Key Points
AML1-ETO-W692A loses interaction between NHR4 and N-CoR, decreases AML1-ETO cellular dysregulation, and promotes leukemia development in mice.
Abstract
AML1-ETO (RUNX1-ETO) fusion proteins are generated by the 8;21 translocation, commonly found in acute myeloid leukemia, which fuses the AML1 (RUNX1) and ETO (MTG8, RUNX1T1) genes. Previous studies have shown that AML1-ETO interferes with AML1 function but requires additional cooperating mutations to induce leukemia development. In mouse models, AML1-ETO forms lacking the C-terminus have been shown to have greatly enhanced leukemogenic potential. Here, we investigate the role of 2 AML1-ETO C-terminal-interacting proteins, N-CoR, a transcriptional corepressor, and SON, a splicing/transcription factor required for cell cycle progression, in AML1-ETO-induced leukemia development. AML1-ETO-W692A loses N-CoR binding at NHR4, displays attenuated transcriptional repression ability and decreased cellular dysregulation, and promotes leukemia in vivo. These results support the importance of the degree of dysregulation by AML1-ETO in cellular transformation and demonstrate that AML1-ETO-W692A can be used as an effective experimental model for determining which factors compromise the leukemogenic potential of AML1-ETO.
Introduction
t(8;21), one of the most common chromosomal abnormalities associated with acute myeloid leukemia (AML),1-4 causes expression of fusion protein AML1-ETO. AML1-ETO alone is insufficient to induce leukemia in mouse models,5 and additional cooperating mutations are required.6 If AML1-ETO is C-terminally truncated through mutation or alternative splicing, it rapidly induces leukemia.7,8
AML1-ETO C-terminus interacts with several factors, including nuclear receptor corepressors N-CoR/SMRT9 and the large nuclear protein SON.10 N-CoR associates with mSin3A and histone deacetylases,11,12 and the N-CoR/AML1-ETO interaction mediates aberrant transcriptional repression of AML1 target genes.13-15 SON is a DNA- and RNA-binding protein, is involved in regulation of gene transcription and RNA splicing, and is required for cell cycle progression.16,17 Blocking protein interactions with AML1-ETO C-terminus by overexpression of a SON fragment is sufficient to rescue cells from AML1-ETO-induced growth arrest.10
C-terminal C663S mutation of AML1-ETO reportedly destroyed the zinc-chelating structure of the NHR4 domain and disrupted N-CoR interaction with NHR4.14,18 We previously showed that AML1-ETO-C663S had similar leukemogenic potential to C-terminally truncated AML1-ETO and that ETO-C663S mutation eliminated SON interaction but retained N-CoR binding.10 The interaction between N-CoR and ETO-C663S is likely a result of N-CoR and Sin3A binding to other ETO domains besides NHR4.14 As stated in our previous report, although we hypothesized that the SON/NHR4 interaction may be one factor inhibiting AML1-ETO leukemogenic potential, we could not exclude the possibility that reduced N-CoR recruitment to AML1-ETO-C663S or other factors also contributes to leukemogenesis.10 As both aberrant transcriptional regulation and cell cycle progression could conceivably attenuate AML1-ETO leukemogenicity, it is important to determine which process, recruitment of N-CoR or inactivation/sequestration of SON, was responsible. Using AML1-ETO mutants to differentiate N-CoR and SON binding, we aim here to delineate the contributions of these 2 proteins in decreasing AML1-ETO leukemogenicity, as they each potentially contribute through very different mechanisms requiring different therapeutic strategies for effective treatment of AML. We demonstrate that AML1-ETO-W692A mutation loses NHR4/N-CoR interaction, attenuates AML1-ETO cellular effects, and promotes leukemogenesis, supporting a new model for study of AML1-ETO transformation and leukemia.
