Key Points
The shortest isoform of C/EBPβ, liver inhibitory protein (LIP), collaborates with Evi1 in leukemogenesis.
Ecotropic viral integration site 1 (Evi1) is one of the master regulators in the development of acute myeloid leukemia (AML) and myelodysplastic syndrome. High expression of Evi1 is found in 10% of patients with AML and indicates a poor outcome. Several recent studies have indicated that Evi1 requires collaborative factors to induce AML. Therefore, the search for candidate factors that collaborate with Evi1 in leukemogenesis is one of the key issues in uncovering the mechanism of Evi1-related leukemia. Previously, we succeeded in making a mouse model of Evi1-related leukemia using a bone marrow transplantation (BMT) system. In the Evi1-induced leukemic cells, we identified frequent retroviral integrations near the CCAAT/enhancer-binding protein β (C/EBPβ) gene and overexpression of its protein. These findings imply that C/EBPβ is a candidate gene that collaborates with Evi1 in leukemogenesis. Cotransduction of Evi1 and the shortest isoform of C/EBPβ, liver inhibitory protein (LIP), induced AML with short latencies in a mouse BMT model. Overexpression of LIP alone also induced AML with longer latencies. However, excision of all 3 isoforms of C/EBPβ (LAP*/LAP/LIP) did not inhibit the development of Evi1-induced leukemia. Therefore, isoform-specific intervention that targets LIP is required when we consider C/EBPβ as a therapeutic target.
Introduction
Ecotropic viral integration site 1 (Evi1), which is located on chromosome 3q26, is one of the critical regulators of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). Several clinical studies have indicated that inappropriate expression of Evi1 is found in 10% of patients with AML regardless of 3q26 abnormalities, and that high expression of Evi1 is associated with poor prognosis.1,-3 Evi1 was originally identified in a common integration site of murine leukemia retrovirus, and enhanced expression of Evi1 by retrovirus integration is thought to be responsible for leukemogenesis in mouse models.4 However, retroviral expression of marker genes such as CD34 has not induced hematologic malignancy even if some clones possessing integration at the Evi1 site have been identified.5 Accordingly, so far we have not found hematologic abnormalities in mice transplanted with bone marrow (BM) cells, which have been transduced with green fluorescent protein (GFP) as a control.
In a case study, 2 patients with X-chronic granulomatous disease (X-CGD), who underwent gene therapy using retrovirus, went on to have myelodysplasia and myeloid leukemia with monosomy 7 as a result of insertional activation of Evi1.6 The cases suggested that overexpression of Evi1 in human cells induced genomic instability and caused additional genetic change, such as monosomy 7 followed by AML.
These reports indicate that Evi1 requires cooperative factors to induce AML, whereas overexpression of Evi1 is enough to lead to clonal expansion of hematopoietic cells. Therefore, identifying genes collaborating with Evi1 in leukemogenesis is one of the key issues in understanding Evi1-related leukemia.
In our previous work, we demonstrated that a point mutation of the transcription factor AML1 (AML1-D171N) induces MDS that progresses to AML in association with overexpression of Evi1 through a mouse BM transplantation (BMT) model.7 Recently, we succeeded in making a mouse model of Evi1-induced leukemia by using retroviral transduction of Evi1 and observed that all of the mice went on to have AML within 6 to 11 months after transplantation.8
We supposed that retroviral enhancers would have some effects on the initiation of leukemia in our Evi1-leukemia model mice. Regarding retroviral integration sites, we identified frequent retroviral integrations at the 3′ side of the C/EBPβ gene in the Evi1-transduced leukemic cells. These results suggest that C/EBPβ is a candidate gene that collaborates with Evi1 in leukemogenesis. C/EBPβ, also known as NF-IL6, is a transcription factor that specifically binds to an interleukin (IL)1-responsive element in the IL-6 gene and has a role in regulation not only for the IL-6 gene but also for several cytokine genes such as tumor necrosis factor, IL-8, and granulocyte colony-stimulating factor (G-CSF).9 The locus of the C/EBPβ gene has been reported as a common integration site in the Retrovirus Tagged Cancer Gene Database (RTCGD),10 which is a database of retroviral insertional mutagenesis in mouse tumors. Furthermore, C/EBPβ collaborates with PRDM16, also named the MDS1/EVI1-like gene, in initiation of a differentiation switch from myoblasts to brown fat cells.11 Therefore, in this study, we examined the effect of C/EBPβ on the development of leukemia and a close collaboration of Evi1 and C/EBPβ in leukemogenesis. Our data raise the possibility that targeting the C/EBPβ pathway may provide a new strategy for the treatment of Evi1-related leukemia. Furthermore, this novel mouse model may be useful for the development of effective treatments of patients with AML/MDS in which Evi1 is highly expressed.
Materials and methods
Vector construction
The pMYs-mouse Evi1-IG was provided by Dr. Takuro Nakamura (the Cancer Institute, Tokyo, Japan), and the pcDNA-mouse Evi1 was provided by Dr. Kazuhiro Morishita (the University of Miyazaki, Miyazaki, Japan). We used 3 isoforms of C/EBPβ: liver activating protein* (LAP*, one of the long isoforms), liver activating protein (LAP, the other long isoform), and liver inhibitory protein (LIP, a short isoform). The coding regions of LAP* and LIP were amplified by reverse transcription (transcriptase)-polymerase chain reaction (RT-PCR) using complementary DNA (cDNA) derived from mouse BM cells. The LAP* cDNA contains all 3 initiation codons (methionine) (ATGs) for LAP*, LAP, and LIP. LAP was obtained from pcDNA 3.1-mouse C/EBPβ (Addgene, Cambridge, MA). The LAP cDNA contains 2 ATGs for LAP and LIP. The LIP cDNA contains 1 ATG for LIP. All of the isoforms were inserted to a pGCDNsam-IRES-Kusabira Orange vector12 to generate pGCDNsam-LAP*, LAP, or LIP-Kusabira Orange, respectively. Plasmid constructs containing the mouse C/EBPβ promoter (−3.5 kb from the first ATG) were prepared by PCR amplification using cDNA derived from C57BL/6 mouse peripheral blood (PB) cells as a template, and the PCR products were cloned into the pGL4.10 [luc2] luciferase vector (Promega, Madison, WI).
Transfection and retrovirus production
Mouse BMT and analysis of the transplanted mice
BM mononuclear cells were isolated from donor mice (age 8 weeks, C57BL/6 (Ly-5.1) or C/EBPβ conditional knockout mice). The BM cells were infected with retroviruses using RetroNectin (Takara Bio, Japan) according to the manufacturer’s recommendations. For double infection, we mixed 2 virus suspensions with an equal ratio. The infected BM cells (derived from Ly-5.1 or C/EBPβ conditional knockout mice) were injected through the tail vein into C57BL/6 (Ly-5.2) recipient mice (age 8-9 weeks), which had been administered a sublethal dose of 5.25 Gy of total-body γ-irradiation (supplemental Methods). Engraftment was confirmed by measuring the percentage of Ly-5.1 and GFP and/or Kusabira Orange–positive cells in the PB. If a recipient mouse were physically inactive such as showing breathing difficulties, exhibiting weight loss, having a low body temperature, or sitting in a hunched position, the mouse was euthanized for analysis. To assess whether the leukemic cells were transplantable, 2 × 105 − 1 × 106 whole BM cells, including blasts, were injected into sublethally irradiated mice through the tail veins. All animal studies were approved by the Animal Care Committee of the University of Tokyo.
Flow cytometric analysis and cell sorting
Flow cytometric analysis was performed on FACSLSRII flow (BD Biosciences), equipped with CellQuest software (BD Biosciences), and analyzed using FlowJo software (Tree Star, San Carlos, CA). We used FACSAria II (BD Biosciences) for cell sorting. Human CD34+ cells were sorted by autoMACS (Miltenyi Biotec, Germany) using CD34-PE conjugated antibody (Beckman Coulter, CA) and anti-PE MicroBeads (Miltenyi Biotec, Germany) according to the manufacturer’s instructions.
