Key Points
Human CD20+CD27+CD43+CD70−CD69− B cells have been described as the human counterpart of murine B-1 B cells, but this is controversial.
Our data demonstrate a pre-plasmablast but not a B-1 phenotype for this population of cells.
Abstract
Controversy has arisen about the nature of circulating human CD20+CD27+CD43+CD70−CD69− B cells. Although originally described as being the human counterpart of murine B-1 B cells, some studies have raised the possibility that these might instead be plasmablasts. In this article, we have further characterized the putative B-1 cells and compared them directly with memory B cells and plasmablasts for several functional characteristics. Spontaneous antibody production of different isotypes as well as the induced production of antigen-specific antibodies after vaccination with a T-cell–dependent antigen did not reveal differences between the putative B-1 cells and genuine CD20− plasmablasts. Gene expression profiling of different B-cell subsets positioned the phenotype of putative B-1 cells closer to CD20− plasmablasts than to memory B cells. Moreover, putative B-1 cells could be differentiated into CD20− plasmablasts and plasma cells in vitro, supporting a pre-plasmablast phenotype. In conclusion, characterization of the putative B-1 cells revealed a functional phenotype and a gene expression profile that corresponds to cells that differentiate into CD20− plasmablasts. Our data offer perspectives for the investigation of differentiation of B cells into antibody secreting cells.
Introduction
In mice, B-1 lymphocytes represent a unique B-cell population distinguished from follicular B cells (B-2 cells) and marginal zone B cells by their surface marker expression, developmental origin, self-renewing capacity, and functions.1 These B-1 cells are identified by cell surface expression of immunoglobulin (Ig)Mhi, IgDlo, CD23−, CD43+, and B220lo. The vast majority is found in peritoneal and pleural cavities, whereas almost no B-1 cells are found in the peripheral blood and lymphoid tissues.1 B-1 cells are responsible for the production of so-called natural antibodies that occur spontaneously in naive “antigen-free” mice.2-4 These are polyspecific antibodies of low affinity and predominantly of the IgM isotype.1 They constitute a first line of defense against microbial antigens, and dysregulation of their antibody production may lead to the development of autoimmunity.5 Moreover, B-1 cells are envisioned as the primary antibody producers in responses to T cell–independent (TI) antigens along with marginal zone B cells.6 Follicular B-2 cells, on the other hand, participate in germinal center reactions and mount antibody responses to T cell–dependent (TD) antigens.7
The identity and existence of the human counterpart of murine B-1 cells have been in doubt for many years, mainly because of the absence of known cell surface markers to identify this population. Recently, Griffin et al8 claimed the discovery of a small subset of B cells in human peripheral and cord blood that specifically express CD20, CD27, and CD43, and that recapitulate several functional characteristics of murine B-1 cells. However, some groups have raised the possibility that the proposed CD20+CD27+CD43+ B-1 cells might be activated cells on their way to plasma cell differentiation.9-11 These concerns were refuted by Griffin et al12 by stating that the proposed B-1 cells are identified on CD20 expression, which is lost during differentiation at the plasmablast stage.13 Importantly, however, CD20+ plasmablasts and plasma cells have been observed in human tonsil tissue and in vitro.14-17
We therefore further characterized the proposed human B-1 cells and compared them directly with circulating CD20− plasmablasts. Spontaneous antibody production of different isotypes as well as the induced production of antigen-specific antibodies after vaccination with a TD antigen did not reveal differences between the proposed B-1 cells and genuine plasmablasts. Gene expression profiling positioned the proposed B-1 cells between CD20+CD27+CD43− memory B cells and CD20−CD27+CD43+ plasmablasts.
Taken together, we reveal a pre-plasmablast phenotype and not a B-1 cell phenotype within the CD20+CD27+CD43+CD70−CD69− B-cell population.
Materials and methods
Donors and samples
Anonymous peripheral blood samples were obtained as buffy coats from the Flemish Red Cross. After signing informed consent, healthy adult volunteers received Tedivax Pro Adulto (GlaxoSmithKline); 7 days later, their blood was collected by venipuncture using heparin as anticoagulant. The study was approved by the Ethics Committee of the University Hospitals of Leuven. All human participants gave written informed consent.
Processing
All samples were treated promptly upon receipt. Peripheral blood mononuclear cells were isolated from buffy coats or whole blood by density gradient separation (Ficoll, Lucron Biopruducts, De Pinte, Belgium). B cells were isolated using CD19+ beads (Miltenyi Biotec, Mönchengladbach, Germany) per manufacturer’s instructions. Unless otherwise noted, cells were incubated in RPMI1640 (Life Technologies, Merelbeke, Belgium) containing 10% fetal calf serum (FCS; Hyclone, Geel, Belgium) plus 2 mM l-glutamine and 50 µg/mL gentamicin (Life Technologies).
Antibodies, flow cytometry, and cell sorting
The following antibodies and reagents were used for flow cytometry and cell sorting: CD19 (HIB19)-Brilliant Violet 421 (Biolegend, Fell, Germany), IgD (IA6-2)-fluorescein isothiocyanate (FITC), IgM-(G20-127)-PerCP-Cy5.5, CD70 (Ki-24)-FITC, CD27 (M-T271)-APC, CD69 (FN50)–Pe-Cy5 (all from BD Biosciences, Erembodegem, Belgium); CD43 (84-3C1)-PE and CD20 (2H7)-APC-eFluor780 (both from eBioscience, Vienna, Austria) and CD3 (BW264/56)-PeVio770 (Miltenyi). Live/Dead Fixable Aqua stain was obtained from Life Technologies.