Study design
Plasmids, immunoprecipitation, and immunoblotting
NHR2-4 constructs expressed from pCMV2 (Sigma-Aldrich). Amino acid mutations generated by site-directed mutagenesis (Agilent). Immunoprecipitation: precleared lysate incubated 4 h-overnight at 4°C with antibody and washed 5 times with lysis buffer.16 Antibodies: N-CoR (Abcam); Flag, α-tubulin (Sigma-Aldrich); CDK4, Cyclin A, Cyclin D3 (Santa Cruz); p27 (BD Biosciences); HA (Covance).
Luciferase reporter assay
Performed as previously described.19 Significance determined by Student t test.
Apoptosis and MTS assays
K562 or primary bone marrow cells transduced with virus produced by cotransfection of packaging vector and MSCV-IRES-Puror containing indicated AML1-ETO constructs in 293T cells, selected 2 to 3 d in 2 μg/mL puromycin, and subjected to Annexin V Apoptosis Kit (BD Pharmingen) or CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega) per manufacturers’ instructions.
Animals
C57BL/6 mice housed in a pathogen-free facility. Procedures approved by Institutional Animal Care and Use Committee of University of California San Diego.
Mouse survival
Kaplan-Meier survival curves generated using GraphPad Prism4 (GraphPad Software, San Diego, CA).
Fetal liver-cell isolation, infection, and transplantation
See supplemental Methods.
Results and discussion
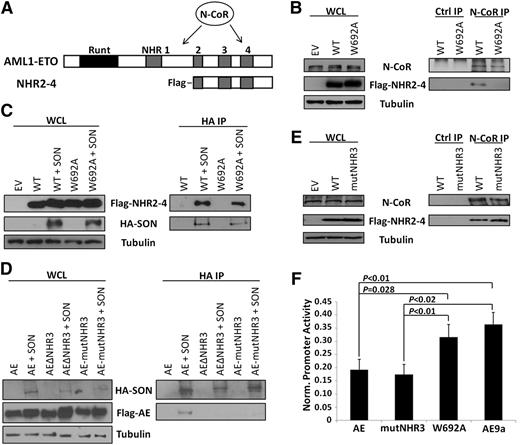
Three groups have reported that N-CoR/SMRT directly interacts with NHR4,13-15 although AML1-ETO without the NHR4 domain can still interact with N-CoR via another site (Figure 1A). C663S mutation is predicted to disrupt the zinc-chelating structure of NHR4 and thereby block all its protein-protein interactions. W692A retains NHR4 structure but is predicted to lose N-CoR interaction.18 To determine whether W692A disrupts N-CoR binding to NHR4, we coimmunoprecipitated endogenous N-CoR and transiently transfected ETO fragments beginning at NHR2, to allow proper tetramerization,20 and continuing through ETO C-terminus (Figure 1A, ‘NHR2-4’), thereby eliminating the additional N-CoR binding site between NHR1 and NHR221 and allowing us to examine specifically the N-CoR interaction at NHR4. Endogenous N-CoR interacts with wild-type, but not W692A-mutant, NHR2-4 (Figure 1B). Conversely, transiently transfected SON retains interaction with W692A (Figure 1C). To substantiate this result further, we confirmed that endogenous SON also retains its interaction with Flag-NHR2-4-W692A (supplemental Figure 1). Combined, these results demonstrate that NHR4-W692A loses N-CoR, but retains SON, binding.
Characterization of NHR4/N-CoR and AML1-ETO/SON binding and effects on gene repression. (A) Structures of AML1-ETO and the NHR2-4 constructs used in the co-IP experiments, with location of N-CoR binding sites indicated. (B) Endogenous N-CoR interacts with WT, but not W692A, NHR2-4. Cell lysates were immunoprecipitated with N-CoR or isotype control (ctrl) antibodies, and N-CoR and NHR2-4 were detected with N-CoR and Flag antibodies, respectively. Tubulin serves as a loading control. (C) SON interacts with both WT and W692A NHR2-4. Cell lysates were immunoprecipitated with HA antibody, and SON and NHR2-4 were detected using HA and Flag antibodies, respectively. (D) SON interacts with full-length AML1-ETO (AE), but not when the NHR3 domain is deleted (ΔNHR3) or when amino acids 629-634 ‘TERAKM’ in NHR3 are mutated to ‘AAAAAA’ (mutNHR3). Immunoprecipitation is as performed in (C). (E) Endogenous N-CoR retains interaction with NHR2-4 containing mutNHR3 alanine mutations. Immunoprecipitation performed as in (B). (F) W692A decreases ability of AML1-ETO to repress a luciferase reporter; mutNHR3 does not. 293T cells were cotransfected with firefly luciferase reporter containing tandem AML1 binding sites upstream of the basal TK promoter, Renilla control luciferase and either empty vector (control) or indicated AML1-ETO construct. Firefly luciferase expression was normalized to Renilla expression and control was set to 1. Equal expression of AML1-ETO constructs was confirmed by immunoblot using HA antibody (data not shown). Data show averages and standard deviations of 3 independent experiments, each performed in duplicate. EV, empty vector; WCL, whole cell lysate.
