To the editor:
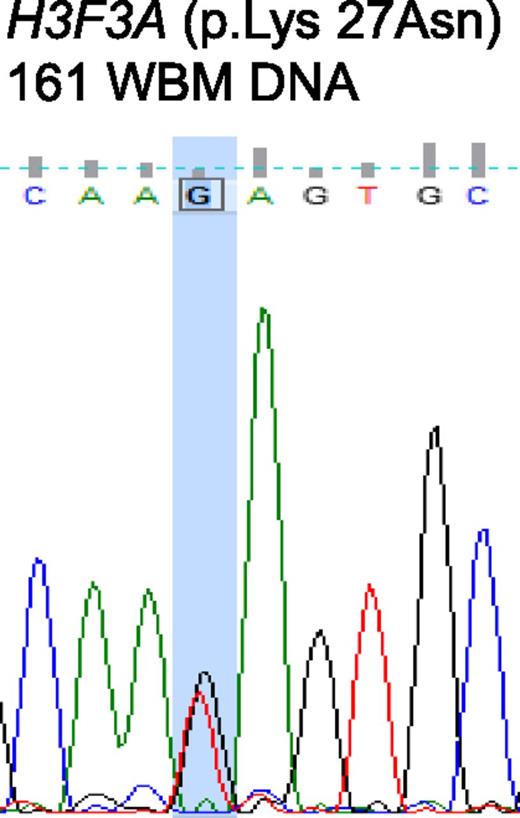
H3.3 represents a replication-independent histone 3 variant. H3.3 associates with alpha thalassemia/mental retardation syndrome X-linked (ATRX) and death domain-associated protein (DAXX) required for H3.3 chromatin assembly at pericentric heterochromatin and telomeres.1,-3 Somatic mutations within H3F3A gene, which encodes the H3.3 histone variant, as well as within ATRX and DAXX, have been reported in solid malignancies, including 44% to 71% of pediatric glioblastoma multiforme,4,-6 suggesting that defects in chromatin architecture can underlie tumor pathogenesis. To study the impact on this pathway in myelodysplastic syndrome (MDS), we evaluated bone marrow aspiration specimens from 77 patients with MDS evaluated at the MD Anderson Cancer Center. Patient characteristics are summarized in supplemental Table 1. Consent for sample collection was obtained in accordance with institutional guidelines. Samples were collected by using Ficoll-Paque density centrifugation. CD34+ cells were sorted by using a MicroBead Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s protocol. Details on mutational, RNA, and methylation analyses are described in supplemental “Methods.” Sequencing of the H3F3A gene in 77 MDS samples identified a mutation at amino acid 27 in only one patient, resulting in a replacement of lysine by asparaginase (K27N) (Figure 1). No other mutations in H3F3A were identified. Another 42 samples were sequenced for ATRX and DAXX, with no mutations identified in either gene. The sole K27N mutant sample occurred in a 76-year-old male with intermediate-1 MDS (International Prognostic Scoring System) with diploid cytogenetics who continues to receive supportive care and transfusion support more than 2 years after diagnosis.
Sanger sequencing chromatogram depicting a mutation of H3F3A at amino acid 27, resulting in a replacement of lysine by asparagine (K27N). Mutations are marked by the column (G > T).
Sanger sequencing chromatogram depicting a mutation of H3F3A at amino acid 27, resulting in a replacement of lysine by asparagine (K27N). Mutations are marked by the column (G > T).
We next analyzed promoter methylation status to evaluate whether aberrant epigenetic patterning was identified within genes of this pathway. Promoter hypermethylation was not identified in either H3F3A (n = 14; median methylation density, 0.92%; range, 0% to 5.89%) or DAXX (n = 42; median methylation density, 3.21%; range, 0% to 6.99%). Pyrosequencing identified 8 (23%) of 35 samples with >10% promoter methylation in ATRX (n = 35; median methylation density, 2.27%; range, 0% to 36.44%). However, of these 8 samples, 6 were from females and, on the basis of reported levels of ATRX methylation in healthy females, it is likely that the hypermethylation seen was a result of normal X-chromosome inactivation. ATRX and DAXX expression in MDS CD34+ cells by real-time polymerase chain reaction identified a tendency toward higher expression (P = .07) of ATRX messenger RNA and significantly upregulated expression of DAXX messenger RNA (P = .003) compared with normal controls (supplemental Figure 1). Neither upregulated ATRX nor DAXX expression levels were significantly associated with patient outcome. No association was observed with expression and World Health Organization or cytogenetic subset. Of interest, DAXX protein was originally identified as a transmembrane death receptor (CD95) FAS-binding protein and has been shown to potentiate FAS-induced apoptosis;7 therefore, it could be involved in dysregulated apoptosis known to occur in MDS patients. In summary, although somatic mutations within this pathway are infrequent, our study suggests that further expression profiling of genes within the chromatin remodeling pathways may provide important clues to the underlying biology of MDS and may ultimately identify new therapeutic targets.
The online version of this article contains a data supplement.
Authorship
Contribution: Y.A., H.Z., Y.J., Z.-H.F., I.G.G. and Y.W. performed experiments; Y.A., Q.G., C.D.D., I.G.G., H.Y., Y.W., H.K., and G.G.-M. analyzed results; H.K. and G.G.-M. designed the research study; and Y.A., Q.G., C.D.D., H.Z., Y.J., Z.-H.F., I.G.G., H.Y., Y.W., H.K., and G.G.-M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guillermo Garcia-Manero, Department of Leukemia, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 428, Houston, TX 77030; e-mail: ggarciam@mdanderson.org.