Human samples
Written informed consent was obtained from all patients whose samples were collected after the ethical guidelines for biomedical research involving human subjects came into force. We used whole BM cells derived from patients diagnosed with primary immune thrombocytopenia or lymphoma without BM invasion as controls, with written informed consent in accordance with the Declaration of Helsinki. Human CD34+ and CD34− cells were obtained from BM or from the residual PB stem cells harvested for autologous transplantation.
Diagnosis
Colony assay
BM cells (1.0 × 104 cells) in methylcellulose M3434 medium (StemCell Technologies, Vancouver, Canada) were plated in duplicate in 35-mm petri dishes. The number of colonies was scored after 7 days, and 1.0 × 104 cells recovered from the cultured plates were replated every 7 days.
Real-time RT-PCR
Total RNA of human BM samples was extracted using ISOGEN (Nippon Gene, Tokyo, Japan). Total RNA of mouse BM cells was extracted using Trizol (Invitrogen, CA). cDNA was prepared with the Superscript III RT kit (Invitrogen) or Transcriptor 1st strand cDNA synthesis kit (Roche Diagnostics, Mannheim, Germany). Real-time RT-PCR was performed using a LightCycler 480 Real-Time PCR System and Light Cycler 2.0 instrument (Roche Diagnostics). cDNA was amplified using a SYBR Premix EX Taq (TAKARA, Japan), LightCycler FastStart Reaction Mix SYBR Green I (Roche Diagnostics), and a LightCycler FastStart DNA Master Hybridization probe (Roche Diagnostics). The primer pairs are shown in supplemental Table 1. Expression levels were normalized by the expression level of 18S ribosomal RNA (rRNA).
Western blot analysis
Spleen cells of leukemic mice, human whole BM cells from patients, and CD34+ and CD34− control cells were lysed with TNE buffer. Equal amounts of total cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA). The following antibodies were used: monoclonal rabbit anti-Evi1 antibody (Cell Signaling, Danvers, MA), a monoclonal mouse anti-Flag M2 antibody (Sigma, St. Louis, MO) for Evi1, a monoclonal rabbit anti-C/EBPβ antibody (C-19; Santa Cruz Biotechnology, Santa Cruz, CA), a monoclonal rabbit anti-C/EBPα antibody (14AA; Santa Cruz Biotechnology), and a monoclonal rabbit anti-β-actin antibody (Cell Signaling). Images were recorded using the luminescent image analyzer (LAS-3000; Fujifilm), and the intensities of the bands were quantitated by densitometry using Multi Gauge Version 3.0 software (Fujifilm). See supplemental Methods for details about the protein complex immunoprecipitation assay.
Southern blot analysis
Genomic DNA was extracted from spleen cells. Enzymatic digestion of 10 µg of DNA with EcoRI was followed by electrophoretic separation and overnight transfer to Hybond N nylon membrane; then proviruses were probed with a GFP probe. To examine the clonality of the leukemic cells with retroviral integration near the C/EBPβ locus, genomic DNA was digested with BamHI, separated by electrophoration, and then transferred to Hybond N nylon membrane overnight. Probes that lie in the vicinity of the integration sites were made by PCR purification and TA cloning. Primer and probe sequences are shown in supplemental Table 2 and 3.
Bubble PCR
The bubble PCR method was performed as described previously7,17,18 (supplemental Methods). Briefly, 10 µg of genomic DNA extracted from spleen cells were digested with EcoRI or BamHI, and the fragments were ligated to a double-stranded bubble linker. Next, PCR was performed on the ligation product using a linker-specific Vectorette primer and an LTR-specific primer. The PCR product was sequenced to identify the retrovirus integration sites. We confirmed the inverse repeated sequence “GGGGGTCTTTCA” as a junction marker between the genomic DNA and the retrovirus sequence. Sequence data are shown in supplemental Table 2.
Luciferase assay
To examine the possibility of transcriptional activation of C/EBPβ by Evi1, a luciferase assay was performed using the reporter plasmid (pGL4-C/EBPβ 3.5k) and the effector plasmid (pcDNA-mouse Evi1). See supplemental Methods for details.
Conditional ablation of C/EBPβ
C/EBPβ conditional knockout mice, which have alleles of C/EBPβ flanking the coding region with LoxP recombination sites, were obtained from Dr. Sterneck at the National Cancer Institute.19 The C/EBPβfl/fl mice were mated to Mx-Cre transgenic mice.20 We use the term “Mx-Cre;C/EBPβfl/fl” for this mice. To excise the C/EBPβ locus, we performed intraperitoneal injection of 400-ug/body pI-pC three or five times. Excision of the C/EBPβ coding region was confirmed in genomic DNA from PB or BM cells by PCR analysis (supplemental Methods). The fragment should be 162 bp for the wild-type allele, 210 bp for the floxed allele, and 72 bp for the deleted allele.
Results
A high incidence of retroviral integration close to the site of the C/EBPβ gene was observed in Evi1-leukemic mice
We succeeded in establishing a mouse model of Evi1-leukemia (Figure 1A).8 As we have reported previously, the Evi1-mice appeared healthy until 5 months after transplantation, but the number of GFP-positive-Evi1−expressing cells gradually increased in the PB, and the mice died at 6 to 11 months after transplantation (Figure 1B). We confirmed that the leukemic cells were transplantable. We wondered if retroviral enhancers might have influenced the induction of leukemia in our model mice, because the long latencies suggested clonal selection in leukemic transformation of primary BM cells.
Retrovirus integrations at 3′ side of the C/EBPβ gene were frequently found in Evi1-leukemia mice. (A) A flowchart of retrovirus infection and BMT. (B) Percentages of GFP/Ly-5.1 double-positive cells in PB. PB was obtained from the tail vein every month after transplantation. (C) Southern blot analysis of Evi1-leukemia mice. DNA samples were digested with EcoRI. Proviruses were probed with a GFP probe. Mice ID are shown at the top of the panel. (D-E) Integration site analysis in BM samples of Evi1-leukemia cells obtained by bubble PCR. Genomic DNAs were digested with EcoRI (D) or BamHI (E). White arrows indicate insertion sites near C/EBPβ. Black arrows indicate retroviral sequence. (F) Schema of integration sites near the C/EBPβ gene. Thick black arrows indicate integration sites. Thin black arrows indicate restriction sites. “B” indicate BamHI site. Black horizontal bars indicate the position of the probes for Southern blotting. (G) Schema of restriction sites on retroviral sequence near the junction of genomic DNA. (H) The leukemic clones with retroviral integration near the C/EBPβ gene were major clones. Black arrows indicate the endogenous band, and white arrows indicate the integrated band.
Retrovirus integrations at 3′ side of the C/EBPβ gene were frequently found in Evi1-leukemia mice. (A) A flowchart of retrovirus infection and BMT. (B) Percentages of GFP/Ly-5.1 double-positive cells in PB. PB was obtained from the tail vein every month after transplantation. (C) Southern blot analysis of Evi1-leukemia mice. DNA samples were digested with EcoRI. Proviruses were probed with a GFP probe. Mice ID are shown at the top of the panel. (D-E) Integration site analysis in BM samples of Evi1-leukemia cells obtained by bubble PCR. Genomic DNAs were digested with EcoRI (D) or BamHI (E). White arrows indicate insertion sites near C/EBPβ. Black arrows indicate retroviral sequence. (F) Schema of integration sites near the C/EBPβ gene. Thick black arrows indicate integration sites. Thin black arrows indicate restriction sites. “B” indicate BamHI site. Black horizontal bars indicate the position of the probes for Southern blotting. (G) Schema of restriction sites on retroviral sequence near the junction of genomic DNA. (H) The leukemic clones with retroviral integration near the C/EBPβ gene were major clones. Black arrows indicate the endogenous band, and white arrows indicate the integrated band.