Cells were stained in PBS with 0.5% bovine serum albumin (BSA) and 2 mM EDTA for 30 minutes at 4°C in the dark. Subsequently, cells were washed with phosphate-buffered saline (PBS) and either fixed for 30 minutes in PBS with 2% formaldehyde and subsequently stored at 4°C in the dark in PBS with 0.5% BSA and 2 mM EDTA (for analysis) or stored in PBS with 0.5% BSA and 2 mM EDTA for sorting. Flow cytometry was performed on a FACSCanto II cytometer (BD Biosciences) or LSRFortessa (BD Biosciences) and cell sorting was performed on a FACSAriaI (BD Biosciences). Compensations were set using BD CompBeads (BD Biosciences) and gates were set using fluorescence minus one controls.18,19 B cells were always gated as CD3−CD19+ cells. The following definitions for the different subpopulations were used: naive B cells: CD3−CD19+CD20+CD43−CD27−CD70−CD69−; memory B cells: CD3−CD19+CD20+CD43−CD27+CD70−CD69−; proposed B-1 cells: CD3−CD19+CD20+CD43+CD27+CD70−CD69−; and plasmablast: CD3−CD19+CD20−CD43+CD27+. Postsort analysis revealed a purity of > 95%. Using this gating strategy, percentages ranged between 1% to 5% in nonvaccinated healthy adult donors.
For some in vitro differentiation experiments, CD43− peripheral blood mononuclear cells (containing mostly B cells) were isolated using CD43-microbeads (Miltenyi) per manufacturer’s instructions. Absence of CD43+ B cells was verified by flow cytometry as described.
ELISPOT
To determine the number of antibody secreting cells, enzyme-linked immunospot (ELISPOT) assays were performed. In brief, individual wells of a nitrocellulose Millititer HA plate (MAHANS 4550, Merck Millipore, Overijse, Belgium) were coated overnight at 4°C with 5 µg/mL of tetanus anatoxin (Merck Millipore), mouse anti-human IgM (BD Biosciences), mouse anti-human IgA (BD Biosciences), or goat anti-human IgG (Life Technologies). After coating, the plates were blocked with PBS containing 1% milk powder for 2 hours at 37°C. Subsequently, cells were cultured overnight in 100 µL RPMI medium supplemented with 10% FCS and 50 µM β-mercaptoethanol (Life Technologies). After washing 5 times with PBS-0.05% Tween20, the wells were incubated with 100 µL of biotinylated goat anti-human IgG, goat anti-human IgM, or goat anti-human IgG (all from ImTec Diagnostics, Antwerpen, Belgium). After 2 hours of incubation at 37°C, wells were washed with PBS and peroxidase-conjugated streptavidin (Jackson Immunoresearch Laboratories, West Grove, PA) was added and incubated for 1 hour at 37°C. Spots were visualized using 3-Amino-9-Ethylcarbazole (Sigma-Aldrich, Bornem, Belgium). After spots appeared, the reaction was stopped with tap water. Analysis was performed using Eli.Analysis software (A.EL.VIS, Hannover, Germany).
In vitro differentiation
For in vitro differentiation of CD43− B cells, 106 sort-purified cells per well of a 24-well plate were cultured in Iscove modified Dulbecco medium (Life Technologies) containing 2 mM glutamax, 10% FCS, 50 µM β-mercaptoethanol, 50 µg/mL gentamicin, 2,5 µg/mL of R-848 (Enzo Life Sciences, Antwerp, Belgium), 1000 U/mL interleukin (IL)-2 (Preprotech, Neuilly Sur Seine, France), 50 µg/mL transferrin (Sigma), and 5 µg/mL insulin (Sigma). For differentiation of CD20+CD27+CD43+CD70− cells, 100.000 sort purified cells per well of a 96-well plate were cultured in Iscove modified Dulbecco medium containing 2 mM glutamax, 10% FCS, 50 µM β-mercaptoethanol, 50 µg/mL gentamicin, 2,5 µg/mL of R-848, 1000 U/mL UL-2, 50 ng/mL IL-10 (Preprotech), 15 ng/mL IL-15 (Preprotech), and 50 ng/mL IL-6 (Preprotech).
Gene expression profiling
Total RNA from sorted cell fractions was extracted using the Absolutely RNA nanoprep kit from Agilent (Walbronn, Germany). The total RNA quantity and quality was determined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and the 2100 Bioanalyzer (Agilent), respectively. Total RNA profiles of all tested samples were similar with sharp 18S and 28S ribosomal RNA peaks on a flat baseline. Total RNA (50-100 ng per sample) was reverse transcribed, subsequently amplified, and fragmented before labeling for analysis of the messenger RNA expression via human Gene 1.0 ST arrays according to manufacturer’s manual 4475209 Rev.B (Life Technologies) and 702808 Rev.6 (Affymetrix, Santa Clara, CA). Arrays were scanned using the Affymetrix 3000 GeneScanner. Image files were generated using the Affymetrix GeneChip command console. Raw data were analyzed and normalized with RMA sketch using the standard settings for Gene 1.0 ST arrays of Expression Console in the Affymetrix GeneChip command software (Affymetrix). Data analysis was performed on log2 transformed absolute values using multiple experiment viewer. Hierarchical clustering was performed using Pearson’s correlation coefficient.
Results
CD20+CD43+CD27+CD69− cells spontaneously secrete IgG, IgA, and IgM
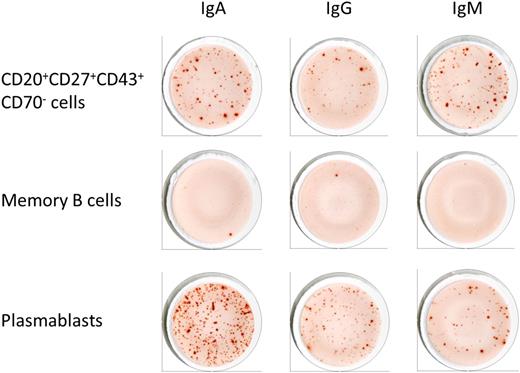
Given the controversies about the nature of the proposed B-1 cells, we determined by ELISPOT whether purified CD3−CD19+CD20+CD43+CD27+CD69− B cells spontaneously secreted IgG and IgA antibodies in addition to IgM. To compare with other B-cell subsets, memory B cells (CD3−CD19+CD20+CD27+CD43−) and plasmablasts (CD3−CD19+CD20−CD27+CD43+) were also isolated and tested for antibody secretion (Figures 1 and 2 and Table 1). We found that the spontaneous secretion of antibodies by unstimulated CD20+CD43+CD27+CD69− B cells was not restricted to IgM, the isotype typically associated with B-1 cells.1 In contrast, IgA was the most abundantly secreted isotype, much like circulating CD20− plasmablasts, which are mainly derived from mucosal immune responses.20 Memory B cells did not produce antibodies of any isotype (Figure 2).