Characterization of NHR4/N-CoR and AML1-ETO/SON binding and effects on gene repression. (A) Structures of AML1-ETO and the NHR2-4 constructs used in the co-IP experiments, with location of N-CoR binding sites indicated. (B) Endogenous N-CoR interacts with WT, but not W692A, NHR2-4. Cell lysates were immunoprecipitated with N-CoR or isotype control (ctrl) antibodies, and N-CoR and NHR2-4 were detected with N-CoR and Flag antibodies, respectively. Tubulin serves as a loading control. (C) SON interacts with both WT and W692A NHR2-4. Cell lysates were immunoprecipitated with HA antibody, and SON and NHR2-4 were detected using HA and Flag antibodies, respectively. (D) SON interacts with full-length AML1-ETO (AE), but not when the NHR3 domain is deleted (ΔNHR3) or when amino acids 629-634 ‘TERAKM’ in NHR3 are mutated to ‘AAAAAA’ (mutNHR3). Immunoprecipitation is as performed in (C). (E) Endogenous N-CoR retains interaction with NHR2-4 containing mutNHR3 alanine mutations. Immunoprecipitation performed as in (B). (F) W692A decreases ability of AML1-ETO to repress a luciferase reporter; mutNHR3 does not. 293T cells were cotransfected with firefly luciferase reporter containing tandem AML1 binding sites upstream of the basal TK promoter, Renilla control luciferase and either empty vector (control) or indicated AML1-ETO construct. Firefly luciferase expression was normalized to Renilla expression and control was set to 1. Equal expression of AML1-ETO constructs was confirmed by immunoblot using HA antibody (data not shown). Data show averages and standard deviations of 3 independent experiments, each performed in duplicate. EV, empty vector; WCL, whole cell lysate.
To find a mutant of AML1-ETO with the opposite interaction pattern, we mapped the SON binding site through deletion and alanine mutation. We previously reported that NHR4 C663S mutation impairs SON interaction (supplemental Figure 1).10 Because disruption of NHR4 structure may also affect protein-protein interactions with neighboring regions of AML1-ETO, we examined whether SON interacts with adjacent domains. Interestingly, deletion of either NHR3 or NHR4 blocks SON interaction with AML1-ETO (supplemental Figure 2). Furthermore, mutation of 6 amino acids (TERAKM) within NHR3 to alanines (AE-mutNHR3) blocks AML1-ETO interaction with SON (Figure 1D), but not N-CoR (Figure 1E).
AML1-ETO downregulates AML1 target genes via interaction with N-CoR/SMRT and associated HDAC complexes,14,15 and loss of N-CoR/NHR4 interaction attenuates the function of AML1-ETO.18 To examine the effect of W692A and NHR3 mutation on AML1-ETO transcriptional repression, we cotransfected AML1-ETO constructs with an AML1 target promoter-luciferase reporter.19 As expected, AML1-ETO strongly repressed luciferase expression up to 80% of control, whereas AML1-ETO9a, which lacks NHR3 and NHR4 domains,8 displayed attenuated repression (Figure 1F). Interestingly, AE-mutNHR3 behaved like full-length AML1-ETO, but AE-W692A displayed impaired repression, similar to AML1-ETO9a (Figure 1F). This same pattern of gene dysregulation was also observed at endogenous AML1-ETO targets in primary murine bone marrow cells (supplemental Figure 3).18 AE-W692A thereby decreases AML1-ETO transcriptional repression, correlating the number of N-CoR binding sites with repression ability.