First, we examined a number of clones in the leukemic mice by Southern blotting using a GFP probe. As shown in Figure1C, the leukemic cells were monoclonal or oligoclonal. Next, we examined the retroviral integration site of these leukemic cells by using the bubble PCR method. Surprisingly, we identified frequent integration near the C/EBPβ gene in 6 of 10 mice transplanted with Evi1-transduced BM cells (Figure 1D-E and Table 1). Because retroviral integration to the C/EBPβ locus has not been observed in our previous study,7,21 Evi1-induced leukemic mice had a high frequency of integration in the C/EBPβ locus with statistical significance compared with other types of leukemic mice (supplemental Table 4; P = .00024, Fisher exact test). The integrations were located at 62.5-86.7 kb downstream from the C/EBPβ gene (Figure 1F). We investigated the integration site in the serially transplanted leukemic cells and confirmed the identical integration sites in the second leukemic cells as in the first ones (Table 1). Furthermore, we performed Southern blot using probes that lie in the vicinity of the integration sites in leukemic cells derived from mice ID2 and 4. The retroviral integrations near the C/EBPβ gene were clearly detected by Southern blotting in the leukemic clones, suggesting that the clones in which Evi1 integrates near the C/EBPβ gene command a majority with proliferative advantage (Figure 1H).
Retrovirus integration sites of leukemic cells
| Transduced gene . | Mouse ID . | Nearest gene . | Chromosome no. . | Distance to gene (start or end) . | Location . | Forward or reverse orientation . | RTCGD hits . |
|---|---|---|---|---|---|---|---|
| Evi1 | 1 | Bcas1 | 2 | 14965 bp | 5′ | R | 2 |
| Evi1* | 2 | C/EBPβ | 2 | 75063 bp | 3′ | F | 25 |
| Evi1 | 3 | C/EBPβ | 2 | 62583 bp | 3′ | F | 25 |
| Evi1 | 4 | C/EBPβ | 2 | 86723 bp | 3′ | F | 25 |
| Evi1 | 5 | Pnp2 | 14 | 11834 bp | 3′ | R | 0 |
| Evi1 | 5 | Rps6ka1 | 4 | 32680 bp | 3′ | R | 4 |
| Evi1 | 5 | Rapgef4 | 2 | — | Intron 6 | R | 0 |
| Evi1 | 6 | C/EBPβ | 2 | 74775 bp | 3′ | F | 25 |
| Evi1 | 7 | C/EBPβ | 2 | 74873 bp | 3′ | F | 25 |
| Evi1 | 7 | E2F2 | 4 | — | Intron 1 | F | 10 |
| Evi1 | 8 | C/EBPβ | 2 | 81088 bp | 3′ | F | 25 |
| Evi1* | 15 | Atp10a | 7 | 2629 bp | 3′ | R | 0 |
| Evi1 | 21 | TLR7 | X | — | Intron 2 | R | 0 |
| Evi1+LAP | 606 | eIF4E3 | 6 | 290630 bp | 3′ | F | 1 |
| Evi1+LAP* | 504 | Ivns1abp | 1 | 64767 bp | 5′ | R | 2 |
| Evi1+LIP+LAP* | 302 | LOC70873 | 2 | — | Intron 9 | R | 0 |
| Evi1+LIP+LAP* | 302 | Prdx3 | 19 | — | Intron 1 | F | 0 |
| LIP | 407 | Ly86 | 13 | 66277 bp | 3′ | F | 2 |
| LIP | 408 | Epb4.1 | 4 | 68817 bp | 5′ | R | 0 |
| Transduced gene . | Mouse ID . | Nearest gene . | Chromosome no. . | Distance to gene (start or end) . | Location . | Forward or reverse orientation . | RTCGD hits . |
|---|---|---|---|---|---|---|---|
| Evi1 | 1 | Bcas1 | 2 | 14965 bp | 5′ | R | 2 |
| Evi1* | 2 | C/EBPβ | 2 | 75063 bp | 3′ | F | 25 |
| Evi1 | 3 | C/EBPβ | 2 | 62583 bp | 3′ | F | 25 |
| Evi1 | 4 | C/EBPβ | 2 | 86723 bp | 3′ | F | 25 |
| Evi1 | 5 | Pnp2 | 14 | 11834 bp | 3′ | R | 0 |
| Evi1 | 5 | Rps6ka1 | 4 | 32680 bp | 3′ | R | 4 |
| Evi1 | 5 | Rapgef4 | 2 | — | Intron 6 | R | 0 |
| Evi1 | 6 | C/EBPβ | 2 | 74775 bp | 3′ | F | 25 |
| Evi1 | 7 | C/EBPβ | 2 | 74873 bp | 3′ | F | 25 |
| Evi1 | 7 | E2F2 | 4 | — | Intron 1 | F | 10 |
| Evi1 | 8 | C/EBPβ | 2 | 81088 bp | 3′ | F | 25 |
| Evi1* | 15 | Atp10a | 7 | 2629 bp | 3′ | R | 0 |
| Evi1 | 21 | TLR7 | X | — | Intron 2 | R | 0 |
| Evi1+LAP | 606 | eIF4E3 | 6 | 290630 bp | 3′ | F | 1 |
| Evi1+LAP* | 504 | Ivns1abp | 1 | 64767 bp | 5′ | R | 2 |
| Evi1+LIP+LAP* | 302 | LOC70873 | 2 | — | Intron 9 | R | 0 |
| Evi1+LIP+LAP* | 302 | Prdx3 | 19 | — | Intron 1 | F | 0 |
| LIP | 407 | Ly86 | 13 | 66277 bp | 3′ | F | 2 |
| LIP | 408 | Epb4.1 | 4 | 68817 bp | 5′ | R | 0 |
bp, base pair.
The same integration sites are identified in the second leukemic cells as in the first ones.
C/EBPβ was overexpressed in the Evi1-leukemic cells
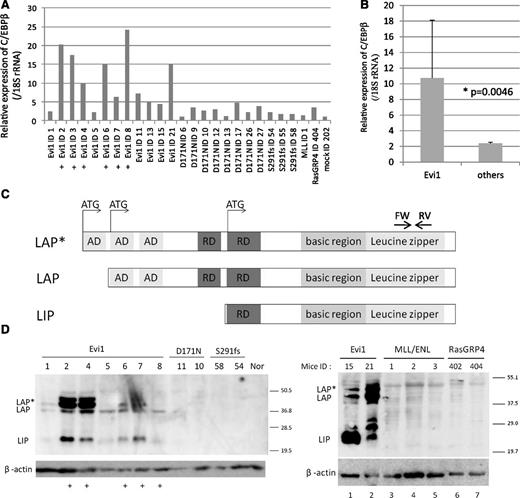
As the locus of integrations seemed to be too far from the genes to influence their expressions, we examined the messenger RNA (mRNA) expression levels of C/EBPβ, Tmem189, LOC10043728, and Ptpn1, all of which are located near the integration site. A high expression level of C/EBPβ, but not Tmem189, LOC10043728, or Ptpn1, was observed in the Evi1-leukemic cells (Figure 2A-B and supplemental Figure 1). High C/EBPβ mRNA levels were observed in 10 mice (ID2, 3, 4, 6, 7, 8, 11, 13, 15, and 21), and the remaining mice showed slightly lower expression levels (ID1 and 5) but still higher than mock. There are 3 isoforms of C/EBPβ, known as LAP*, LAP, and LIP, that arise from unique C/EBPβ mRNA by differential initiation of translation (Figure 2C). To distinguish 3 isoforms, we performed western blotting for C/EBPβ and confirmed high expression of all C/EBPβ isoforms in the Evi1-leukemic cells (Figure 2D). High expression of C/EBPβ was also observed in the Evi1-leukemic cells without retroviral integration near C/EBPβ (ID 1, 5, 15, and 21). In contrast, C/EBPβ expression was not observed in other leukemic cells expressing AML1-D171N, AML1-S291fs,7 MLL/ENL, or RasGRP4.21 Thus, high expression of C/EBPβ was a characteristic feature of Evi1-leukemia (Figure 2B; P = .0046). To examine the possibility of transcriptional activation of C/EBPβ by Evi1, we performed luciferase assays using 3.5 kb upstream of the C/EBPβ gene body. This area includes 3 predictive sequences as Evi1 binding sites (supplemental Figure 2A). However, Evi1 did not activate the C/EBPβ promoter in 293T and HeLa cell lines (supplemental Figure 2B).