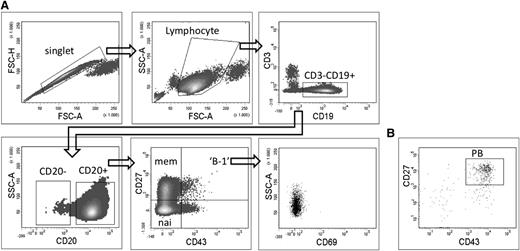
Sorting strategy of different B-cell subpopulations starting from CD19+-enriched B cells. CD19+-enriched B cells were stained with CD19-PE, CD3-Pe-Vio770, CD20-APC-eFluor780, CD43-FITC, CD27-APC, and CD69-PeCy5. (A) Doublets were excluded based on forward scatter (FSC)-H and FSC-A profile, cells with lymphocyte morphology were gated on FSC-A vs SSC-A profile, T cells were excluded by gating CD3− cells. Naive B cells (nai) are defined as CD3−CD19+CD20+CD27−CD43−, memory B cells (mem) as CD3−CD19+CD20+CD27+CD43−, the proposed B-1 cells as CD3−CD19+CD20+CD27+CD43+CD69−. (B) Plasmablasts (PB) were gated as CD3−CD19+CD20−CD27+CD43+ with the plot showing CD3−CD19+CD20− cells as gated on the lower left plot of (A).
Sorting strategy of different B-cell subpopulations starting from CD19+-enriched B cells. CD19+-enriched B cells were stained with CD19-PE, CD3-Pe-Vio770, CD20-APC-eFluor780, CD43-FITC, CD27-APC, and CD69-PeCy5. (A) Doublets were excluded based on forward scatter (FSC)-H and FSC-A profile, cells with lymphocyte morphology were gated on FSC-A vs SSC-A profile, T cells were excluded by gating CD3− cells. Naive B cells (nai) are defined as CD3−CD19+CD20+CD27−CD43−, memory B cells (mem) as CD3−CD19+CD20+CD27+CD43−, the proposed B-1 cells as CD3−CD19+CD20+CD27+CD43+CD69−. (B) Plasmablasts (PB) were gated as CD3−CD19+CD20−CD27+CD43+ with the plot showing CD3−CD19+CD20− cells as gated on the lower left plot of (A).
Spontaneous antibody production by different subsets of B cells. CD3−CD19+CD20+CD27+CD43+CD69− B cells as well as memory B cells (CD3−CD19+CD20+CD27+CD43−) and plasmablasts (CD3−CD19+CD20−CD27+CD43+) were analyzed for their capacity to spontaneously secrete IgA, IgM, and IgG antibodies by ELISPOT. Cells were plated at 2 × 104 cells per well and incubated for 12 hours at 37°C. Spot numbers are shown in Table 1.
Spontaneous antibody production by different subsets of B cells. CD3−CD19+CD20+CD27+CD43+CD69− B cells as well as memory B cells (CD3−CD19+CD20+CD27+CD43−) and plasmablasts (CD3−CD19+CD20−CD27+CD43+) were analyzed for their capacity to spontaneously secrete IgA, IgM, and IgG antibodies by ELISPOT. Cells were plated at 2 × 104 cells per well and incubated for 12 hours at 37°C. Spot numbers are shown in Table 1.
Spontaneous antibody production by different B-cell subsets
| . | CD3−CD19+ CD20+CD27+CD43− . | CD3−CD19+CD20+ CD27+CD43+CD69− . | CD3−CD19+ CD20−CD27+CD43+ . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | IgA . | IgG . | IgM . | IgA . | IgG . | IgM . | IgA . | IgG . | IgM . |
| Donor 1 | 0 | 0 | 0 | 120 | 65 | 95 | ND | ND | ND |
| Donor 2 | 1 | 1 | 3 | 38 | 30 | 29 | 137 | 54 | 25 |
| Donor 3 | 0 | 0 | 0 | 59 | 30 | 28 | 57 | 30 | 33 |
| . | CD3−CD19+ CD20+CD27+CD43− . | CD3−CD19+CD20+ CD27+CD43+CD69− . | CD3−CD19+ CD20−CD27+CD43+ . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | IgA . | IgG . | IgM . | IgA . | IgG . | IgM . | IgA . | IgG . | IgM . |
| Donor 1 | 0 | 0 | 0 | 120 | 65 | 95 | ND | ND | ND |
| Donor 2 | 1 | 1 | 3 | 38 | 30 | 29 | 137 | 54 | 25 |
| Donor 3 | 0 | 0 | 0 | 59 | 30 | 28 | 57 | 30 | 33 |
Memory B cells (CD3−CD19+CD20+CD27+CD43−), CD3−CD19+CD20+CD27+CD43+CD69− B cells, and plasmablasts (CD3−CD19+CD20−CD43+CD27+) from 3 healthy donors were purified by a combination of magnetic cell isolation and cell sorting (according to the gating strategy shown in Figure 1). The cells were plated at 2.5 × 103 cells per well, incubated at 37°C for 12 h, and analyzed for spontaneous antibody secretion by ELISPOT. ND, not done.
We thus show here that the proposed B-1 cells isolated from adult peripheral blood cannot be functionally distinguished from plasmablasts with regard to spontaneous antibody production.
CD20+CD43+CD27+CD69− cells secrete antibodies against TD antigen
We next examined the capacity of the proposed B-1 cells to produce antigen-specific IgA, IgG, and IgM antibodies in response to the vaccine Tedivax Pro Adulto, a vaccine that induces a secondary TD antibody response. We compared these responses to those of memory B cells and CD20− plasmablasts. Healthy adult volunteers were vaccinated with the Tedivax Pro Adulto vaccine; 7 days after vaccination, the different B-cell subsets were isolated and their capacity to secrete antibodies to tetanus anatoxin was analyzed by ELISPOT. As shown in Table 2, we found that, as with plasmablasts, the proposed B-1 cells were able to secrete tetanus anatoxin–specific antibodies, with a higher frequency found among plasmablasts. Moreover, we observed that for the proposed B-1 cells as well as for the plasmablasts the antibodies were mainly of the IgG isotype, which for a recall TD antigen as tetanus toxin, is reminiscent of memory B-2 cells undergoing differentiation.
Tetanus toxin–specific antibody production by different B-cell subsets 7 d postvaccination with Tedivax Pro Adulto
| . | CD3−CD19+ CD20+CD27+CD43− . | CD3−CD19+CD20+ CD27+CD43+CD69− . | CD3−CD19+ CD20−CD27+CD43+ . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | IgA . | IgG . | IgM . | IgA . | IgG . | IgM . | IgA . | IgG . | IgM . |
| Donor 1 | 0 | 0 | 0 | 0 | 40 | 0 | 0 | 82 | 1 |
| Donor 2 | ND | ND | ND | 5 | 47 | 0 | 15 | 84 | 0 |
| Donor 3 | 0 | 1 | 0 | 0 | 13 | 0 | 5 | 45 | 0 |
| Donor 4 | 0 | 0 | 0 | 1 | 61 | 0 | 3 | 53 | 1 |
| . | CD3−CD19+ CD20+CD27+CD43− . | CD3−CD19+CD20+ CD27+CD43+CD69− . | CD3−CD19+ CD20−CD27+CD43+ . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | IgA . | IgG . | IgM . | IgA . | IgG . | IgM . | IgA . | IgG . | IgM . |
| Donor 1 | 0 | 0 | 0 | 0 | 40 | 0 | 0 | 82 | 1 |
| Donor 2 | ND | ND | ND | 5 | 47 | 0 | 15 | 84 | 0 |
| Donor 3 | 0 | 1 | 0 | 0 | 13 | 0 | 5 | 45 | 0 |
| Donor 4 | 0 | 0 | 0 | 1 | 61 | 0 | 3 | 53 | 1 |
Memory B cells (CD3−CD19+CD20+CD27+CD43−), CD3−CD19+CD20+CD27+CD43+CD69− B cells, and plasmablasts (CD3−CD19+CD20−CD43+CD27+) from 3 healthy donors were purified by a combination of magnetic cell isolation and cell sorting (according to the gating strategy shown in Figure 1). The cells were plated at 2.5 × 103 cells per well, incubated at 37°C for 12 h, and analyzed for tetanus toxin–specific antibody secretion by ELISPOT. ND, not done.
These results reveal striking similarities between the proposed B-1 cells and plasmablasts with regard to antibody production after vaccination.
Different B-cell subsets can differentiate to CD20+CD43+CD27+CD70−CD69− cells
Abundant evidence has been accumulating that murine B-1 and B-2 B cells derive from distinct progenitors and belong to a separate lineage of B cells.21 Therefore, we wanted to verify whether the proposed B-1 cells are a separate lineage of B cells different from conventional B-2 cells. We cultured isolated CD43− B cells (which do not include the proposed B-1 cells) in the presence of R-848 and IL-2 and monitored their surface phenotype daily from day 3 to day 5 of culture. Culturing memory B cells with both R-848 and IL-2 induces differentiation into plasmablasts.22 If the human B-1 cells were a distinct lineage different from conventional B cells, the culture conditions should not induce these cells from CD43− B cells.
Flow cytometric analysis of the cells after 5 days of culture in the presence of R-848 and IL-2 revealed a significant induction of CD20+CD43+CD27+CD70−CD69− B cells from CD43− B cells. This population reached 20% of all CD20+ B cells by day 5 of culture (data not shown).
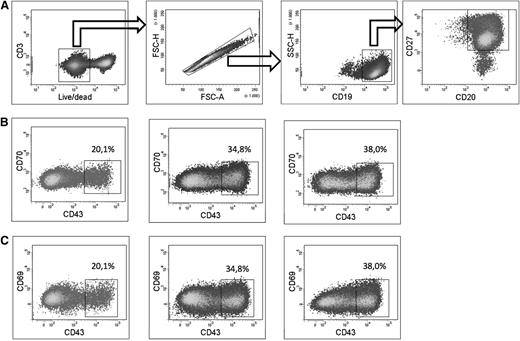
A caveat is the potential existence of an unidentified precursor in the CD43− B-cell population that may be responsible for the induction of the proposed B-1 cells. To exclude this possibility, different subsets of CD43− B cells were sorted and subsequently cultured in presence of R-848 and IL-2. Irrespective of culturing “IgM-only memory” (CD20+CD27+CD43−IgD−IgM+), “switched memory” (CD20+CD27+CD43−IgD−IgM−), or “natural effector” (CD20+CD27+CD43−IgD+IgM+) B cells, we were able to generate CD20+CD27+CD43+CD70−CD69− B cells (Figure 3). The amount of such cells formed in vitro was comparable (less than 2-fold differences) among the different starting B-cell subpopulations.
In vitro generation of CD20+CD27+CD43+CD70−CD69−cells from CD43−B cells. (A) On day 0, CD19+-enriched B cells were sorted into CD3−CD19+CD20+CD27+CD43−IgD+IgM+ (natural effector), CD3−CD19+CD20+CD27+CD43−IgD−IgM+ (IgM only memory), and CD3−CD19+CD20+CD27+CD43−IgD−IgM− (switched memory) B cells. Subsequently, sorted cells were put in culture in the presence of R-848 and IL-2. Cells were analyzed by flow cytometry on day 5. Live CD3−CD19+CD20+CD27+ cells of the 3 cultured populations were gated as shown. (B) Plots showing expression of CD43 and CD70 of live CD3−CD19+CD20+CD27+ cells starting from natural effector (left), IgM only memory (middle), and switched memory B cells (right). (C) Plots showing expression of CD43 and CD69 of live CD3-CD19+CD20+CD27+ cells starting from natural effector (left), IgM only memory (middle), and switched memory B cells (right). Plots from a representative experiment of 3 are shown.
In vitro generation of CD20+CD27+CD43+CD70−CD69−cells from CD43−B cells. (A) On day 0, CD19+-enriched B cells were sorted into CD3−CD19+CD20+CD27+CD43−IgD+IgM+ (natural effector), CD3−CD19+CD20+CD27+CD43−IgD−IgM+ (IgM only memory), and CD3−CD19+CD20+CD27+CD43−IgD−IgM− (switched memory) B cells. Subsequently, sorted cells were put in culture in the presence of R-848 and IL-2. Cells were analyzed by flow cytometry on day 5. Live CD3−CD19+CD20+CD27+ cells of the 3 cultured populations were gated as shown. (B) Plots showing expression of CD43 and CD70 of live CD3−CD19+CD20+CD27+ cells starting from natural effector (left), IgM only memory (middle), and switched memory B cells (right). (C) Plots showing expression of CD43 and CD69 of live CD3-CD19+CD20+CD27+ cells starting from natural effector (left), IgM only memory (middle), and switched memory B cells (right). Plots from a representative experiment of 3 are shown.
In summary, we were able to differentiate the proposed B-1 cells starting from distinct subpopulations of CD43− B cells using stimuli that are known to induce expansion and differentiation of memory B cells into plasmablasts22 ; therefore, the explanation that as-yet unidentified B-1 precursor cells are among the memory B cells can be ruled out. Our results show that the proposed B-1 cells represent activated CD27+ B cells that can be derived from different B-cell subsets.
Gene expression profiling confirms resemblance to plasmablasts
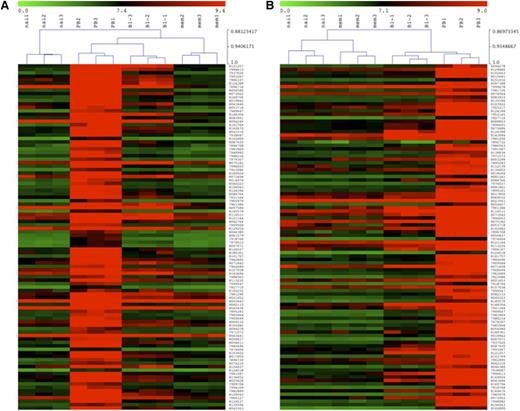
So far, our data indicate that the proposed B-1 cell closely resemble CD20− plasmablasts. To more thoroughly compare these cells with other B-cell subsets, gene expression analysis was performed. RNA was isolated from sort-purified naive (CD3−CD19+CD20+CD27−CD43−CD69−) and memory (CD3−CD19+CD20+CD27+CD43−CD69−) B cells as well as from the proposed B-1 cells (CD3−CD19+CD20+CD27+CD43+CD69−) and circulating plasmablasts (CD3−CD19+CD20−CD27+CD43+) obtained from 3 healthy adults. Given the outcome of the other experiments, we tested the hypothesis that that the proposed B-1 cells are an intermediate population between memory B cells and plasmablasts. Therefore, we first considered the 100 genes that were most upregulated in plasmablasts vs memory B cells and we compared their expression profile with that of the proposed B-1 cells. Hierarchical cluster analysis (Figure 4A) revealed that for these genes the proposed B-1 cells cluster closer to the plasmablasts than to the memory B cells. When we next considered the 100 genes that were most upregulated in plasmablasts relative to the proposed B-1 cells, we again observed that the proposed B-1 cells cluster closer to plasmablasts than to memory B cells (Figure 4B). Table 3 shows the top 100 genes that are overexpressed in plasmablasts compared with memory B cells and the top 100 genes that are overexpressed in plasmablasts compared with the proposed B-1 cell population. All genes shown in both comparisons had higher expression levels in the proposed B-1 cells than in the memory B cells.
Gene expression profiles of naive, memory, B-1 cells, and plasmablasts. The gene expression profile of purified B-cell subsets as defined in Figure 1 was determined and clustering analysis on log2-transformed normalized expression values is shown (red/green scale bar). (A) Dendrogram of hierarchical clustering of different B-cell subpopulations based on the top 100 overexpressed genes discriminating plasmablasts from memory B cells on a red/green map display for over- and underexpressed genes. The scale ranges from 0 (green) to 9.4 (red). (B) Dendrogram of hierarchical clustering of different B-cell subpopulations based on the top 100 overexpressed genes discriminating plasmablasts from the proposed B-1 cells on a red/green map display for over- and underexpressed genes. The scale ranges from 0 (green) to 9.0 (red).
Gene expression profiles of naive, memory, B-1 cells, and plasmablasts. The gene expression profile of purified B-cell subsets as defined in Figure 1 was determined and clustering analysis on log2-transformed normalized expression values is shown (red/green scale bar). (A) Dendrogram of hierarchical clustering of different B-cell subpopulations based on the top 100 overexpressed genes discriminating plasmablasts from memory B cells on a red/green map display for over- and underexpressed genes. The scale ranges from 0 (green) to 9.4 (red). (B) Dendrogram of hierarchical clustering of different B-cell subpopulations based on the top 100 overexpressed genes discriminating plasmablasts from the proposed B-1 cells on a red/green map display for over- and underexpressed genes. The scale ranges from 0 (green) to 9.0 (red).
Top 100 genes overexpressed in plasmablasts relative to memory B cells and proposed B-1 cells
| ranking according to overexpression . | Plasmablast vs memory . | Plasmablast vs B1 . |
|---|---|---|
| 1 | PRDM1 | FNDC3B |
| 2 | SLAMF7 | SLC7A11 |
| 3 | MKi-67 | PSAT1 |
| 4 | TNFRSF17 | HRASLS2 |
| 5 | SEMA4A | APOBEC3B |
| 6 | HIST1H3B | ALDH1L2 |
| 7 | BUB1 | AQP3 |
| 8 | APOBEC3B | DENND2C |
| 9 | DCAF12 | DCAF12 |
| 10 | TYMS | IGKV6-21 |
| 11 | IGKV6-21 | GLDC |
| 12 | IGKV2-40 | SLAMF7 |
| 13 | KIAA0101 | NUCB2 |
| 14 | TRIB1 | ITGA6 |
| 15 | FNDC3B | SLC1A4 |
| 16 | CD38 | FKBP11 |
| 17 | HPGD | HPGD |
| 18 | AQP3 | PRDM1 |
| 19 | SLC1A4 | TNFRSF17 |
| 20 | NUCB2 | NT5DC2 |
| 21 | SLC7A11 | MKI-67 |
| 22 | NT5DC2 | CDC6 |
| 23 | CCNB2 | TYMS |
| 24 | HRASLS2 | BMP8B |
| 25 | SLCO3A1 | BUB1 |
| 26 | DLGAP5 | SLC41A2 |
| 27 | XBP1 | DLGAP5 |
| 28 | SPN | SLCO3A1 |
| 29 | ASPM | CCNB2 |
| 30 | GLDC | KIAA0101 |
| 31 | V3-3 | ELL2 |
| 32 | TOP2A | TRIB1 |
| 33 | RRM2 | ESCO2 |
| 34 | PSAT1 | RRM2 |
| 35 | HIST1H2BB | PDIA5 |
| 36 | TP63 | TNFRSF17 |
| 37 | ELL2 | SLC44A1 |
| 38 | ALDH1L2 | AMPD1 |
| 39 | GPRC5D | SEC11C |
| 40 | GAB1 | ASPM |
| 41 | ESCO2 | VDR |
| 42 | MGC29506 | CDC20 |
| 43 | MANEA | IGLV7-46 |
| 44 | MYBL2 | DTL |
| 45 | DTL | C14orf145 |
| 46 | MAN1A1 | GPRIN3 |
| 47 | ITGA6 | HIST1H2AJ |
| 48 | TPX2 | SEMA4A |
| 49 | DENND2C | ELL2 |
| 50 | SKA3 | MANEA |
| 51 | CDC6 | CDKN3 |
| 52 | HIST1H1B | GPR155 |
| 53 | BMP8B | SEC24D |
| 54 | GPRIN3 | IGKV2-40 |
| 55 | FKBP11 | XBP1 |
| 56 | IGLV6-57 | SPN |
| 57 | VDR | IGLV6-57 |
| 58 | SLC44A1 | MGC29506 |
| 59 | PDIA4 | GPRC5D |
| 60 | GRCh37:15 | SEPT10 |
| 61 | ELL2 | LMAN1 |
| 62 | TNFRSF17 | HSPA13 |
| 63 | CDK1 | WIPI1 |
| 64 | LEF1 | CNKSR1 |
| 65 | WARS | CHAC2 |
| 66 | SEC11C | TSHR |
| 67 | IGKV2-24 | TP63 |
| 68 | PDIA5 | FAM172B |
| 69 | κ constant | C17orf28 |
| 70 | SLC4A12 | BHLHA15 |
| 71 | CDC20 | IL6ST |
| 72 | HSPA13 | SLC7A5 |
| 73 | SEC24D | RCC1 |
| 74 | NCAPG | FKBP14 |
| 75 | RCC1 | SELS |
| 76 | GRCH37:2 | GRCH37:15 |
| 77 | GPR155 | WARS |
| 78 | NCRNA00152 | PDIA4 |
| 79 | C14orf145 | HIST1H3B |
| 80 | CDKN3 | IL2RB |
| 81 | CASP3 | EIF4E3 |
| 82 | WIPI1 | CDK1 |
| 83 | MGC:178074 | HYOU1 |
| 84 | HIST1H3F | HIST1H2BB |
| 85 | HIST1H2AJ | SPATS2 |
| 86 | SELS | CASP3 |
| 87 | BHLA15 | CAV1 |
| 88 | LDLR | GRCh37e:3 |
| 89 | CENPF | DERL3 |
| 90 | PLK1 | GRCH37:14 |
| 91 | NUSAP1 | TXNDC11 |
| 92 | RUNX2 | LARP1B |
| 93 | SELPLG | GGH |
| 94 | HIST1H3J | CCR10 |
| 95 | CAV1 | CCNA2 |
| 96 | NCRNA00152 | PERP |
| 97 | C17orf28 | NCAPG |
| 98 | KIF23 | TOP2A |
| 99 | CNKSR1 | IGKV2OR22-4 |
| 100 | IL2RB | ADA |
| ranking according to overexpression . | Plasmablast vs memory . | Plasmablast vs B1 . |
|---|---|---|
| 1 | PRDM1 | FNDC3B |
| 2 | SLAMF7 | SLC7A11 |
| 3 | MKi-67 | PSAT1 |
| 4 | TNFRSF17 | HRASLS2 |
| 5 | SEMA4A | APOBEC3B |
| 6 | HIST1H3B | ALDH1L2 |
| 7 | BUB1 | AQP3 |
| 8 | APOBEC3B | DENND2C |
| 9 | DCAF12 | DCAF12 |
| 10 | TYMS | IGKV6-21 |
| 11 | IGKV6-21 | GLDC |
| 12 | IGKV2-40 | SLAMF7 |
| 13 | KIAA0101 | NUCB2 |
| 14 | TRIB1 | ITGA6 |
| 15 | FNDC3B | SLC1A4 |
| 16 | CD38 | FKBP11 |
| 17 | HPGD | HPGD |
| 18 | AQP3 | PRDM1 |
| 19 | SLC1A4 | TNFRSF17 |
| 20 | NUCB2 | NT5DC2 |
| 21 | SLC7A11 | MKI-67 |
| 22 | NT5DC2 | CDC6 |
| 23 | CCNB2 | TYMS |
| 24 | HRASLS2 | BMP8B |
| 25 | SLCO3A1 | BUB1 |
| 26 | DLGAP5 | SLC41A2 |
| 27 | XBP1 | DLGAP5 |
| 28 | SPN | SLCO3A1 |
| 29 | ASPM | CCNB2 |
| 30 | GLDC | KIAA0101 |
| 31 | V3-3 | ELL2 |
| 32 | TOP2A | TRIB1 |
| 33 | RRM2 | ESCO2 |
| 34 | PSAT1 | RRM2 |
| 35 | HIST1H2BB | PDIA5 |
| 36 | TP63 | TNFRSF17 |
| 37 | ELL2 | SLC44A1 |
| 38 | ALDH1L2 | AMPD1 |
| 39 | GPRC5D | SEC11C |
| 40 | GAB1 | ASPM |
| 41 | ESCO2 | VDR |
| 42 | MGC29506 | CDC20 |
| 43 | MANEA | IGLV7-46 |
| 44 | MYBL2 | DTL |
| 45 | DTL | C14orf145 |
| 46 | MAN1A1 | GPRIN3 |
| 47 | ITGA6 | HIST1H2AJ |
| 48 | TPX2 | SEMA4A |
| 49 | DENND2C | ELL2 |
| 50 | SKA3 | MANEA |
| 51 | CDC6 | CDKN3 |
| 52 | HIST1H1B | GPR155 |
| 53 | BMP8B | SEC24D |
| 54 | GPRIN3 | IGKV2-40 |
| 55 | FKBP11 | XBP1 |