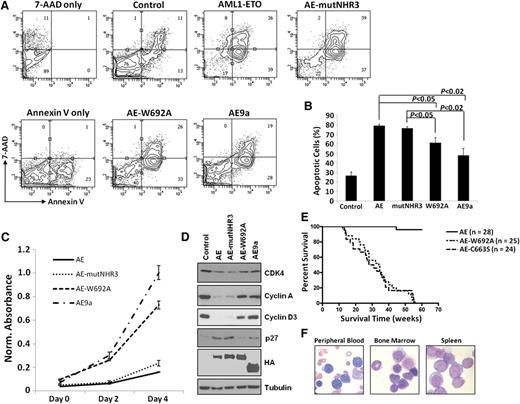
We next tested the functional effects of these mutants on cellular dysregulation. AE-mutNHR3 increased apoptosis and decreased proliferation in both primary murine bone marrow (Figures 2A-C) and K562 cells (supplemental Figure 4). AE-mutNHR3 also increased p27 levels and decreased expression of CDK4 and cyclins A and D3 (Figure 2D). These results mirror those obtained with full-length AML1-ETO and demonstrate the negative effect of these proteins on survival and cell cycle. AE-W692A, however, behaved similarly to the leukemogenic AE9a, demonstrating significantly reduced defects in the previously mentioned assays (Figures 2A-D; supplemental Figure 4). These results clearly indicate stronger negative cellular effects of AE-mutNHR3 compared to AE-W692A.
Effects of AML1-ETO–mutNHR3 and –W692A on cellular dysregulation. (A) Effects of AML1-ETO mutants on apoptosis in primary bone marrow cells. Murine bone marrow cells transduced with the indicated AML1-ETO constructs were harvested after 2 d of selection, stained with 7-AAD and Annexin V-APC, and analyzed by flow cytometry. Gating controls are shown at left. Data are representative of 3 independent experiments. (B) Averages and standard deviations of 3 independent experiments from (A). (C) Effects of AE constructs on cellular metabolic activity. Primary bone marrow cells were transduced and selected as in (A), seeded in duplicate at 10 000 cells/well in a 96-well plate, and assayed every 2 d by MTS assay. Data are representative of 3 independent experiments. (D) Immunoblot analysis of CDK4, cyclins A and D3, and p27 expression in K562 cells infected and selected as in supplemental Figure 4A. Data are representative of 4 independent experiments. (E) W692A increases leukemogenic potential of AML1-ETO. Kaplan-Meier survival curves of mice transplanted with hematopoietic cells expressing the indicated AML1-ETO construct. Data combined from 3 independent transplantations. (F) Presence of hematopoietic blast cells in tissues of mice transplanted with AML1-ETO-W692A expressing cells. Peripheral blood smear and cytocentrifugation of bone marrow and spleen cells were stained with Wright-Giemsa solutions.
Effects of AML1-ETO–mutNHR3 and –W692A on cellular dysregulation. (A) Effects of AML1-ETO mutants on apoptosis in primary bone marrow cells. Murine bone marrow cells transduced with the indicated AML1-ETO constructs were harvested after 2 d of selection, stained with 7-AAD and Annexin V-APC, and analyzed by flow cytometry. Gating controls are shown at left. Data are representative of 3 independent experiments. (B) Averages and standard deviations of 3 independent experiments from (A). (C) Effects of AE constructs on cellular metabolic activity. Primary bone marrow cells were transduced and selected as in (A), seeded in duplicate at 10 000 cells/well in a 96-well plate, and assayed every 2 d by MTS assay. Data are representative of 3 independent experiments. (D) Immunoblot analysis of CDK4, cyclins A and D3, and p27 expression in K562 cells infected and selected as in supplemental Figure 4A. Data are representative of 4 independent experiments. (E) W692A increases leukemogenic potential of AML1-ETO. Kaplan-Meier survival curves of mice transplanted with hematopoietic cells expressing the indicated AML1-ETO construct. Data combined from 3 independent transplantations. (F) Presence of hematopoietic blast cells in tissues of mice transplanted with AML1-ETO-W692A expressing cells. Peripheral blood smear and cytocentrifugation of bone marrow and spleen cells were stained with Wright-Giemsa solutions.