High expression of C/EBPβ was observed in Evi1-leukemia cells. (A) Relative expression of C/EBPβ in whole BM cells derived from Evi1-leukemia mice, AML1-related leukemia mice (D171N and S291fs), a MLL/ENL-leukemia mouse, a RasGRP4-leukemia mouse, and a mock vector control mouse. Expression levels were normalized by the expression level of 18S rRNA. The expression level of the control mouse (mock ID 202) is set to 1. Plus marks indicate retroviral integration near C/EBPβ. (B) Comparison of C/EBPβ expression level between Evi1-leukemia and other leukemia. “Others” include mice transplanted with Runx1-D171N (n = 8), S291fs (n = 3), MLL-ENL (n = 1), RasGRP4 (n = 1), or mock (n = 1) transduced BM cells. (C) Schematics of 3 isoforms of C/EBPβ. AD stands for transactivating domain, and RD indicates repression domain. The horizontal arrows point to the location of the oligonucleotide primer pairs used for RT-PCR amplification. (D) Total cell lysates of the spleen cells were immunoblotted with anti-C/EBPβ Ab (Santa Cruz, C-19) and β-actin (Cell Signaling). C/EBPβ antibody (C-19) is a polyclonal antibody raised against a peptide mapping at the C-terminus of the C/EBPβ of rat origin. The spleen cells were derived from Evi1-leukemia mice, AML1-related leukemia mice (AML1-D171N and AML1-S291fs), MLL/ENL-leukemia mice, RasGRP4-leukemia mice, and a control normal mouse. Mice ID numbers are shown at the top of panels. Plus marks indicate retroviral integration near the C/EBPβ.
High expression of C/EBPβ was observed in Evi1-leukemia cells. (A) Relative expression of C/EBPβ in whole BM cells derived from Evi1-leukemia mice, AML1-related leukemia mice (D171N and S291fs), a MLL/ENL-leukemia mouse, a RasGRP4-leukemia mouse, and a mock vector control mouse. Expression levels were normalized by the expression level of 18S rRNA. The expression level of the control mouse (mock ID 202) is set to 1. Plus marks indicate retroviral integration near C/EBPβ. (B) Comparison of C/EBPβ expression level between Evi1-leukemia and other leukemia. “Others” include mice transplanted with Runx1-D171N (n = 8), S291fs (n = 3), MLL-ENL (n = 1), RasGRP4 (n = 1), or mock (n = 1) transduced BM cells. (C) Schematics of 3 isoforms of C/EBPβ. AD stands for transactivating domain, and RD indicates repression domain. The horizontal arrows point to the location of the oligonucleotide primer pairs used for RT-PCR amplification. (D) Total cell lysates of the spleen cells were immunoblotted with anti-C/EBPβ Ab (Santa Cruz, C-19) and β-actin (Cell Signaling). C/EBPβ antibody (C-19) is a polyclonal antibody raised against a peptide mapping at the C-terminus of the C/EBPβ of rat origin. The spleen cells were derived from Evi1-leukemia mice, AML1-related leukemia mice (AML1-D171N and AML1-S291fs), MLL/ENL-leukemia mice, RasGRP4-leukemia mice, and a control normal mouse. Mice ID numbers are shown at the top of panels. Plus marks indicate retroviral integration near the C/EBPβ.
Expression of C/EBPβ in murine hematopoietic stem cells and human leukemic patient samples
To examine the expression pattern of C/EBPβ in normal hematopoietic cells, we performed real-time PCR for C/EBPβ in c-Kit–positive, Sca1-positive, lineage-negative (KSL) cells, common myeloid progenitor (CMP) cells, megakaryocyte erythroid progenitor (MEP) cells, and granulocyte/macrophage progenitor (GMP) cells. Interestingly, expression of C/EBPβ was higher in KSL cells than in CMP, MEP, or GMP cells (Figure 3A). In addition, we examined the Evi1 deficiency effect on mouse hematopoietic stem cells by analyzing the Evi1-deficient mice that we had made22 (GSE11557. 1418901_at. The gene chip data are available on the GEO Web site). Although there was no statistical significance, expression of C/EBPβ was decreased in the hematopoietic stem cells of the Evi1-deficient mice (P = .11; supplemental Figure 3).
C/EBPβ expression in mouse HSC and human AML patients. (A) Expression of C/EBPβ and Evi1 in the BM of normal mice. The expression level of KSL is set to 1. KSL indicates c-Kit–positive, Sca1-positive and lineage-negative cells; CMP, common myeloid progenitor cells; MEP, megakaryocyte erythroid progenitor cells; and GMP, granulocyte/macrophage progenitor cells. Expression levels were normalized by the expression level of 18S rRNA. (B-C) Total cell lysates of the human whole BM cells, CD34+ cells, and CD34− cells were immunoblotted with anti-C/EBPβ Ab (top panel, Santa Cruz, C-19), anti-Evi1 Ab (second panel, Cell Signaling), anti-C/EBPα Ab (third panel, Santa Cruz, 14AA), and anti-β actin (bottom panel, Cell Signaling). CD34+/− cells of control sample ID 6 are derived from the residual PB stem cells for autologous transplantation. CD34+/− cells of control sample ID 10 and 11 are derived from BM. (D) The intensities of the bands of LAP*, LIP, Evi1, and β-actin in AML samples were quantitated by densitometry using Multi Gauge Version 3.0 software (Fujifilm). The intensities of the bands of LAP*, LIP, and Evi1 were normalized by the intensities of the bands of β-actin (n = 10). Spearman rank correlation coefficient of Evi1 and LAP* = 0.744; P = .0032. Spearman rank correlation coefficient of Evi1 and LIP = 0.679; P = .0128.
C/EBPβ expression in mouse HSC and human AML patients. (A) Expression of C/EBPβ and Evi1 in the BM of normal mice. The expression level of KSL is set to 1. KSL indicates c-Kit–positive, Sca1-positive and lineage-negative cells; CMP, common myeloid progenitor cells; MEP, megakaryocyte erythroid progenitor cells; and GMP, granulocyte/macrophage progenitor cells. Expression levels were normalized by the expression level of 18S rRNA. (B-C) Total cell lysates of the human whole BM cells, CD34+ cells, and CD34− cells were immunoblotted with anti-C/EBPβ Ab (top panel, Santa Cruz, C-19), anti-Evi1 Ab (second panel, Cell Signaling), anti-C/EBPα Ab (third panel, Santa Cruz, 14AA), and anti-β actin (bottom panel, Cell Signaling). CD34+/− cells of control sample ID 6 are derived from the residual PB stem cells for autologous transplantation. CD34+/− cells of control sample ID 10 and 11 are derived from BM. (D) The intensities of the bands of LAP*, LIP, Evi1, and β-actin in AML samples were quantitated by densitometry using Multi Gauge Version 3.0 software (Fujifilm). The intensities of the bands of LAP*, LIP, and Evi1 were normalized by the intensities of the bands of β-actin (n = 10). Spearman rank correlation coefficient of Evi1 and LAP* = 0.744; P = .0032. Spearman rank correlation coefficient of Evi1 and LIP = 0.679; P = .0128.
Next, we examined the relationship between Evi1 and C/EBPβ in human samples. The information regarding patient samples is shown in supplemental Table 5. Consistent with other reports,2 we found that 14.6% of patients expressed a high level of Evi1. However, we did not find any correlation between Evi1 and C/EBPβ mRNA expression in patients with AML (supplemental Figure 4). Additional studies using published microarray data of larger cohorts showed no statistical correlation between Evi1 and C/EBPβ expression in transcriptional levels (supplemental Figure 5).
Therefore, we next performed western blotting for CEBPβ on human AML samples. The BM cells of patients with lymphoma without BM invasion or primary immune thrombocytopenia were analyzed as controls (Figure 3B). The information about patient samples is shown in supplemental Table 6. The protein analysis allowed us to evaluate expression of the 3 isoforms of C/EBPβ individually. Interestingly, expression of these isoforms was high in some AML samples compared with control samples. Among the C/EBPβ isoforms, LAP* and LIP were predominantly expressed in human AML samples. Expression of C/EBPβ was not detected in CD34+ cells from BM and PB of the control cells (Figure 3C). We also performed western blotting for Evi1 protein expression and C/EBPα expression in these samples. We quantitated the intensities of the bands of LAP*, LIP, Evi1, and β-actin by densitometry using Multi Gauge Version 3.0 software (Fujifilm). Expression of Evi1 closely correlated with that of LAP* and LIP in patients with AML at a protein level (Figure 3D).
LIP induces leukemic transformation of primary BM cells in vitro and AML in vivo
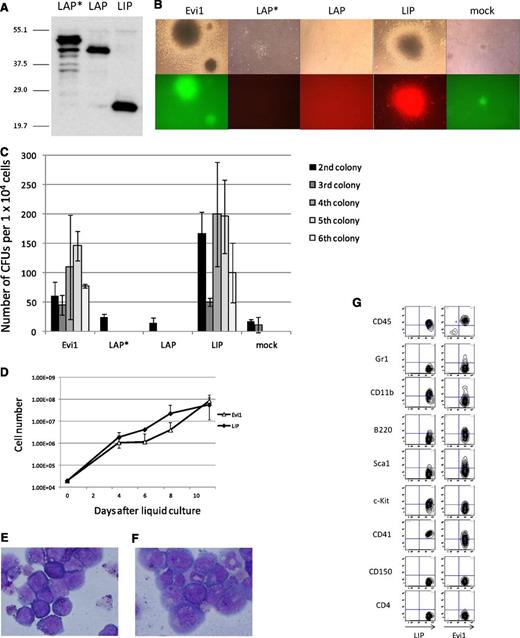
To examine the transforming activity of the 3 isoforms (LAP*, LAP and LIP), we performed a colony-replating assay. The expression plasmids were transfected into PLAT-E cells, and protein expression was examined by immunoblotting (Figure 4A). Although the LAP* cDNA contains 3 ATGs for all isoforms, it leads to expression of LAP* with a scarce amount of LAP and no LIP. The 3 isoforms were transduced in primary BM cells by retroviral transduction, and the transduced cells were cultured in methylcellulose with the defined cytokines. Although neither LAP* nor LAP showed the ability to transform primary BM cells, LIP induced transformation of the primary BM cells (Figure 4B-C). Furthermore, we succeeded in making a LIP-expressing cell line, which was grown in a liquid culture medium, RPMI supplemented with 10% fetal calf serum and 5 ng/mL of IL-3 (Figure 4D-E). We also established an Evi1-expressing cell line under the same culture condition for the LIP-expressing cell line (Figure 4D,F). Marker profiles of LIP and Evi1 leukemic cells in the liquid culture are shown in Figure 4G, both of which coincide with that of myeloid leukemia.
LIP induced transformation of primary BM cells in vitro in the same way as Evi1; LAP* and LAP did not. (A) Total cell lysates of PLAT-E cells transfected with pGCDNsam-LAP*, LAP, or LIP-Kusabira Orange were immunoblotted with the anti-C/EBPβ antibody (Santa Cruz, C-19). (B) Third colonies of the primary BM cells transduced with Evi1, LAP*, LAP, LIP, and mock (top panels). The Evi1-colonies were GFP positive, and the LIP-colonies were Kusabira Orange–positive (bottom panels). (C) Numbers of CFUs per cell plated in each generation. Mean from 2 independent experiments are depicted with standard deviation, each in duplicate. (D) Growth curves of the Evi1-expressing cells and the LIP-expressing cells in liquid culture (medium conditions: RPMI containing 10% fetal calf serum and 5 ng/mL of IL-3). (E-F) The liquid culture cells transformed by LIP (E) or Evi1 (F) were stained with Wright-Giemsa. Microscope, BH-2, Olympus; camera module, DP20, Olympus; objective lens, NC SPlan, Olympus; magnification, ×1000. (G) Flow cytometric analysis of the Evi1-liquid culture cells and the LIP-liquid culture cells.
LIP induced transformation of primary BM cells in vitro in the same way as Evi1; LAP* and LAP did not. (A) Total cell lysates of PLAT-E cells transfected with pGCDNsam-LAP*, LAP, or LIP-Kusabira Orange were immunoblotted with the anti-C/EBPβ antibody (Santa Cruz, C-19). (B) Third colonies of the primary BM cells transduced with Evi1, LAP*, LAP, LIP, and mock (top panels). The Evi1-colonies were GFP positive, and the LIP-colonies were Kusabira Orange–positive (bottom panels). (C) Numbers of CFUs per cell plated in each generation. Mean from 2 independent experiments are depicted with standard deviation, each in duplicate. (D) Growth curves of the Evi1-expressing cells and the LIP-expressing cells in liquid culture (medium conditions: RPMI containing 10% fetal calf serum and 5 ng/mL of IL-3). (E-F) The liquid culture cells transformed by LIP (E) or Evi1 (F) were stained with Wright-Giemsa. Microscope, BH-2, Olympus; camera module, DP20, Olympus; objective lens, NC SPlan, Olympus; magnification, ×1000. (G) Flow cytometric analysis of the Evi1-liquid culture cells and the LIP-liquid culture cells.
Next, we examined the role of C/EBPβ in leukemogenesis in vivo. Mouse BMT with C/EBPβ were performed in the same way as for the Evi1-leukemia BMT model.8 Infection efficiency and transplanted cell numbers are shown in supplemental Table 7. Interestingly, transduction of LIP alone induced AML with long latencies of ∼6 to 9 months (Figure 5A-B). On the other hand, LAP*- or LAP-expressing cells showed no engraftment.
Chimerisms of LIP-, LAP*-, or LAP-positive cells in PB. (A,C) Time courses of the percentage of donor chimerism in PB. LAP* (n = 7), LAP (n = 6), LIP (n = 14), Evi1/LAP* (n = 6), Evi1/LAP (n = 6), Evi1/LIP (n = 16), and Evi1/LIP/LAP* (n = 5). KO indicates Kusabira Orange. (B,D) Representative flow cytometric analysis data of transduction efficiencies at the day of the BMT (top panel). Representative flow cytometric analysis data of the PB cells at 5 months after the BMT (bottom panel).
Chimerisms of LIP-, LAP*-, or LAP-positive cells in PB. (A,C) Time courses of the percentage of donor chimerism in PB. LAP* (n = 7), LAP (n = 6), LIP (n = 14), Evi1/LAP* (n = 6), Evi1/LAP (n = 6), Evi1/LIP (n = 16), and Evi1/LIP/LAP* (n = 5). KO indicates Kusabira Orange. (B,D) Representative flow cytometric analysis data of transduction efficiencies at the day of the BMT (top panel). Representative flow cytometric analysis data of the PB cells at 5 months after the BMT (bottom panel).
LIP collaborates with Evi1 to induce AML in vivo
To clarify the relationship between Evi1 and C/EBPβ in leukemogenesis in vivo, we performed Evi1-C/EBPβ coexpressing BMT. For double infection, we mixed the virus carrying Evi1 (GFP) and one of the 3 viruses carrying LIP, LAP, or LAP* (Kusabira Orange) with an equal volume. The number of Evi1 and LIP double-positive cells gradually increased in the PB of the recipient mice (Figure 5C-D). Finally, the mice that had received the BM cells cotransduced with Evi1 and LIP went on to have AML ∼4 to 5 months after the transplantation. The latencies were shorter than those of the mice that had received the BM cells expressing Evi1 alone or LIP alone (Figure 6A and Table 2). This finding indicates that LIP accelerates the transforming capacity of Evi1 in vivo.
LIP collaborates with Evi1 to induce AML in BMT model. (A) Kaplan-Meier analysis for the mice that had received BMT. LIP synergized with Evi1 in inducing AML. Evi1 (n = 21), LIP (n = 14), LAP (n = 6), LAP* (n = 7), Evi1/LIP (n = 16), Evi1/LAP (n = 6), Evi1/LAP* (n = 6), and Evi1/LAP*/LIP (n = 5). (B) Total cell lysates of the spleen cells and PLAT-E cells transfected with pGCDNsam-LAP*, LAP, or LIP-Kusabira Orange were immunoblotted with anti-Flag Ab (top panel, SIGMA), anti-C/EBPβ Ab (middle panel, Santa Cruz, C-19), and anti-β actin (bottom panel, Cell Signaling). The spleen cells were derived from Evi1/LIP-leukemia mice and Evi1/LIP/LAP*-leukemia mice. Mice ID numbers are shown at the top of the panels. Lane numbers are shown at the bottom of the panels. Vertical lines have been inserted to indicate repositioned gel lanes. M, size markers. (C) The Evi1-leukemic cells and the Evi1/LIP-leukemic cells showed a similar surface marker profile. Flow cytometric analysis of Evi1, LIP-, Evi1/LIP-, and the Evi1/LIP/LAP*-leukemia cells. KO indicates Kusabira Orange.
LIP collaborates with Evi1 to induce AML in BMT model. (A) Kaplan-Meier analysis for the mice that had received BMT. LIP synergized with Evi1 in inducing AML. Evi1 (n = 21), LIP (n = 14), LAP (n = 6), LAP* (n = 7), Evi1/LIP (n = 16), Evi1/LAP (n = 6), Evi1/LAP* (n = 6), and Evi1/LAP*/LIP (n = 5). (B) Total cell lysates of the spleen cells and PLAT-E cells transfected with pGCDNsam-LAP*, LAP, or LIP-Kusabira Orange were immunoblotted with anti-Flag Ab (top panel, SIGMA), anti-C/EBPβ Ab (middle panel, Santa Cruz, C-19), and anti-β actin (bottom panel, Cell Signaling). The spleen cells were derived from Evi1/LIP-leukemia mice and Evi1/LIP/LAP*-leukemia mice. Mice ID numbers are shown at the top of the panels. Lane numbers are shown at the bottom of the panels. Vertical lines have been inserted to indicate repositioned gel lanes. M, size markers. (C) The Evi1-leukemic cells and the Evi1/LIP-leukemic cells showed a similar surface marker profile. Flow cytometric analysis of Evi1, LIP-, Evi1/LIP-, and the Evi1/LIP/LAP*-leukemia cells. KO indicates Kusabira Orange.
Average number of survival days of the recipient mice
| . | n . | Average no. of survival days . | P value (log-rank test) . |
|---|---|---|---|
| Evi1 | 21 | 282.9 | |
| LIP | 14 | 279.2 | .98 |
| Evi1+LAP* | 6 | 257.3 | .343 |
| Evi1+LAP | 6 | 321.5 | .809 |
| Evi1+LIP | 16 | 156.9 | 4.37E-07 |
| Evi1+LAP*+LIP | 5 | 112.4 | 1.13E-06 |
| . | n . | Average no. of survival days . | P value (log-rank test) . |
|---|---|---|---|
| Evi1 | 21 | 282.9 | |
| LIP | 14 | 279.2 | .98 |
| Evi1+LAP* | 6 | 257.3 | .343 |
| Evi1+LAP | 6 | 321.5 | .809 |
| Evi1+LIP | 16 | 156.9 | 4.37E-07 |
| Evi1+LAP*+LIP | 5 | 112.4 | 1.13E-06 |
In contrast, the Evi1 and LAP double-positive cells showed no engraftment. Cotransduction of Evi1 and LAP into primary BM cells resulted in the development of Evi1-leukemia without Kusabira Orange expression in the recipient mice (Figure 5C). Although the latencies were slightly longer than those of the mice that had received the BM cells expressing Evi1 alone, the results were not statistically significant (Table 2). Only 1 mouse showed increased double-positive cells in the PB, and the mouse died at 251 days after BMT. Therefore, acceleration of the Evi1-leukemia by LAP was not identified in our model.
Cotransduction of Evi1 and LAP* into primary BM cells showed diverse results. Three of the mice died of the Evi1 and LAP* double-positive leukemia, 2 of the mice died of the Evi1-leukemia without Kusabira Orange expression, and the remaining mouse died of B-lineage acute lymphoblastic leukemia derived from recipient cells (Figure 5C). Although the average survival duration was slightly shorter than that of the mice which had received the BM cells expressing Evi1 alone, the result was not statistically significant (Table 2).
LIP and LAP* collaborate with Evi1 to induce AML in vivo
Because the human AML with activated Evi1 predominantly expressed LIP and LAP* (Figure 3C), we tried triple induction of Evi1, LAP*, and LIP into primary BM cells and performed BMT. For triple infection, we mixed the viruses carrying Evi1 (GFP), LIP (Kusabira Orange), and LAP* (Kusabira Orange) with an equal volume. Interestingly, the triple induction induced AML with shorter latencies than that of Evi1 alone or Evi1/LIP double induction (Figure 6A and Table 2). We confirmed simultaneous expression of Evi1, LAP*, and LIP in the Evi1/LAP*/LIP-leukemic cells by immunoblotting (Figure 6B). These results suggest that LAP* provides a synergistic effect on the development of leukemia with high Evi1 expression in the presence of LIP. Although the direct interaction of Evi1 and C/EBPβ have been reported in adipocytes,23 the protein complex immunoprecipitation assay was negative for direct interaction between Evi1 and C/EBPβ in the leukemic cells (data not shown).
To identify the key factors in Evi1-LIP leukemogenesis, we examined the expression of several genes in the leukemic cells induced by Evi1 alone, LIP alone, and Evi1 plus LIP (supplemental Figure 6). We identified a high expression of GATA2 and Pbx1 in the Evi1-leukemia, consistent with previous reports.24,25 Interestingly, expression of Dnmt3a increased in the LIP-leukemic cells. LIP did not inhibit expression of C/EBPα.
C/EBPβ excision does not inhibit the development of Evi1-induced leukemia in vivo
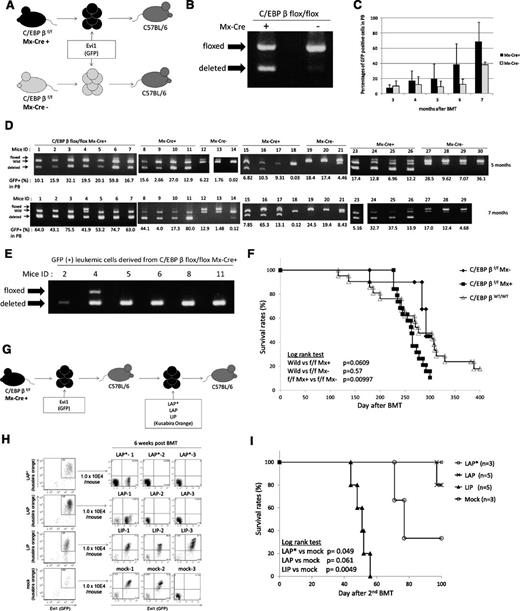
To assess the effect of C/EBPβ excision in Evi1-related leukemia, we transduced Evi1 into the BM cells derived from the Mx-Cre;C/EBPβfl/fl mice and then performed BMT (Figure 7A). At the startup of this experiment, we planned to delete C/EBPβ by injecting pIpC after development of Evi1-leukemia. However, we confirmed that excision of the C/EBPβ locus had already occurred in a part of the BM cells on infection of the Evi1 retrovirus before transplantation (Figure 7B), which suggests partial activation of the Mx promoter by retrovirus infection–induced interferon secretion without pI-pC injection. We transplanted all of the infected cells, including both C/EBPβfl/fl cells and C/EBPβ−/− cells, into the recipient mice. The percentages of GFP-positive cells in the PB were higher in the mice that received the BM cells derived from the Mx-Cre;C/EBPβfl/fl mice than the mice that received the BM cells derived from the C/EBPβfl/fl mice (Figure 7C). Interestingly, the C/EBPβ−/− cells increased without pIpC injection, parallel with the percentages of GFP-positive cells (Figure 7D). Most of the Evi1-leukemia derived from the Mx-Cre;CEBPβfl/fl cells lost the C/EBPβ locus (Table 3). As all of the genotyping PCR contained a wild-type band by contamination of the wild-type cells derived from the recipient, we sorted GFP-positive BM cells and performed genotyping PCR to avoid contamination of wild-type cells (Figure 7E). There were 5 of 6 examined leukemic cells that had only deleted alleles. The mean survival duration of the mice that received the BM cells derived from the Mx-Cre;C/EBPβfl/fl mice was slightly shorter than that of the mice which had received the wild-type BM cells expressing Evi1 (261.8 days vs 282.9 days); however, this was not statistically significant (Figure 7F). These results indicate that excision of C/EBPβ does not inhibit the development of Evi1-induced leukemia and that C/EBPβ is not essential for Evi1-induced transformation. Because a previous study from another group has shown the relevance of diminished C/EBP activity to leukemia with an erythroid phenotype, in which bcr-abl induces erythroleukemia in CEBPα−/− mice,26 we stained the Evi1-induced leukemic cells with Ter119 and confirmed that the leukemic cells were negative for Ter119 regardless of C/EBPβ genotype (supplemental Figure 7).