| 56 | IGLV6-57 | SPN |
| 57 | VDR | IGLV6-57 |
| 58 | SLC44A1 | MGC29506 |
| 59 | PDIA4 | GPRC5D |
| 60 | GRCh37:15 | SEPT10 |
| 61 | ELL2 | LMAN1 |
| 62 | TNFRSF17 | HSPA13 |
| 63 | CDK1 | WIPI1 |
| 64 | LEF1 | CNKSR1 |
| 65 | WARS | CHAC2 |
| 66 | SEC11C | TSHR |
| 67 | IGKV2-24 | TP63 |
| 68 | PDIA5 | FAM172B |
| 69 | κ constant | C17orf28 |
| 70 | SLC4A12 | BHLHA15 |
| 71 | CDC20 | IL6ST |
| 72 | HSPA13 | SLC7A5 |
| 73 | SEC24D | RCC1 |
| 74 | NCAPG | FKBP14 |
| 75 | RCC1 | SELS |
| 76 | GRCH37:2 | GRCH37:15 |
| 77 | GPR155 | WARS |
| 78 | NCRNA00152 | PDIA4 |
| 79 | C14orf145 | HIST1H3B |
| 80 | CDKN3 | IL2RB |
| 81 | CASP3 | EIF4E3 |
| 82 | WIPI1 | CDK1 |
| 83 | MGC:178074 | HYOU1 |
| 84 | HIST1H3F | HIST1H2BB |
| 85 | HIST1H2AJ | SPATS2 |
| 86 | SELS | CASP3 |
| 87 | BHLA15 | CAV1 |
| 88 | LDLR | GRCh37e:3 |
| 89 | CENPF | DERL3 |
| 90 | PLK1 | GRCH37:14 |
| 91 | NUSAP1 | TXNDC11 |
| 92 | RUNX2 | LARP1B |
| 93 | SELPLG | GGH |
| 94 | HIST1H3J | CCR10 |
| 95 | CAV1 | CCNA2 |
| 96 | NCRNA00152 | PERP |
| 97 | C17orf28 | NCAPG |
| 98 | KIF23 | TOP2A |
| 99 | CNKSR1 | IGKV2OR22-4 |
| 100 | IL2RB | ADA |
Different B cell subsets were sorted and gene expression analysis was performed on sorted populations. The top 100 genes that were overexpressed in plasmablasts compared with memory B cells (left column) and in plasmablasts compared with proposed B-1 cells (right column) are listed, ranked in descending order of difference in expression. Genes depicted in bold have been associated with plasma cell differentiation23-25 or have been associated with a plasma cell–specific biological network, as defined by Jourdan et al, 2009.25
In addition to the genes listed in Table 3, we observed that the proposed B-1 cells (and plasmablasts) had upregulated gene expression relative to memory B cells of the IRF4 transcription factor that is involved in the differentiation of memory B cells to plasma cells and that has been shown not to be upregulated in murine peritoneal B-1 cells.26,27 These results are at odds with the proposed B-1 cells being the genuine human counterpart of murine B-1 cells.
In vitro differentiation of CD20+CD43+CD27+CD70−CD69− cells to plasmablasts and plasma cells
Thus far, our data revealed a phenotype intermediate between memory B cells and CD20− plasmablasts for the proposed B-1 cells. To further validate their intermediate nature, CD20+CD43+CD27+CD70−CD69− proposed B1 cells, CD20+CD43−CD27− memory B cells, and CD20+CD27−CD43− naive B cells were cultured in vitro in the presence of R-848, IL-2, IL-10, IL-15, and IL-6 for 5 days and analyzed by flow cytometry.
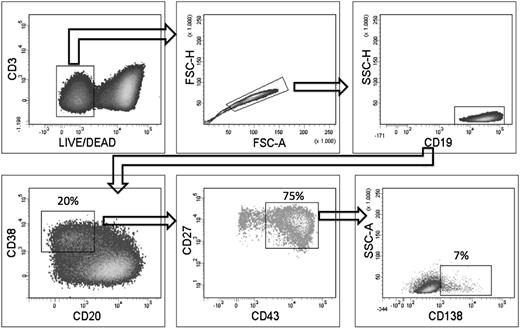
Starting from the CD20+CD43+CD27+CD70−CD69− proposed B-1 cells, 15% to 20% of the cells displayed the CD19+CD20−CD38+CD27+CD43+ plasmablast phenotype after 5 days of culture (Figure 5). A small percentage of CD138+ plasma cells was also formed (ie, 7% of the CD19+CD20−CD38+CD27+CD43+ cells). By culturing memory B cells under these conditions, no more than 5% plasmablasts and no CD138+ plasma cells were detected (data not shown). Naive B cells did not differentiate into cells of the plasmablast or plasma cell phenotype (data not shown).
In vitro generation of CD19+CD20−CD38+CD27hiCD43hiCD138+/−cells from proposed B-1 cells. The proposed B-1 cells were isolated as described in Figure 1 and were cultured for 5 days in the presence of R-848, IL-2, IL-10, IL-15, and IL-6. After culture, cells were stained for Live/Dead Fixable Aqua, CD19, CD3, CD20, CD27, CD43 CD38, and CD138, and analyzed by flow cytometry. Plots shown are from a representative experiment of 3.
In vitro generation of CD19+CD20−CD38+CD27hiCD43hiCD138+/−cells from proposed B-1 cells. The proposed B-1 cells were isolated as described in Figure 1 and were cultured for 5 days in the presence of R-848, IL-2, IL-10, IL-15, and IL-6. After culture, cells were stained for Live/Dead Fixable Aqua, CD19, CD3, CD20, CD27, CD43 CD38, and CD138, and analyzed by flow cytometry. Plots shown are from a representative experiment of 3.