SON is also expressed as an isoform lacking exon 3 (SONΔE3), which is expressed at very low levels in blood cell lines, is not detectable by Northern blot, and loses interaction with both AE-W692A and AE-mutNHR3 (supplemental Figure 5). Because these two mutants have very different effects on AML1-ETO function (Figure 2), it is unlikely that loss of SONΔE3 binding contributes to AE-W692A phenotypes.
Because AML1-ETO-W692A behaved similarly to AE9a in vitro, we tested whether it also induced leukemia in a mouse model. AML1-ETO-C663S rapidly induced leukemia, as previously reported,10 and AML1-ETO-W692A induced oligoclonal leukemia with indistinguishable latency (Figure 2E; supplemental Figure 6), with increased numbers of blast cells in peripheral blood, bone marrow, and spleen (Figure 2F), confirming that single W692A substitution is sufficient to enable AML1-ETO-induced leukemogenesis. These phenotypes are similar to those of AML1-ETO9a-induced leukemia.8
In conclusion, we have shown that W692A substitution disrupts N-CoR/NHR4 binding, but interacts with SON. Conversely, AE-mutNHR3 disrupts SON binding, but not N-CoR. W692A substitution correlates with attenuated transcriptional repression and reduced negative effects of AML1-ETO in vitro. Loss of SON binding causes no such change in in vitro phenotypes. Finally, AML1-ETO-W692A is sufficient to induce leukemia in vivo. Therefore, between SON and N-CoR, SON is not the factor that interacts with NHR4 and diminishes the leukemogenic potential of AML1-ETO via NHR4 domain. In contrast, it is more likely that N-CoR binding to NHR4 enhances the transcription repression activity of AML1-ETO and reduces the leukemogenic potential of AML1-ETO. However, it is also possible that other N-CoR unrelated factor(s) interact with AML1-ETO but not with AML1-ETO-W692A, and they regulate the leukemogenic potential of AML1-ETO.
Attenuation of AML1-ETO-driven transcriptional repression and cellular dysregulation correlates with enhanced leukemogenic potential. Although seemingly counterintuitive, this is not unprecedented. Mice with 50% to 80% (but not complete) loss of Pu.1 expression developed a precancerous state predisposing them to leukemia.22 Similarly, histone acetyltransferase mutations correlated with leukemia and lymphoma development were usually monoallelic or otherwise did not completely block HAT function, indicating that decreased dosage or attenuated function is sufficient for disease progression.23,24 We propose a similar model for the contribution of AML1-ETO to leukemia development, in which the attenuation of AML1-ETO function, through mutations that either directly affect the interaction of N-CoR or other proteins at W692 or reduce the downstream effects of these interactions, allows cells to overcome the negative effects of AML1-ETO and to promote leukemia development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr John Bushweller and all members of the D.-E.Z. laboratory for valuable discussions.
This work was supported by National Institutes of Health grants CA096735 (D.-E.Z.) and 5T32GM007240.
Authorship
Contribution: R.C.D. designed and performed the research and wrote the paper; M.Y., E.-Y.A., and W.-J.S. designed and performed the research; N.A.S. provided critical reagents and aided in manuscript preparation; and D.-E.Z. supervised experimental design, data analysis, and manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for E.-Y.A. is Mitchell Cancer Institute, University of South Alabama, Mobile, AL.
Correspondence: Dong-Er Zhang, Mail Code 0815, Room 5328, Moores UCSD Cancer Center, University of California San Diego, 3855 Health Sciences Dr, La Jolla, CA 92093; e-mail: d7zhang@ucsd.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal