Key Points
Endothelial cells secrete exosomes containing miR-214, which suppress senescence and stimulates an angiogenic program in target cells.
Exosomal miR-214 regulates ataxia telangiectasia mutated expression in recipient endothelial cells.
Signaling between endothelial cells, endothelial progenitor cells, and stromal cells is crucial for the establishment and maintenance of vascular integrity and involves exosomes, among other signaling pathways. Exosomes are important mediators of intercellular communication in immune signaling, tumor survival, stress responses, and angiogenesis. The ability of exosomes to incorporate and transfer messenger RNAs (mRNAs) encoding for “acquired” proteins or micro RNAs (miRNAs) repressing “resident” mRNA translation suggests that they can influence the physiological behavior of recipient cells. We demonstrate that miR-214, an miRNA that controls endothelial cell function and angiogenesis, plays a dominant role in exosome-mediated signaling between endothelial cells. Endothelial cell–derived exosomes stimulated migration and angiogenesis in recipient cells, whereas exosomes from miR-214–depleted endothelial cells failed to stimulate these processes. Exosomes containing miR-214 repressed the expression of ataxia telangiectasia mutated in recipient cells, thereby preventing senescence and allowing blood vessel formation. Concordantly, specific reduction of miR-214 content in exosome-producing endothelial cells abolishes the angiogenesis stimulatory function of the resulting exosomes. Collectively, our data indicate that endothelial cells release miR-214–containing exosomes to stimulate angiogenesis through the silencing of ataxia telangiectasia mutated in neighboring target cells.
Introduction
Signaling between endothelial cells, as well as signaling to bone marrow–residing endothelial progenitor cells and stromal cells,1,-3 depends both on intercellular contact and the exchange of secretory proteins.4,5 Additionally, endothelial cells have been demonstrated to secrete exosomes and capture exosomes from various cell types.6,,-9 The transfer of signaling molecules by exosomes may thus provide a third way to regulate endothelial function.9,10 Exosomes, small vesicles secreted by a multitude of cell types, have gained much attention for their role in intercellular communication,11,12 immune signaling mediation,13,14 tumor survival,15,16 stress responses,17,18 and angiogenesis.9,10,19,20 The ability of exosomes to incorporate and transfer functional RNA suggests that exosomes influence the physiological phenotype of recipient cells by introducing messenger RNAs (mRNAs) encoding “acquired” proteins10,20 or through the delivery of micro RNAs (miRNAs) that repress “resident” mRNA translation.21,-23
Several miRNAs have been demonstrated to affect endothelial function and angiogenesis,24,25 and exosomal transfer of such miRNAs may thus be instrumental for exosomes to elicit their effects on endothelial cells. One miRNA that has been identified in angiogenesis-stimulating tumor exosomes, potentially representing an miRNA important in the angiogenesis-stimulating properties of these exosomes, is miR-214.26,-28 Here we demonstrate that endothelial cells secrete miR-214–enriched exosomes that promote endothelial cell migration and angiogenesis in vitro and in vivo. These exosomes repress the expression of ataxia telangiectasia mutated (ATM) in recipient cells in an miR-214–dependent manner, thereby avoiding senescence and allowing the stimulation of blood vessel formation.
Materials and methods
Cell culture
Cells from the human microvascular endothelial cell line (HMEC-1)29 (Centers for Disease Control and Prevention, Atlanta, GA) were maintained up to passage 27 at 37°C, 5% carbon dioxide in MCDB 131 medium (Life Technologies, Grand Island, NY) containing 10% fetal calf serum (FCS), 100 U/mL penicillin, 100 µg/mL streptomycin, 50 nM hydrocortisone, and 10 mM l-glutamine (all from Life Technologies). Exosome-free medium was prepared using FCS centrifuged for at least 1 hour at 200 000 × g followed by filter-sterilization (0.20 µm).
Exosome isolation
Exosomes were collected by differential centrifugation.30 Briefly, cells were grown to subconfluency (approximately 80%) before culturing in exosome-free medium for 24 hours. Then culture medium was centrifuged subsequently for 15 minutes at 1500 × g, 30 minutes at 10 000 × g, and 60 minutes at 100 000 × g. Peletted exosomes were resuspended in basal medium, pelleted again, and finally resuspended in appropriate medium. Exosomes from 1 × 106 cells were resuspended in 150 μL medium.
Sucrose gradient analysis
Exosomes were resuspended in 250 µL 2.5 M sucrose, 20 mM Tris-HCl (Tris-Hydrochloride), pH 7.4, and floated into a linear sucrose gradient (2.0-0.25 M sucrose, 20 mM Tris-HCl, pH 7.4) for 16 hours at 190 000 × g. Gradient fractions (250 μL) were collected and analyzed by immunoblotting.
Immunoblotting
Exosome samples were diluted 1:1 in exosome sample buffer (5% sodium dodecyl sulfate, 9 M urea, 10 mM EDTA, 120 mM Tris-HCl, pH 6.8, and 2.5% β-mercaptoethanol) and heated (95°C, 5 minutes). After sodium dodecyl sulfate–polyacrylamide gel electrophoresis, proteins were blotted onto immobilon polyvinyl membranes (Millipore, Bedford, NY); blocked for 1 hour with 5% nonfat dried milk (ELK; Campina, Amersfoort, The Netherlands) in TBST (TBS with Tween 20; 0.2% [vol/vol] Tween 20); and incubated with rabbit anti–flottilin-1 (1:500; Santa Cruz Biotechnologies, Santa Cruz, CA), goat anti– platelet endothelial cell adhesion molecule-1 (goat anti–PECAM-1; 1:1000; Santa Cruz Biotechnologies), mouse anti-ATM (1:500; Santa Cruz Biotechnologies), or mouse anti–β-actin (1:20 000; Sigma) in TBST with 5% nonfat dried milk. As secondary antibodies, 1:2000 diluted affinity-purified swine anti-rabbit, rabbit anti-mouse, or donkey anti-goat coupled to horseradish peroxidase (Dako, Glostrup, Denmark) were used. Antigen-antibody reactions were visualized with enhanced chemiluminescence according to the manufacturer’s guidelines (chemiluminescent peroxidase substrate; Sigma) and imaged using a Gel Doc XR+ System (Bio-Rad, Hercules, CA).
Electron microscopy
Transmission electron microscopy was performed as described in Slot and Geuze.31 Briefly, carbon-coated Formvar filmed grids were placed on a 5-μL exosome suspension for 20 minutes and washed 3 times with 0.15% glycin in phosphate-buffered saline (PBS), and once with 0.1% bovine serum albumin in PBS. Vesicles were fixed in 1% glutaraldehyde in PBS for 5 minutes and washed twice with PBS. After washing with distilled water, grids were placed on a drop of ice-cold 1.8% methylcellulose (25 Ctp)/0.4% uranyl acetate for 5 minutes and air-dried. Exosomes were visualized using a Tecnai 12 transmission electron microscope (FEI, Hillsboro, OR).
Exosome labeling
Exosomes were labeled with the green fluorescent dye PKH67 (Sigma) as described in Pegtel et al.21 Exosomes from 150 million cells were resuspended in 180 µL PBS with 20 µL of 1:50 diluted PKH67 (in Diluent C). After 3 minutes of incubation at room temperature (RT), 3.8 mL exosome-free medium was added to terminate the labeling reaction, and then the exosomes were harvested and washed twice with PBS by centrifugation (100 000 × g for 1 hour). Exosomes were resuspended in 9.6 mL basal medium, and 250 μL was added to a subconfluent layer of HMEC cells and incubated for 2 hours at 37°C. Cells were washed twice with PBS, fixed with 4% paraformaldehyde in PBS for 30 minutes at RT, and stained with DAPI (4,6 diamidino-2-phenylindole). After embedding (Vectashield; Vector Laboratories, Burlingame, CA), cells were analyzed using an Olympus CX60 microscope (Tokyo, Japan).
Matrigel angiogenesis assay
The angiogenic capacity of HMEC-1 cells was tested by seeding 10 000 cells (in 10 µL basal medium) onto 10 µL solidified Matrigel (ECMatrix, Millipore) in a µ-Slide Angiogenesis (Ibidi, Martinsried, Germany). Fifty µL of test medium (with/without exosomes) was added, following incubation at 37°C for 18 hours. Images were recorded using an Olympus CX60 microscope converted to black and white and analyzed using AngioQuant software.32 Tube length increase (relative to basal medium) was regarded as a measure for angiogenesis.
Migration assay
EC migration was assessed by scratching a confluent layer of HMEC-1 cells in a 24-well plate using a 20 to 200 µL pipette tip. Loose cells were removed by a PBS wash, and 200 µL test medium (with/without exosomes) was added, followed by incubation at 37°C. Images were recorded at t = 0 and t = 6 hours, after which wound area reduction was determined using Image-Pro Plus software (version 3.0; Media Cybernetics).
Sprouting assay
Angiogenic sprouting was determined by seeding HMEC-1 cells onto Cytodex beads (Sigma) and embedding them in a 1:1:4 mixture of basal medium/test medium/Growth Factor Reduced Matrigel (Becton, Dickinson, Franklin Lake, NJ). Solidified gels were overlaid with basal medium and incubated at 37°C, 5% carbon dioxide, for 72 hours. Images were recorded, and sprout lengths were determined using AngioQuant software.
Transfections
Anti–miR-214, pre–miR-214, and nontargeting (NT) control RNAs (Life Technologies) were transfected with siPORT NeoFX (Life Technologies) using 100 nM oligonucleotide concentrations according to the manufacturer’s guidelines. After 16 hours, medium was refreshed. After 72 hours, cells were passaged, and exosomes were harvested another 72 hours later. ATM and control small interfering RNA (siRNA) (Promega) transfections were performed using 50 nM oligos at t = 0 in migration assays. Knockdown at t = 6 was confirmed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR).
Quantitative polymerase chain reaction (qPCR)
Total RNA was isolated using the mirVana RNA Isolation Kit (Ambion). miR-214 and RNU19 levels were determined using TaqMan MicroRNA Assays (Life Technologies) by qRT-PCR amplification in a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad). Cycle threshold (Ct) values were extracted, and ΔΔCt values were calculated to determine relative abundances.
In vivo Matrigel plug assay
Cell infiltration and blood vessel formation in vivo were analyzed by subcutaneous injection of 200 µL Growth Factor Reduced Matrigel containing exosomes from 107 cells into C57BL6 mice (1 plug per mouse). After 2 weeks, plugs were excised (n = 7 [no exosomes], n =6 [control miR-214 exosomes], n = 5 [anti–miR-214 exosomes]) and embedded in paraffin. Six-µm sections were cut (3 depths, 0.7-mm intervals). Cell infiltration was determined by counting nuclei in >6 hematoxilin/eosin (HE)–stained high-power fields. Endothelial cells were stained for CD31,33 and relative CD31+ cell amounts were determined by the counting of CD31-positive cells in at least 1000 DAPI-stained nuclei. Blood vessel surfaces were determined by measuring the surfaces of red blood cell–containing areas in HE-stained sections. All analyses were performed using ImageJ software. All animal experiments were approved by and performed in accordance to the guidelines of the Utrecht University Internal Review Board.
Gene expression analysis
Total RNA was isolated after migration assays (t = 6 hours) using the mirVana RNA Isolation Kit (Ambion) and quantified using an ND1000 spectrophotometer (NanoDrop Technologies). RNA integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA) and an RNA 6000 Nano LabChip kit, accepting RNA integrity numbers of >9.0. Samples were labeled using the Agilent Low RNA Input Linear Amplification Kit Plus (5188-5340) according to the manufacturer’s guidelines. Briefly, 500 ng RNA was amplified, reverse-transcribed using T7 polymerase, and labeled with Cy3 or Cy5. Dye incorporation was measured using an ND1000 spectrophotometer. Subsequently, complementary RNA was hybridized using the Agilent Gene Expression Hybridization Kit. Cy3- and Cy5-labeled complementary RNA were mixed 1:1 (825 ng each), fragmented (60°C, 30 minutes), and hybridized on Agilent hybridization chamber gasket slides (G2534-60011) while rotating (65°C, 17 hours). Slides were scanned using a microarray scanner (Agilent) (accession number GSE45375). Image analysis was performed using Feature Extraction software version 9.5 (Agilent), applying the GE2-v5_95 protocol with default settings. Data preprocessing and analysis was performed using the R-Bioconductor package Limma,34 using robust Edwards background correction. Within-array and between-array normalizations were performed using loess and scale standardization. Significantly differentially expressed transcripts (P < .05) from recipient anti–miR-214 and recipient control miR-214 cells compared with control miR-214 cells were selected.
Gene ontology and pathway enrichment analysis
Gene Ontology Term-for-Term analysis was performed using Ontologizer.35 P values for significant overrepresentation of Gene Ontology (GO) terms in identified gene sets as compared with human genes was calculated using a modified Fisher exact test (1-tailed) applying Benjamini-Hochberg multiple testing correction, with the significance threshold at P = .05.
Senescence assay
Subconfluent layers of HMEC-1 cells were incubated with 100 µL medium with or without exosomes. After 6 hours, cells were fixed and stained using the Senescence Cells Histochemical Staining Kit (Sigma) according to the manufacturer’s guidelines. Images were recorded, and the percentage of positive cells was determined.
Luciferase reporter assay
HEK293 cells were cotransfected with 100 ng pRL-ATM-3′UTR36 (kind gift from Hailiang Hu, University of California, Los Angeles) and 50 nM pre–miR-214 or NT oligonucleotides (both from Ambion) using Lipofectamine 2000 (Invitrogen). pMIR-REPORT (firefly luciferase; Promega) was cotransfected to normalize Renilla expression levels expressed from pRL-ATM-3′UTR. After 24 hours, cells were lysed (reporter lysis buffer; Promega), and luciferase activity was measured using a luminometer (Lumat LB 9507; EG&G Berthold).
Statistics
Data were normalized to means of each experiment with the reference condition set at 1. Normal distribution was tested using the Shapiro-Wilk test, and statistical significance was determined using (paired) Student t test or analysis of variance (ANOVA) with Student-Newman-Keuls posthoc correction where appropriate. α for all tests was 0.05; all values are expressed as mean ± standard deviation (SD). Blood vessel surfaces were 10log transformed to obtain a gaussian distribution.
Results
Endothelial cells produce migration- and angiogenesis-stimulating exosomes
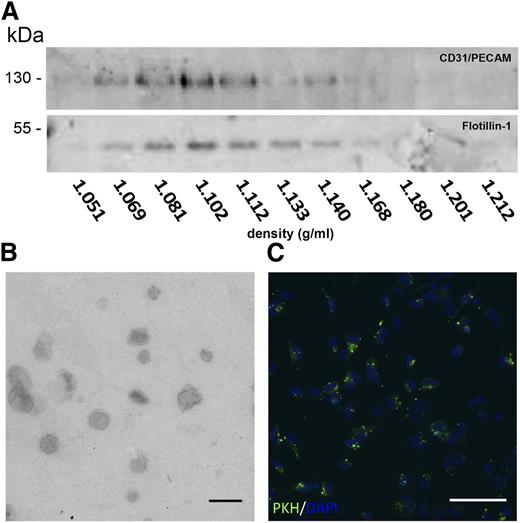
As a physiologically relevant in vitro model system, we used HMEC-1 cells.29 Established cultures received fresh medium, from which, after 24 hours of culture, exosomes were isolated using a well-established differential centrifugation scheme.30 When pelleted vesicles were floated into sucrose density gradients, the endothelial marker protein CD31/PECAM-1 comigrated with the exosomal marker flotillin-1 to a density of approximately 1.11 g/mL (Figure 1A), characteristic for exosomes (1.09-1.18 g/mL).12 Transmission electron microscopy analysis showed vesicle diameters around 120 nm, consistent with our previous nanoparticle tracking analysis,37 and a cup-like shape, typical for exosomes in such analyses (Figure 1B). The capacity of endothelial exosomes to be transferred to recipient endothelial cells was studied by examining the uptake of isolated exosomes labeled with PKH67. After 2 hours, allowing for sufficient exosome uptake, a prerequisite for subsequent RNA transfer,10,21 recipient cells were washed to remove unbound exosomes and fixed, and DNA was stained using DAPI. Fluorescence microscopy analysis demonstrated that the PKH67 label had been taken up and was transferred to perinuclear compartments, presumably representative of late endocytic compartments (Figure 1C).
Isolation of HMEC exosomes. (A) Immunoblots of sucrose gradient samples for the endothelial marker CD31/PECAM-1 (upper panel) and exosome marker flotillin-1 (lower panel) are shown. (B) Transmission electron microscopic analysis of exosomes is shown. Bar represents 250 nm. (C) Endothelial cells after 2 hours of incubation with PKH67 fluorescently labeled exosomes are shown (PKH67 in green, DAPI in blue). Bar represents 50 μm. Images were recorded at RT on an Olympus CX60 microscope using an Olympus UPlan Fl 20×/0.05 objective lens that was coupled to an Olympus DP71 camera operated using CellP software. Brightness was enhanced using Adobe Photoshop software.
Isolation of HMEC exosomes. (A) Immunoblots of sucrose gradient samples for the endothelial marker CD31/PECAM-1 (upper panel) and exosome marker flotillin-1 (lower panel) are shown. (B) Transmission electron microscopic analysis of exosomes is shown. Bar represents 250 nm. (C) Endothelial cells after 2 hours of incubation with PKH67 fluorescently labeled exosomes are shown (PKH67 in green, DAPI in blue). Bar represents 50 μm. Images were recorded at RT on an Olympus CX60 microscope using an Olympus UPlan Fl 20×/0.05 objective lens that was coupled to an Olympus DP71 camera operated using CellP software. Brightness was enhanced using Adobe Photoshop software.
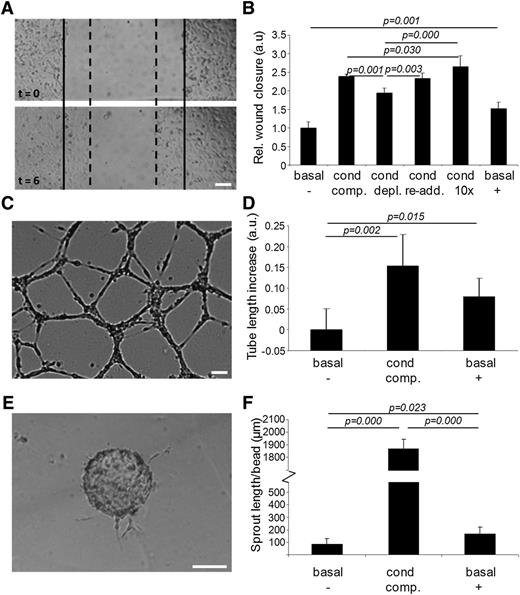
The functional relevance of exosome transfer between endothelial cells was investigated in a scratch wound migration assay (Figure 2A). Migration in basal medium (MCDB 131 without FCS, hydrocortisone, and human epidermal growth factor) was set at 1 (Figure 2B, basal,−). Migration increased 2.5-fold when conditioned medium (medium exposed to an established HMEC-1 culture for 24 hours) was used (cond, comp.). Migration was slightly but significantly impaired when the conditioned medium was depleted from exosomes by ultracentrifugation (cond, depl.). The re-addition of isolated exosomes to exosome-depleted conditioned medium fully restored cell migration (cond, re-add.). This stimulating effect was enhanced further by adding exosomes at a 10-fold higher concentration (cond,10×). Cell migration could also be stimulated 1.5-fold by isolated exosomes resuspended in basal medium (basal,+).
Endothelial exosomes stimulate migration of endothelial cells and angiogenesis. (A) Shown is an example of the cell migration assay. Borders of the scratch at t = 0 are indicated with solid lines, borders after migration at t = 6 hours with dotted lines. (B) Quantitation of HMEC cell migration assay after incubation with, respectively, (1) basal medium (basal,−); (2) conditioned medium (cond, comp.); (3) exosome-depleted conditioned medium (cond, depl.); (4) exosome-depleted conditioned medium after re-addition of exosomes (cond, re-add.); (5) reconstituted conditioned medium with 10 × concentrated exosomes (cond, 10×); and (6) purified exosomes resuspended in basal medium (basal, +). Values are normalized to condition 1 (n = 5 ± SD, ANOVA). Shown are (C) an example image of the in vitro angiogenesis assay, and (D) quantitation of mean tube lengths in the endothelial cell network formed after incubation with (1) basal medium (basal,−); (2) conditioned medium (cond, comp.); and (3) purified exosomes resuspended in basal medium (basal, +). Tubule lengths are indicated relative to condition 1 (n = 5 ± SD, ANOVA). Shown are (E) an illustrative image of a cell-seeded bead with angiogenic sprouts and (F) quantitation of average total sprout length per bead upon incubation with (1) basal medium (basal,−); (2) conditioned medium (cond, comp.); and (3) purified exosomes resuspended in basal medium (basal, +). Bars = 100 μm.
Endothelial exosomes stimulate migration of endothelial cells and angiogenesis. (A) Shown is an example of the cell migration assay. Borders of the scratch at t = 0 are indicated with solid lines, borders after migration at t = 6 hours with dotted lines. (B) Quantitation of HMEC cell migration assay after incubation with, respectively, (1) basal medium (basal,−); (2) conditioned medium (cond, comp.); (3) exosome-depleted conditioned medium (cond, depl.); (4) exosome-depleted conditioned medium after re-addition of exosomes (cond, re-add.); (5) reconstituted conditioned medium with 10 × concentrated exosomes (cond, 10×); and (6) purified exosomes resuspended in basal medium (basal, +). Values are normalized to condition 1 (n = 5 ± SD, ANOVA). Shown are (C) an example image of the in vitro angiogenesis assay, and (D) quantitation of mean tube lengths in the endothelial cell network formed after incubation with (1) basal medium (basal,−); (2) conditioned medium (cond, comp.); and (3) purified exosomes resuspended in basal medium (basal, +). Tubule lengths are indicated relative to condition 1 (n = 5 ± SD, ANOVA). Shown are (E) an illustrative image of a cell-seeded bead with angiogenic sprouts and (F) quantitation of average total sprout length per bead upon incubation with (1) basal medium (basal,−); (2) conditioned medium (cond, comp.); and (3) purified exosomes resuspended in basal medium (basal, +). Bars = 100 μm.
To investigate the effect of exosomes on angiogenic tubule formation, HMECs were seeded on a Matrigel substratum and incubated with either basal medium, conditioned medium, or basal medium supplemented with isolated exosomes. After 18 hours, endothelial networks had formed (Figure 2C), and average tubule lengths were determined. Compared with basal medium (Figure 2D, basal,−), conditioned medium (cond, comp.) and basal medium supplemented with isolated exosomes (basal,+) slightly but significantly increased the mean tube length. Also the formation of angiogenic sprouts, investigated using a bead-based sprouting assay (Figure 2E), was stimulated by both conditioned medium (Figure 2F, cond, comp.) and, to a lesser extent, by isolated exosomes (basal,+) as compared with basal medium (basal,−).
Depletion of miR-214 reduces functional effects in recipient cells
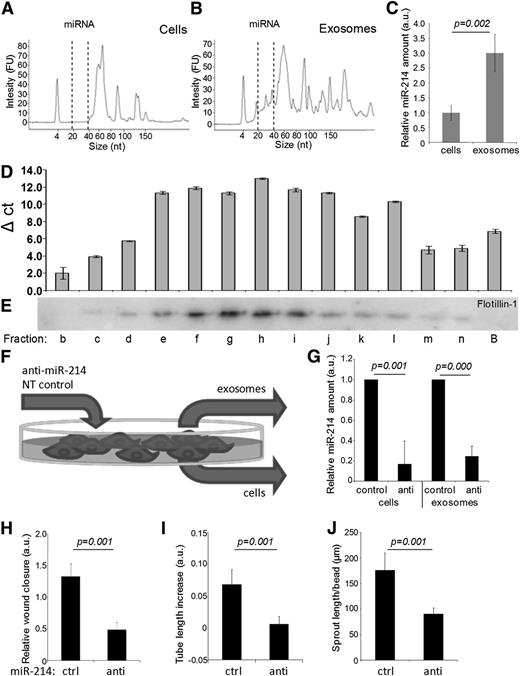
Recent studies indicate that, besides proteins and lipids, functional RNAs are also packaged into and transported by various types of exosomes.10,20,,-23 To explore the RNA content of endothelial exosomes, we compared the RNA content of endothelial cells and endothelial cell–derived exosomes. The presence of high-molecular RNA (see supplemental Figure 1A-B on the Blood website) and more-abundant small RNAs (Figure 3A-B) in endothelial exosomes was demonstrated using Agilent Bioanalyzer total-RNA and small-RNA chips. Consistent with studies on exosomes from other cell types,10,21 the relative abundance of ribosomal RNA in endothelial exosomes was smaller than in corresponding cells (16.50 ± 5.26% vs 50.65 ± 7.39%; supplemental Figure 1C), whereas the relative amount of miRNAs was markedly higher in exosomes (38.75 ± 4.57% vs 7.25 ± 4.65%; supplemental Figure 1D). qRT-PCR analysis showed a threefold higher abundance of miR-214 in endothelial exosomes compared with cells (normalized to RNU19; Figure 3C), and ∼sixfold to eightfold higher when normalized to the common mRNA housekeeping genes p0 (5.95-fold, P = .012), β-actin (6.14-fold, P = .017), and glyceraldehyde-3-phosphate dehydrogenase (7.68-fold, P = .0495). Analysis of sucrose gradient fractions confirmed that at least 92% (fractions e-j) of the miR-214 in the 100 000 × g pellet is associated with exosomes and not with membrane-free high molecular weight complexes (Figure 3D-E). To investigate whether miR-214 is involved in the observed in vitro effects of endothelial exosomes, we reduced miR-214 levels by transfecting cells with modified single-strand RNA molecules that specifically bind and reduce miR-214 (anti–miR-214; Figure 3F). As determined by qRT-PCR (Figure 3G), this yielded “anti–miR-214 cells,” producing exosomes with reduced miR-214 levels (“anti–miR-214 exosomes”). As a control, control miR-214 cells, producing control miR-214 exosomes, were obtained using NT control RNA for transfection. Transfection of endothelial cells with miR-214 inhibitors did not affect endothelial cells with respect to cell number, endothelial identity, or angiogenic capacity, but it reduced migration (supplemental Figure 2A-D). The amount of exosomes released from control and anti–miR-214–transfected cells did not differ, although differences in exosome content other than miR-214 cannot be excluded (supplemental Figure 2E-F).
Exosomal miR-214 induces migration and angiogenesis in vitro. Bioanalyzer profiles of small RNA from (A) cells and (B) exosomes are shown with miRNA indicated. (C) qPCR analysis of miR-214 in endothelial cells and exosomes is shown relative to RNU19 (relative to cell content, n = 6 ± SD, Student t test). (D) miR-214 analysis on RNA from sucrose density gradient fractions is shown. Ct values were subtracted from the fraction showing the highest Ct value (fraction a: Ct = 39.49; B, bottom fraction). (E) Western blot detected flotillin-1 in the corresponding sucrose density gradient fractions; B, bottom fraction. (F) This schematic overview shows the procedure for generating anti–miR-214 and control miR-214 cells and exosomes. The graph shows (G) qPCR analysis of miR-214 content in anti–miR-214 and control-miR-214 cells and exosomes. (Values are plotted relative to samples from cells that were transfected with NT negative control RNA (n = 4 ± SD, Student t test). Quantification of (H) migration (n = 5, ± SD, Student t test), (I) angiogenesis (n = 5, ± SD, Student t test), and (J) sprouting assays (n = 3, ± SD, Student t test) in which endothelial cells were treated with anti–miR-214 or control miR-214 exosomes.
Exosomal miR-214 induces migration and angiogenesis in vitro. Bioanalyzer profiles of small RNA from (A) cells and (B) exosomes are shown with miRNA indicated. (C) qPCR analysis of miR-214 in endothelial cells and exosomes is shown relative to RNU19 (relative to cell content, n = 6 ± SD, Student t test). (D) miR-214 analysis on RNA from sucrose density gradient fractions is shown. Ct values were subtracted from the fraction showing the highest Ct value (fraction a: Ct = 39.49; B, bottom fraction). (E) Western blot detected flotillin-1 in the corresponding sucrose density gradient fractions; B, bottom fraction. (F) This schematic overview shows the procedure for generating anti–miR-214 and control miR-214 cells and exosomes. The graph shows (G) qPCR analysis of miR-214 content in anti–miR-214 and control-miR-214 cells and exosomes. (Values are plotted relative to samples from cells that were transfected with NT negative control RNA (n = 4 ± SD, Student t test). Quantification of (H) migration (n = 5, ± SD, Student t test), (I) angiogenesis (n = 5, ± SD, Student t test), and (J) sprouting assays (n = 3, ± SD, Student t test) in which endothelial cells were treated with anti–miR-214 or control miR-214 exosomes.
Similar to exosomes from nontransfected endothelial cells, isolated exosomes from control miR-214 cells stimulated the migration of endothelial cells in vitro. This stimulatory effect was significantly less when anti–miR-214 exosomes were used (Figure 3H). Similarly, in the angiogenesis and sprouting assays, mean tubule length and sprout length were stimulated less with anti–miR-214 exosomes compared with control miR-214 exosomes (Figure 3I-J). Exosomes from endothelial cells in which miR-214 levels were increased by pre–miR-214 transfection did not show significant effects in either functional assay (supplemental Figure 3), suggesting that an optimal miR-214 concentration was already present in control miR-214 cells and their exosomes.
Together, these results indicate that exosome-mediated stimulation of in vitro endothelial migration and angiogenesis is dependent on miR-214 expression in exosome-producing cells.
Control miR-214 exosomes prevent cell cycle arrest
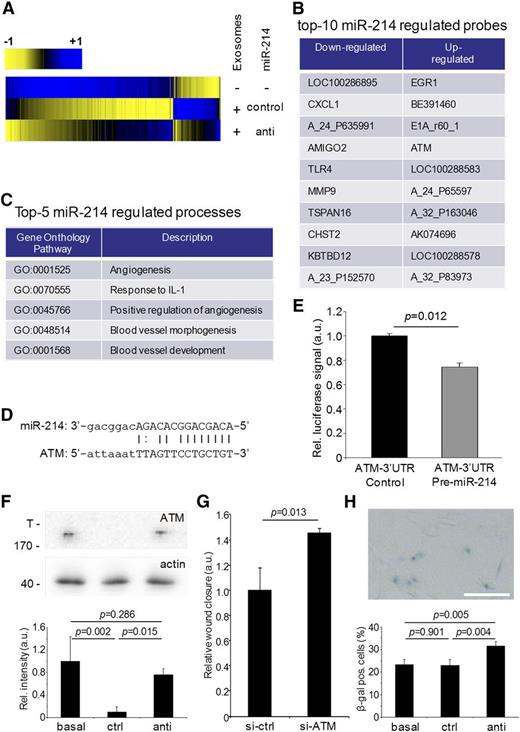
To elucidate the mechanism by which exosomal miR-214 stimulates the angiogenic program in recipient endothelial cells, we searched for early changes in gene expression by gene expression analysis on exosome-recipient cells. In particular, we were interested in immediate repression of potential miR-214 target genes. Hereto, cellular RNA, isolated from endothelial cells exposed to control miR-214 or anti–miR-214 exosomes for 6 hours, was used for microarray analysis. Although miR-214 levels in recipients cells were not significantly affected by the amount of miR-214 delivered by exosomes (supplemental Figure 2G), 146 genes that were differentially expressed in endothelial cells that were exposed to control miR-214 exosomes, in comparison with control cells (not exposed to exosomes), were identified (Figure 4A). Importantly, 28 differentially expressed genes were observed in cells incubated with control miR-214 vs anti–miR-214 exosomes (Figure 4B; supplemental Tables 1 and 2).
Gene expression analysis of exosomal miR-214 effects on recipient endothelial cells. (A) Gene expression analysis was performed on endothelial cells exposed to control miR-214 or anti–miR-214 exosomes relative to endothelial cells that were incubated in the absence of purified exosomes and are presented in a heat map displaying genes that were upregulated (blue) or downregulated (yellow). (B) The list identifies the top 10 downregulated and upregulated probes in recipient cells treated with anti–miR-214 exosomes compared with cells subjected to control miR-214 exosomes. (C) The list identifies the top 5 biological processes (with accompanying GO terms) in recipient cells regulated by exosomal miR-214. (D) The predicted miR-214 seed sequence in the ATM 3′UTR region is shown. (E) The graph shows the results of miR-214 ATM luciferase reporter assay (n = 3, Student t test). (F) ATM protein levels in lysates from cells incubated either without (basal) or with control miR-214 (ctrl) or anti–miR-214 exosomes (anti) were determined by immunoblotting (upper panel; T, top of gel; 170 indicates MW of the highest MW marker band), and quantitation using β-actin was used as a loading control (n = 3, ± SD, ANOVA). (G) Quantitation of a migration assay with cells transfected with control or ATM-siRNA is shown (n = 3, ± SD, Student t test). (H) β-galactosidase staining (upper panel) was used to quantify the percentage of senescent cells (lower panel; n = 4, ± SD, ANOVA).
Gene expression analysis of exosomal miR-214 effects on recipient endothelial cells. (A) Gene expression analysis was performed on endothelial cells exposed to control miR-214 or anti–miR-214 exosomes relative to endothelial cells that were incubated in the absence of purified exosomes and are presented in a heat map displaying genes that were upregulated (blue) or downregulated (yellow). (B) The list identifies the top 10 downregulated and upregulated probes in recipient cells treated with anti–miR-214 exosomes compared with cells subjected to control miR-214 exosomes. (C) The list identifies the top 5 biological processes (with accompanying GO terms) in recipient cells regulated by exosomal miR-214. (D) The predicted miR-214 seed sequence in the ATM 3′UTR region is shown. (E) The graph shows the results of miR-214 ATM luciferase reporter assay (n = 3, Student t test). (F) ATM protein levels in lysates from cells incubated either without (basal) or with control miR-214 (ctrl) or anti–miR-214 exosomes (anti) were determined by immunoblotting (upper panel; T, top of gel; 170 indicates MW of the highest MW marker band), and quantitation using β-actin was used as a loading control (n = 3, ± SD, ANOVA). (G) Quantitation of a migration assay with cells transfected with control or ATM-siRNA is shown (n = 3, ± SD, Student t test). (H) β-galactosidase staining (upper panel) was used to quantify the percentage of senescent cells (lower panel; n = 4, ± SD, ANOVA).
Analysis of enriched GO terms using Ontologizer35 confirmed that endothelial control miR-214 exosomes stimulated general cellular functions (supplemental Table 3a) and inhibited the response to cell cycle/death regulation. In contrast, anti–miR-214 exosomes specifically inhibited processes like angiogenesis, blood vessel development, blood vessel morphology, and response to interleukin-1 in recipient cells (Figure 4C; supplemental Table 3b). Interestingly, among the genes that were more effectively downregulated in cells exposed to control miR-214 exosomes compared with anti–miR-214 exosomes, the ATM gene contained a predicted miR-214 target sequence in its 3′ untranslated region (UTR) (Figure 4D).38 We confirmed that this miR-214 target sequence is functional by using luciferase reporter assays,36 and we found that exposure of endothelial cells to control miR-214, but not to anti–miR-214 exosomes, downregulates ATM protein expression (Figure 4E-F). A key role for ATM was demonstrated using a migration assay, showing that siRNA-mediated repression of ATM stimulated migration (Figure 4G). Induced ATM expression may explain the observed negative effects on endothelial function via 2 different mechanisms: first, through downregulation of the angiogenesis-stimulating hypoxia-inducible factor-1α (HIF-1α) pathway,39 and second, through the induction of cell cycle arrest and senescence.40,41 qRT-PCR analysis of HIF-1α and 2 HIF-1α target genes (VEGFA, MMP9) revealed that expression of these genes did not differ between cells exposed to control miR-214 or anti–miR-214 exosomes (supplemental Figure 3), indicating that the HIF-1α pathway is not regulated through ATM in recipient cells. The alternative pathway, resulting in cellular senescence,40,41 was investigated using β-galactosidase staining for senescent cells (Figure 4H). The percentage of senescent cells did not differ between cells incubated with basal medium or control miR-214 exosomes but was significantly higher in cells exposed to anti–miR-214 exosomes.
Suppression of cell cycle arrest by control miR-214 exosomes allows stimulation of blood vessel formation in vivo
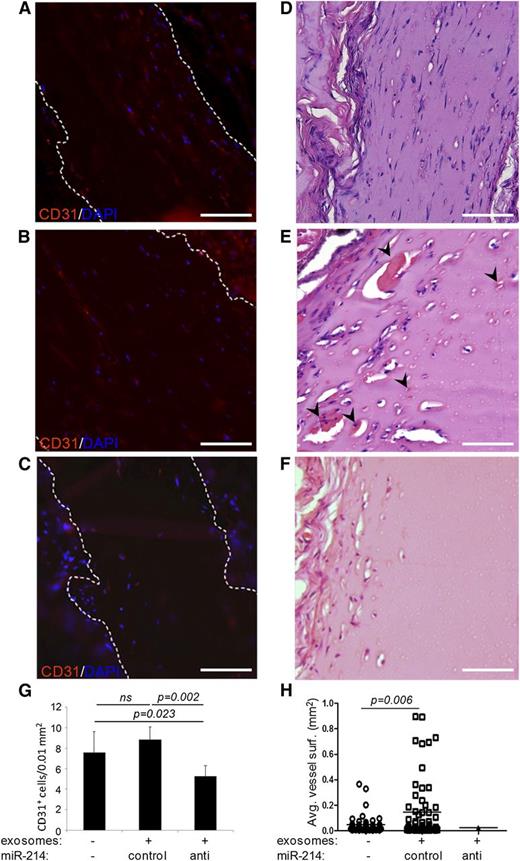
As the mature mouse mmu-miR-214 and human hsa-miR-214 are identical, we could study the effects of control miR-214 and anti–miR-214 exosomes in vivo using a Matrigel plug assay in mice. Matrigel was mixed with vehicle (PBS) and control miR-214 or anti–miR-214 exosomes and subsequently grafted subcutaneously in mice. Plugs were excised after 2 weeks to determine infiltration of endothelial cells and formation of functional blood vessels, as described by Niemistö et al.32 Interestingly, plugs containing anti–miR-214 exosomes showed significantly reduced numbers of endothelial cells that infiltrated into the Matrigel plugs compared with vehicle and control miR-214–containing plugs (Figure 5). In plugs containing vehicle or control miR-214 exosomes, infiltrated cells organized into functional, red blood cell–containing blood vessels. Comparing blood vessel morphology revealed that average blood vessel areas were approximately threefold larger in Matrigel plugs containing control miR-214 exosomes compared with plugs without exosomes. Furthermore, the few functional blood vessels that were detected in plugs containing anti–miR-214 exosomes showed an even smaller blood vessel area (Figure 5H).
Functional analysis of endothelial anti–miR-214 or control miR-214 exosomes in vivo. Shown are sections of dissected Matrigel plugs that contained either no exosomes (A,D), control miR-214 exosomes (B,E), or anti–miR-214 exosomes (C,F) and had subcutaneously been grafted in mice. Sections were stained with hematoxylin and eosin (D-F) (red blood cell–containing blood vessel indicated with arrowheads), or CD31 (red) and DAPI (blue) (A-C). The graphs show (G) quantitation of murine endothelial cells that had infiltrated into the plugs and (H) blood vessel surfaces. Images were recorded at RT on an Olympus CX60 microscope using an Olympus UPlan Fl 20×/0.05 objective lens that was coupled to an Olympus DP71 camera operated using CellP software. Brightness of fluorescent images was enhanced using Adobe Photoshop software. Bars = 100 μm.
Functional analysis of endothelial anti–miR-214 or control miR-214 exosomes in vivo. Shown are sections of dissected Matrigel plugs that contained either no exosomes (A,D), control miR-214 exosomes (B,E), or anti–miR-214 exosomes (C,F) and had subcutaneously been grafted in mice. Sections were stained with hematoxylin and eosin (D-F) (red blood cell–containing blood vessel indicated with arrowheads), or CD31 (red) and DAPI (blue) (A-C). The graphs show (G) quantitation of murine endothelial cells that had infiltrated into the plugs and (H) blood vessel surfaces. Images were recorded at RT on an Olympus CX60 microscope using an Olympus UPlan Fl 20×/0.05 objective lens that was coupled to an Olympus DP71 camera operated using CellP software. Brightness of fluorescent images was enhanced using Adobe Photoshop software. Bars = 100 μm.
Discussion
In the study presented here, we used in vitro and in vivo approaches to elucidate the role of miR-214 in exosome-mediated crosstalk between endothelial cells. In recent years, it has become clear that, besides growth factors and cytokines, exosomes also provide signaling cues for endothelial cells, stimulating activation, differentiation, and angiogenesis.8,9,42,43 We used an endothelial cell line that produces protein- and RNA-containing exosomes37 as an in vitro model to investigate the autocrine effects of endothelial exosomes. As described for tumor exosomes in Hood et al,8 exosomes from these cells also contribute to conditioned medium-stimulated endothelial function, demonstrated by the reduced stimulation of migration by exosome-depleted conditioned medium. Importantly, the re-addition of exosomes to exosome-depleted medium completely restored the stimulatory effect of conditioned medium, demonstrating that the exosome isolation procedure did not affect the functional properties of exosomes. Not all stimulatory effects could be attributed to exosomes, and growth factors, cytokines, and soluble (mi-)RNAs that are also secreted by endothelial cells are likely to contribute alongside exosomes. Together, our in vitro functional assays demonstrate that endothelial exosomes stimulate the migratory and angiogenic potential of recipient endothelial cells.
Several miRNAs have already been assigned a role in endothelial function and angiogenesis (reviewed in Kuehbacher et al24 and Urbich et al25 ). We focused on miR-214, which is highly expressed in endothelial cells,44 has a role in angiogenesis33,45 and cell migration,46 and is present in angiogenesis-stimulating ovarian- and lung cancer–derived exosomes.26,-28 Like miR-214 in tumor exosomes, miR-214 was enriched in endothelial cell-derived exosomes, which may already indicate its importance in endothelial exosome function. By transfecting exosome-producing endothelial cells with miR-214 inhibitors, we could reduce exosome miR-214 levels without affecting the cell number or exosome secretion. Anti–miR-214 exosomes showed a reduced capacity to stimulate migration, angiogenesis, and sprouting. Accordingly, genes involved in angiogenesis and blood vessel formation were significantly downregulated in anti–miR-214 recipient cells compared with control miR-214 exosome–recipient cells. miR-214 levels in exosome-recipient cells were not significantly affected after exposure to control miR-214 or anti–miR-214 exosomes, indicating that the absolute miR-214 amount added through exosomes is minute compared with the total cellular RNA amount. These data suggest that senescent cells with reduced miR-214 levels may be rescued from entering senescence by incorporating miR-214 from neighboring miR-214–producing cells via the exosomal shuttle.
Furthermore, this shows that in our approach, exosomes should not be considered to act as transfection agents but rather as delicate, multicomponent signaling devices. Nevertheless, exosome-delivered miR-214 appeared functional, as the expression of the miR-214 target gene ATM appeared upregulated upon exposure to anti–miR-214 exosomes.
Regulation of ATM appears pivotal in the effects of exosomal miR-214. ATM prevents cell cycle progression,40,41 and upregulation of ATM in anti–miR-214 exosome–recipient cells correlated to increased cellular senescence. The HIF-1α pathway, another described ATM target,39 remained unaffected. As downregulation alone of ATM in endothelial cells enhanced cell migration, it appears that exosomal miR-214 primarily represses senescence through its effect on ATM suppression. The induction of an angiogenic program may depend on other exosome-induced pathways, that is, Notch-signaling9 resulting in the observed exosome-stimulatory effects.
Our data indicate that negative effects on cell cycle progression are most profound in cells exposed to anti–miR-214 exosomes, suggesting that exosomal miR-214 may stimulate the formation of blood vessels by preventing cell cycle arrest in recipient endothelial cells.
Our in vivo studies, showing fewer cells and blood vessels in plugs containing anti–miR-214 exosomes, underline the physiological relevance of our in vitro observations. Because mouse and human miR-214 are identical, and the 3′-UTR of the mouse atm gene contains a predicted miR-214 target site,38 effects on cell numbers can be explained by effects on cell cycle arrest, in line with our in vitro analyses. Here basal-medium and control miR-214 exosomes did not affect the amount of senescent cells, explaining the presence of similar amounts of CD31+ cells in the Matrigel plugs. Increased senescence in anti–miR-214 exosome–recipient cells, observed in vitro, corresponds to reduced cell amounts detected in corresponding Matrigel plugs. The in vivo effects on blood vessel number and size demonstrate that exosomal miR-214 is also functional in inducing an angiogenic program.
Previous studies showed that nonendothelial exosomes can stimulate angiogenesis.10,19 Also, miR-214 has been previously shown to affect the proliferation and differentiation of myoblasts and T cells46,47 and ginsenoside-Rg1–induced angiogenesis.45 Furthermore, our findings agree with the observed role for miR-214 in the stimulation of the migration of melanoma cells46 and neurons48 and the prevention of cell cycle arrest and apoptosis in cardiomyocytes.49 miR-214 appeared downregulated in endothelial cells exposed to hypoxia or tumor necrosis factor-α (supplemental Figure 2H); however it remains to be investigated whether this is reflected in exosomes. Additionally, the effects on exosome mRNA and protein content likely contribute to exosome function, providing a low signal-to-noise ratio studying specific miR-214 effects.37 The antiangiogenic effects of miR-214 have been reported in human umbilical vein endothelial cells, but these cells may be considered “young” endothelial cells, while HMECs are more “mature” endothelial cells,33,45 with subsequent different miR-214 levels.50 For exosome function, miR-214 levels thus may provide paradoxal information. Exosomal components have been described to provide clues that are in contrast with their function in the context of cells, potentially as they are provided to target cells through a pathway different from the normal, (intra-)cellular pathway, with the inhibition of Notch signaling by Notch ligand Δ-like 4–containing exosomes in endothelial cells9 as a prime example.
In conclusion, our results indicate an intricate exosome-mediated crosstalk interface at the vascular endothelium that prevents cell cycle arrest and regulates the angiogenic program, at least partly, via miR-214. More generally, our data indicate that individual species of miRNA play a crucial role in exosome function.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Professor Dr E. Cuppen and E. de Bruijn (Hubrecht Institute, Utrecht, The Netherlands); H. Gremmels and C. Tilburgs (Department of Nephrology and Hypertension, University Medical Center Utrecht, Utrecht, The Netherlands); Dr P. Vader (Department of Pharmaceutics, Utrecht University, Utrecht, The Netherlands); and Dr G. Posthuma (Department of Cell Biology, University Medical Center Utrecht, Utrecht, The Netherlands) for excellent assistance. The authors also thank Dr H. Hu (Department of Pathology and Laboratory Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA) for kindly providing the ATM luciferase reporter plasmid.
This work was supported by The Netherlands Organization for Scientific Research (NGI/ZonMW Horizon project 935190280) (B.W.M.v.B.) and UMC Utrecht Focus and Massa (grant DIGD-DGK-DHL) (M.C.V.). T.W. and M.C.V. are both supported by The Netherlands Organization for Scientific Research (NWO-VIDI).
Authorship
Contribution: B.W.M.v.B., O.G.d.J., M.S., K.d.O., P.M.d.B., M.A.J.v.E., and J.B. performed, analyzed, and interpreted experiments. B.W.M.v.B., W.S., T.W., D.M.P., and M.C.V. supervised the project and prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bas W.M. van Balkom, Department of Nephrology and Hypertension, Heidelberglaan 100, 3584CX Utrecht, The Netherlands; e-mail: b.w.m.vanbalkom@umcutrecht.nl.
References
Author notes
O.G.d.J. and M.S. contributed equally to this study.