Discussion
In this study, we functionally characterized the human CD20+CD27+CD43+CD70−CD69− proposed B-1 cells and identified a B-cell phenotype that has characteristics inconsistent with murine B-1 cells but that are compatible with pre-plasmablasts.
First, we found that the spontaneous antibody secretion by the proposed B-1 cells is in striking accordance with that of circulating plasmablasts. Both for the proposed B-1 cells and the plasmablasts isolated from peripheral blood of healthy adults, antibodies of the IgA isotype were the most frequently secreted. These circulating IgA secreting plasmablasts have previously been shown to be mainly derived from mucosal immune responses.20 In mice, many of the IgA+ plasma cells in the mucosa are believed to be derived from B-1 cells.28 However, this does not hold true for humans, in which intestinal IgA+ plasmablast are generated in germinal centers29 and carry a number of somatic hypermutations in their Ig variable regions (a hallmark of selection of follicular B-2 cells through germinal center reactions), comparable to IgG+ plasmablasts.30 The results of spontaneous antibody secretion by the proposed B-1 cells are at odds with the phenotype of murine B-1 cells, for which IgM is expected to be the dominant isotype produced.1 In addition to spontaneous antibody secretion, we also found striking similarities between the proposed B-1 cells and plasmablasts with regard to antibody production induced by vaccination. We have shown previously11 that the proposed B-1 cells are capable of secreting antibodies against TI-2 antigens after vaccination (as do plasmablasts and plasma cells). Because B-1 cells have in fact been shown to participate in TI-2 responses, these results were in line with a potential B-1 cell phenotype. The corresponding circulating plasmablasts could descend from these cells because differentiation of B-1 cells to plasma cells after TI-2 priming has been described in mice. In the current study, however, we found that the proposed B-1 cells also produced antibodies against a TD antigen after vaccination (as do plasmablasts). Moreover, not only did we find that the proposed B-1 cells produced antibodies to the TD antigen, the antigen-specific antibodies were predominantly of the IgG isotype, as expected after a TD recall response.7 These results are not compatible with a B-1 cell phenotype because TD antibodies are known to derive from B-2 cells that participate in germinal center reactions.7
In contrast to Griffin et al,8 we did not find surface expression of IgD on the majority of the proposed B-1 cells (data not shown). However, our results of spontaneous and induced antibody production of different isotypes are in agreement with the presence of IgG or IgA on these cells as described by Descatoire et al.10
Our in vitro differentiation experiments further question the phenotype of the proposed B-1 cells being the genuine counterpart of murine B-1 B cells. We found that the proposed B-1 cell phenotype could be generated from distinct B-cell populations (as defined by Berkowska et al31 ); ie, IgM memory B cells, switched memory B cells, and natural effector B cells. These findings are in contrast to studies from which there is accumulating evidence that B-1 and B-2 B cells derive from distinct progenitors and belong to a separate lineage of B cells.21 Because we were able to differentiate the proposed B-1 cells from distinct B cell subsets, we can rule out the possibility that these cells were derived from an as-yet unidentified precursor hidden in 1 of these subsets. Interestingly, not only could we generate the proposed B-1 cells starting from different B cell subsets, we also were able to further differentiate the proposed B-1 cells to plasmablasts and plasma cells, suggesting a pre-plasmablast phenotype.
Finally, our microarray data positioned the gene expression of the proposed B-1 cells between that of memory B cells and plasmablasts. Moreover, hierarchical cluster analysis considering the 100 genes that were most upregulated in plasmablasts vs memory B cells revealed that B-1 cells clustered closer to plasmablasts than to memory B cells.
The study of Griffin et al8 in which the CD20+CD27+CD43+ were claimed as being human B-1 cells stated that these cells are not in any way related to plasmablasts and plasma cells because of CD20 expression (which should be lost during differentiation at the plasmablast stage; Jego et al13 ). However, it is important to note that CD20+ plasmablasts have been described in tonsils as well as after in vitro differentiation experiments.14-16 Moreover, in agreement with the study of Sushanek et al,32 we observed low expression of CD20 in the proposed B-1 cells relative to both naive and memory B-cell subsets (as measured by the median fluorescence intensities by flow cytometry; data not shown). The higher expression levels of both CD43 and CD27 and the lower expression levels of CD20 relative to memory B cells (and comparable to plasmablasts, this work, and Verbinnen et al11 ) indicate a pre-plasmablastic phenotype.
Altogether, our data demonstrate a pre-plasmablast phenotype within the proposed B-1 cells because these cells (1) spontaneously produce antibodies of different isotypes, (2) are involved in the induced specific antibody secretion to TD as well as TI antigen, (3) have a gene expression profile that is intermediate between memory B cells and CD20− plasmablasts, (4) can be generated in vitro from distinct CD43− B cells, and (5) can be further differentiated into CD20− plasmablasts and plasma cells.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE42724).
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Greet Wuyts, Renaud Lavend'homme, Boukje Hoekman, and Petra Vandervoort for excellent technical assistance.
This study was supported by a research grant from the Fund for Scientific Research-Flanders (Krediet aan Navorsers) and by a research grant from the Research Council of the Catholic University of Leuven. X.B. is a senior clinical investigator of the Fund for Scientific Research-Flanders; K.C., B.V., and N.G. are supported through a grant from the Flemish Institute for Innovation through Science and Technology; and I.M. is supported by a KOF (Klinisch Onderzoeksfonds) grant from the University of Leuven.
Authorship
Contribution: K.C. and B.V. designed and performed research, analyzed and interpreted data, and wrote the manuscript; X.B. designed experiments and interpreted data; I.M., N.G., and M.J. assisted in experimental design; and F.S. and L.V.L. contributed reagents and analytical tools and performed microarray experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xavier Bossuyt, Experimental Laboratory Immunology, Department of Microbiology and Immunology, Katholieke Universiteit Leuven, University Hospitals Gasthuisberg, Herestraat 49, 3000 Leuven, Belgium; e-mail: xavier.bossuyt@uzleuven.be.
References
Author notes
K.C. and B.V. contributed equally to the study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal