and/or the use of array-based sequencing platforms (the Roche NimbleGen and Agilent Capture Array being 2 examples) may be the best candidates for initiating comprehensive genetic profiling of patient samples in clinical practice. The current limitations which prevent implementation of such sequencing in clinical practice include the high cost, slow turnaround time, and lack of clinical validation. Efforts to limit the sequencing to panels of candidate target genes may improve all of these limitations, however. Changes in treatment which may be made based on the identification of these mutations includes use of dose-escalated daunorubicin (90 mg/m2) during induction for DNMT3A and MLL mutant AML. Clinical development of mutant-specific inhibitors of IDH1 and/or IDH2, EglN, and translocated MLL may further improve the need to identify these alterations in the clinical management of AML patients.
Our current understanding of the spectrum of somatic genetic alterations in AML has already outpaced the range of therapeutic approaches currently available for the treatment of patients with AML. However, the identification of frequent somatic mutations in the epigenetic modifiers described here has informed several novel potential therapeutic approaches in the treatment of AML. A number of these compounds are already in phase 1 trials for patients with AML and a larger number of compounds are in preclinical development. In some cases novel compounds targeting posttranslational modifications of histones and DNA cytosine modifications appear to result in potent and specific inhibition of specific enzymatic activities. However, the preclinical and clinical development of these agents may be influenced by the identification that these agents may require longer exposure to have therapeutic efficacy than has been the case with traditional cytotoxic therapies or small-molecule tyrosine kinase inhibitors.54,55,60 In addition, preclinical studies of several of these compounds in AML cells suggest they may result in alterations in phenotype and gene expression characteristic of restoration of hematopoietic differentiation rather than direct cytotoxicity of AML cells.56,58,60 Further preclinical studies using in vitro and in vivo systems will improve of the role of mutations in epigenetic modifiers in AML pathogenesis, and the effects of inhibiting epigenetic modulators on normal hematopoiesis.
Acknowledgments
This work was supported by grants from the Gabrielle’s Angel Fund (R.L.L. and O.A.-W.) and a grant from the Anna Fuller Fund (R.L.L.). O.A.-W. is an American Society of Hematology Basic Research Fellow and is supported by a grant from the National Institutes of Health K08 Clinical Investigator Award (1K08CA160647-01).
Authorship
Contribution: O.A.-W. and R.L.L. reviewed the literature, co-wrote the manuscript, and made the tables and figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ross L. Levine, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10065; e-mail: leviner@mskcc.org.