C/EBPβ excision does not inhibit the development of Evi1-induced leukemia in vivo. (A) Schema of the BMT model using Mx-Cre;C/EBPβfl/fl mice and C/EBPβfl/fl mice. (B) Genotyping PCR of BM cells from Mx-Cre;C/EBPβfl/fl mice and C/EBPβfl/fl mice 3 days after retrovirus infection. Excision of C/EBPβ occurred without pI-pC injection. (C) Time courses of the percentage of donor chimerism in PB. Mx-Cre;C/EBPβfl/fl (n = 19) and C/EBPβfl/fl (n = 10). (D) The genomic DNA of PB derived from the recipient mice were analyzed for recombination events at 5 months (top panel) and 7 months (bottom panel) after the BMT. The intensity of the shortest band for the deleted allele was higher at 7 months than at 5 months. (E) The GFP-positive BM cells derived from the leukemic mice were sorted and analyzed for recombination events by genotyping PCR. (F) Kaplan-Meier analysis for the mice that had received the Evi1-transduced BM cells of the Mx-Cre;C/EBPβfl/fl mice (n = 19), C/EBPβfl/fl mice (n = 10), and wild-type mice (n = 21). (G) Schema of the second BMT model. LAP*, LAP, or LIP were transduced into the Evi1-induced leukemic cells derived from C/EBPβ−/− cells by using retroviral infection. The double-positive cells (Evi1-GFP/C/EBPβ–Kusabira Orange) were sorted and then were transplanted into the recipient mice. (H) Left panels show flow cytometric data of C/EBPβ–deleted Evi1 (GFP) leukemia cells with forced expression of each C/EBPβ isoform (Kusabira Orange) or empty vector (Kusabira Orange). The double-positive cells were sorted and transplanted to recipient mice. Right panels show engraftment of the double-positive (Evi1+, LIP+, or mock+) transplanted cells but no engraftment of the LAP*- or LAP-expressing cells at 6 weeks after transplantation. (I) Survival curves of the second BMT recipient mice. LAP* (n = 3), LAP (n = 5), LIP (n = 5), and mock (n = 3). The triangles and circles on the curves are inserted to distinguish each curve but do not show the number of samples.
C/EBPβ excision does not inhibit the development of Evi1-induced leukemia in vivo. (A) Schema of the BMT model using Mx-Cre;C/EBPβfl/fl mice and C/EBPβfl/fl mice. (B) Genotyping PCR of BM cells from Mx-Cre;C/EBPβfl/fl mice and C/EBPβfl/fl mice 3 days after retrovirus infection. Excision of C/EBPβ occurred without pI-pC injection. (C) Time courses of the percentage of donor chimerism in PB. Mx-Cre;C/EBPβfl/fl (n = 19) and C/EBPβfl/fl (n = 10). (D) The genomic DNA of PB derived from the recipient mice were analyzed for recombination events at 5 months (top panel) and 7 months (bottom panel) after the BMT. The intensity of the shortest band for the deleted allele was higher at 7 months than at 5 months. (E) The GFP-positive BM cells derived from the leukemic mice were sorted and analyzed for recombination events by genotyping PCR. (F) Kaplan-Meier analysis for the mice that had received the Evi1-transduced BM cells of the Mx-Cre;C/EBPβfl/fl mice (n = 19), C/EBPβfl/fl mice (n = 10), and wild-type mice (n = 21). (G) Schema of the second BMT model. LAP*, LAP, or LIP were transduced into the Evi1-induced leukemic cells derived from C/EBPβ−/− cells by using retroviral infection. The double-positive cells (Evi1-GFP/C/EBPβ–Kusabira Orange) were sorted and then were transplanted into the recipient mice. (H) Left panels show flow cytometric data of C/EBPβ–deleted Evi1 (GFP) leukemia cells with forced expression of each C/EBPβ isoform (Kusabira Orange) or empty vector (Kusabira Orange). The double-positive cells were sorted and transplanted to recipient mice. Right panels show engraftment of the double-positive (Evi1+, LIP+, or mock+) transplanted cells but no engraftment of the LAP*- or LAP-expressing cells at 6 weeks after transplantation. (I) Survival curves of the second BMT recipient mice. LAP* (n = 3), LAP (n = 5), LIP (n = 5), and mock (n = 3). The triangles and circles on the curves are inserted to distinguish each curve but do not show the number of samples.
Genotype of the Evi1-induced leukemic cells derived from C/EBPβ (f/f) Mx1-Cre (+ or −) mice and average of survival days
| . | N . | Development of leukemia . | Genotype . | Average no. of survival days . | ||
|---|---|---|---|---|---|---|
| Deleted . | Deleted + floxed . | Floxed . | ||||
| C/EBPβ f/f Mx+ | 19 | 18 | 14 | 4 | 0 | 267.1 |
| C/EBPβ f/f Mx− | 10 | 9 | 0 | 0 | 9 | 307.8 |
| . | N . | Development of leukemia . | Genotype . | Average no. of survival days . | ||
|---|---|---|---|---|---|---|
| Deleted . | Deleted + floxed . | Floxed . | ||||
| C/EBPβ f/f Mx+ | 19 | 18 | 14 | 4 | 0 | 267.1 |
| C/EBPβ f/f Mx− | 10 | 9 | 0 | 0 | 9 | 307.8 |
Next, we attempted to delete the C/EBPβ locus by pI-pC injection before transplantation. However, the C/EBPβ locus was not completely ablated (supplemental Figure 8). In this experiment, we confirmed that the deleted band was enhanced after cytokine stimulation and retroviral infection. These results suggest that a successful retroviral infection induced interferon secretion and C/EBPβ depletion, and then the C/EBPβ–deleted clones generated Evi1-leukemia. Consistent with these findings, the percentages of GFP-positive cells correlated with enhancement of the deleted band in the genotyping PCR of PB cells (Figure 7D).
LAP* and LAP inhibit the development of Evi1-induced leukemia
To examine the role of the 3 isoforms of C/EBPβ (LAP*, LAP, and LIP) in the development of Evi1-induced leukemia, we transduced each isoform into the Evi1-induced leukemic cells derived from C/EBPβ−/− cells (Figure 7G). For this experiment, we used BM cells derived from mice ID5 and ID2 (Figure 7E). After retroviral infection, we sorted the double-positive cells (Evi1-GFP/C/EBPβ-Kusabira Orange) and transplanted the cells into recipient mice. Interestingly, only recipients of Evi1-induced leukemia cells with LIP overexpression showed leukemia development with a double-positive (Evi1+, LIP+) population in the recipients, whereas LAP* or LAP transduction did not yield such a population, and the recipient mice were healthy for 3 months (Figure 7H-I). In 2 mice (ID LAP*-1 and ID LAP-1), we confirmed engraftment of GFP (Evi1)–positive but Kusabira Orange (LAP* or LAP)–negative leukemic cells, and LAP-1 died with Evi1-single–positive leukemia at day 97. These results indicate that LIP accelerates the development of Evi1-induced leukemia, whereas LAP* and LAP inhibit such development.
Discussion
The cooperation of Evi1 and LIP that we showed in this study is a novel mechanism in Evi1-induced leukemogenesis. In leukemia research, it is known that C/EBPβ is recurrently targeted by IGH translocations in B-cell precursor acute lymphoblastic leukemia,27 and it regulates transcription factors critical for the proliferation and survival of multiple myeloma cells.28 Furthermore, expression of LIP is increased in FMS-like tyrosine kinase 3 (FLT3)-internal tandem duplication (ITD)–positive myeloid leukemic cells.29 On the other hand, C/EBPβ was shown to be upregulated during ATRA-induced differentiation of acute promyelocytic leukemia cells, and it was considered as a potential therapeutic target in leukemia for granulocytic differentiation.30 Taken together, C/EBPβ has leukemia suppressor–like activity in acute promyelocytic leukemia, and it also has critical oncogenic functions in FLT3-ITD leukemia, B-lineage acute lymphoblastic leukemia, and multiple myeloma.
To make clear the role of C/EBPβ in leukemogenesis, we need to distinguish 3 isoforms of C/EBPβ: LAP*, LAP, and LIP. All of these isoforms arise from unique C/EBPβ mRNA by differential initiation of translation.31,32 LAP* and LAP are transactivators, and the former has additional 23 N-terminal amino acids. LIP, a short isoform, lacks the transactivation domain and represses the transcriptional activity of LAP* and LAP.33
In our study, we demonstrated that overexpression of LIP alone caused the transformation of primary BM cells in vitro and induced the development of AML in vivo with long latencies. Our study is the first that provides direct evidence of the role of LIP in leukemogenesis in vivo. On the other hand, neither LAP*- nor LAP-expressing cells engrafted in recipient mice. These findings imply that overexpression of LAP* or LAP does not contribute to leukemic transformation of the BM cells, which may be related to their potential to induce granulocytic differentiation.34,35 However, we observed that the Evi1-induced leukemic cells expressed not only LIP but also LAP* and LAP in the mouse model. Furthermore, expression of Evi1 correlated closely with expression of both LAP* and LIP in patients with AML. Consistent with these findings, triple induction of Evi1, LAP*, and LIP induced AML with shorter latencies than that of double induction of Evi1 and LIP. These results suggest that LIP may not always antagonize but may cooperate with LAP* depending on the cellular milieu, as is the case with the development of Evi1-leukemia.
LIP can act as an inhibitor of the other C/EBPs, including C/EBPα, by forming nonfunctional heterodimers. C/EBPα-p30, a short isoform, also has a dominant-negative function on C/EBPα-p42. Recent studies have suggested that the loss of function or expression of C/EBPα promotes the development of AML.36 Furthermore, the ability of C/EBPα-p30 to induce AML has been shown in a mouse model.37,38 In the same way as C/EBPα-p30, LIP may dominantly inhibit C/EBPα function and subsequently contribute to the development of AML.
Two-step leukemogenesis has been suggested in several clinical investigations39,,-42 and murine BMT models.43,-45 According to these reports, leukemia-related mutations are classified into 2 groups: class I and class II mutations. Class I mutations include proliferative or antiapoptotic gene mutations such as c-Kit, BCR/ABL, or FLT3-ITD. Class II mutations include dominant negative mutations of transcription factors involved in the differentiation of hematopoietic cells, such as AML1/ETO, PML/RARa, or C/EBPα. As LIP has a dominant negative effect on LAP* and LAP, LIP might be included in a class II gene, which blocks differentiation of hematopoietic cells. Therefore, in this context, Evi1 may act as a class I gene. Consistently, Evi1 also collaborates with AML1-D171N, which is recognized as a class II mutation in leukemogenesis.7 Although mutations of C/EBPβ are rare events in human samples,46 extensive and careful research including that of translational control factors47 may yield valuable information that will allow us to clarify the role of C/EBPβ in leukemogenesis. We also identified a subgroup of patients with AML, which shows undetectable Evi1 expression but exhibits high C/EBPβ expression. C/EBPβ may collaborate with other factors in this type of leukemia.
In our experiments, C/EBPβ excision did not inhibit the development of Evi1-induced leukemia. On the other hand, serial transduction of LAP* and LAP inhibits the development of Evi1-induced leukemia, whereas LIP accelerates such development. Given a potential tumor-suppressive property of LAP*/LAP on which the transforming activity of LIP may depend, one possibility is that loss of LAP*/LAP dominates the phenotype of C/EBPβ–deleted mice when all of the 3 isoforms are deleted by excision of C/EBPβ alleles. Therefore, we conclude that translational control to reduce expression of LIP is required when we consider C/EBPβ as a therapeutic target in the setting of translation to clinical application. Conditional excision of each isoform may be informative to clarify the role of C/EBPβ in leukemogenesis.
Interestingly, we observed higher mRNA expression of C/EBPβ in hematopoietic stem cells than in progenitor cells. On the other hand, it has been shown that a significant number of C/EBPβ–deficient mice die perinatally and that hematopoietic progenitor cells of surviving C/EBPβ–deficient mice respond imperfectly to GM-CSF and G-CSF,34 which suggests an important role of C/EBPβ in both hematopoietic stem cells and progenitor cells in mice. However, we did not detect C/EBPβ protein in human CD34+ cells. Therefore, the role of C/EBPβ in human hematopoietic stem cells remains to be elucidated.
It is a puzzle to us why the mice showed a high expression of C/EBPβ without retroviral integration near its locus. Although we examined the possibility of transcriptional activation of C/EBPβ by Evi1, Evi1 had no potential to activate the C/EBPβ promoter region. Sauvageau and colleagues48 used common retroviral integration sites to find out that Gfi1 was highly expressed in 14 leukemic mice but that only 7 of those showed retroviral integrations near Gfi1. The mechanism of Gfi1 expression in 7 mice without retroviral integration remains to be elucidated. However, their approach succeeded in identification of the candidate genes as cell type–specific cancer genes in leukemia.48 We and another group also observed similar results.7,49 Therefore, we believe that insertional mutagenesis is one of the powerful tools to predict a cell-specific region containing genes responsible for initiation or progression of leukemia. Overexpression of the genes in the absence of nearby retroviral integrations might be caused by the activation of some upstream regulators in the same pathway or might be associated with the higher-order architecture of genome DNA.
C/EBPβ forms a transcriptional complex with PRDM16, also named a MDS1/EVI1–like gene, in the transmission of myoblasts to brown fat.11 PRDM16 is a zinc finger transcription factor with an N-terminal PR domain and belongs to the gene family of Evi1. PRDM16 is also involved in chromosomal translocations in human leukemias.50 Similar to Evi1, this gene is expressed exclusively in the hematopoietic stem and progenitor cells.51 Although we identified the collaboration of Evi1 with C/EBPβ in leukemogenesis, we did not detect the direct interaction of the 2 factors in the leukemic cells. Therefore, the precise mechanism of how Evi1 collaborates with C/EBPβ in leukemogenesis is currently unclear and needs to be examined.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Takuro Nakamura and Dr Kazuhiro Morishita for providing the pMYs-mouse Evi1-IG and the pcDNA-mouse Evi1. The authors are grateful to Dr Masafumi Onodera for providing the pGCDNsam-IRES-Kusabira Orange vector. The authors also extend their thanks to Dr Esta Sterneck and Dr Toru Minamino for providing the C/EBPβ conditional knockout mice. Dr Hideyo Hirai gave his time for supporting the C/EBPβ knockout mouse model. Dr Takayuki Takahashi, Dean Charles, and Jamie Lesley gave their time for excellent language assistance. The authors also thank Fumi Kaminaga and Mayumi Kobayashi for technical assistance. Finally, the authors give special thanks to the late Yuko Sawamoto for her excellent technical support.
This study was partially supported by the Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (JSPS), a grant from the Japan Leukemia Research Fund, and a grant from the Takeda Science Foundation.
Authorship
Contribution: N.W.-O. conducted all of the experiments and wrote the manuscript; A.Y. made cDNAs derived from patients with AML and actively participated in writing the manuscript; T.S. assisted with the cell-sorting experiments and participated in writing the manuscript; T.I. assisted with the luciferase assay; K.K. participated in writing the manuscript; K.T. assisted with the experiments in cell sorting of CD34+ cells from human BM cells; Y.S. made cDNAs derived from patients with AML; A.S. made cDNAs derived from the hematopoietic cells of mice; T.T. assisted with the BMT model; J.K. provided experimental guidance and assisted with the BMT model; T.K. provided experimental guidance on the BMT model; A.M., H.Y., Y.Y., and K.T. are mediators for this project; and M.K. conceived and directed the project, secured funding, and actively participated in writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mineo Kurokawa, Department of Hematology & Oncology, Graduate School of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8655, Japan; e-mail: kurokawa-tky@umin.ac.jp.