Key Points
A new histo-blood group system was discovered, based on the identification of Forssman glycolipid antigen on human red blood cells.
A newly described polymorphism in the GBGT1 gene activates the encoded enzyme to synthesize Forssman antigen.
Abstract
In analogy with histo-blood group A antigen, Forssman (Fs) antigen terminates with α3-N-acetylgalactosamine and can be used by pathogens as a host receptor in many mammals. However, primates including humans lack Fs synthase activity and have naturally occurring Fs antibodies in plasma. We investigated individuals with the enigmatic ABO subgroup Apae and found them to be homozygous for common O alleles. Their erythrocytes had no A antigens but instead expressed Fs glycolipids. The unexpected Fs antigen was confirmed in structural, serologic, and flow-cytometric studies. The Fs synthase gene, GBGT1, in Apae individuals encoded an arginine to glutamine change at residue 296. Gln296 is present in lower mammals, whereas Arg296 was found in 6 other primates, > 250 blood donors and Apae family relatives without the Apae phenotype. Transfection experiments and molecular modeling showed that Agr296Gln reactivates the human Fs synthase. Uropathogenic E coli containing prsG-adhesin–encoding plasmids agglutinated Apae but not group O cells, suggesting biologic implications. Predictive tests for intravascular hemolysis with crossmatch-incompatible sera indicated complement-mediated destruction of Fs-positive erythrocytes. Taken together, we provide the first conclusive description of Fs expression in normal human hematopoietic tissue and the basis of a new histo-blood group system in man, FORS.
Introduction
Carbohydrate histo-blood group antigens, first recognized on red blood cells (RBCs) in 1900,1 have been suggested to be part of our innate immune response.2 Major carbohydrate histo-blood groups in man include the ABO, P1PK, H, Lewis, I, and GLOB systems in which glycoproteins and glycolipids carry immunodominant terminal sugars,3 defining polymorphic antigens. Other mammals also express carbohydrate histo-blood groups, such as ABO,4 fucose-less B antigen (Galili),5 and Forssman (Fs)6,7 but their expression on RBCs varies among species. Although the biologic function of polymorphic carbohydrates on RBCs is unresolved, these antigens can be used as receptors by pathogens8-11 and their expression in tissues and bodily secretions are thus believed to reflect microbial selection.8 In response to blood-group-mimicking glycans on bacterial surfaces, naturally occurring antibodies with the capacity to neutralize various microorganisms are formed. However, these antibodies also constitute substantial transfusion and transplantation barriers.3,12
In 1987, 3 families with a supposed ABO subgroup named Apae were reported.13 Although Helix pomatia lectin reacted strongly and polyclonal anti-A weakly with RBCs from some family members, monoclonal (MAb) anti-A reagents were later shown to be nonreactive, thus presenting an apparent paradox. The biochemical and genetic background of this enigmatic phenotype has remained unknown, as has its biologic consequences. We hypothesized that an explanation may be found by studying the glycolipids of this phenotype.14 Here we report the identification of Fs glycolipids, normally found only on RBCs of selected nonprimate mammals, are strongly expressed on human Apae RBCs. In nonprimates, Fs antigen is synthesized by Fs synthase (globoside 3-α-N-acetyl-D-galactosaminyltransferase, EC2.4.1.88),7 an enzyme homologous to the ABO transferase. We also reveal a genetic polymorphism in the human Fs gene (GBGT1) that alters the enzymatically inactive human protein15 to its active nonprimate counterpart. Overall, these findings qualify Fs as a new histo-blood group system with potential implications for both transfusion/transplantation medicine and pathogen susceptibility.
Methods
Samples
Five and 3 RBC units were collected from each of 2 unrelated Apae individuals (Apae#1 and Apae#2, respectively) from 2 of the originally reported families.13 Saliva samples were provided by 14 relatives. Informed consent was obtained according to the Helsinki Declaration after approval from the North Sheffield Local Research Ethics Committee (2008) or Sheffield Research Ethics Committee, National Research Ethics Service (2011), United Kingdom. Blood units were leukocyte-depleted, washed free of plasma with phosphate-buffered saline and frozen at −20°C until processed. Aliquots of plasma, RBCs, and leukocytes were kept for analysis. Approvals from the Regional Ethics Review Board at Lund University were obtained for bone marrow collection from healthy volunteers and genetic blood group analysis on samples from blood donors with known ABO phenotypes. Blood from anonymized Swedish donor samples was used for screening and control purposes.
Glycolipids
Glycolipid preparation.
Lysed blood units were thawed and total neutral glycolipids with < 20 sugar residues were isolated (see supplemental Methods, where control glycolipid preparations are also described; available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Open-column chromatography.
Total glycolipids (∼ 110 mg) from each of Apae#1 and #2 were fractionated by silica chromatography column (5g silica/100 mg lipid; Silica 60, Merck) in a system of chloroform (C) methanol (M) solvent mixes (supplemental Methods).
Thin-layer chromatography-enzyme immunoassay (TLC-EIA).
Structural analysis
Capillary liquid chromatography: electrospray ionization tandem mass spectrometry (LC-ESI/MS-MS).
Endoglycoceramidase II from Rhodococcus spp. (TakaraBio Europe) was used to release saccharides from ceramide for analysis by LC-ESI/MS-MS as described.18
Proton NMR spectroscopy.
1H-NMR spectra were acquired on a Varian 600-MHz spectrometer at 30°C. Samples were dissolved in dimethylsulfoxide/D2O (98:2 vol/vol) after deuterium exchange. Two-dimensional double quantum-filtered correlated spectroscopy (2D-DQF-COSY) spectra were recorded using the standard pulse sequence.19
Genetic analysis of ABO and GBGT1
ABO genotyping was performed.20 GBGT1 exons 1-7 were sequenced in 11 humans, including 2 Apae propositi, and compared with Ensembl entry ENST00000372040. Exon 7 was sequenced in 6 DNA samples from different nonhuman primates. Two allele-specific primer (ASP) PCR assays were designed to detect 887G > A and 363C > A. RNA extraction, cDNA preparation, and transcript analysis were performed (for details see supplemental Methods and supplemental Table 2).
Transfection
A GBGT1 (EMBL database; HE583599) wild-type transcript was used as template and 887G > A introduced by site-directed mutagenesis. Wild-type and mutant fragments were cloned into pEF1α-IRES+ZsGreen1 vectors (Clontech Laboratories). A B3GALNT1 wild-type fragment (GenBank; AB050855.1) cloned into the same vector was cotransfected to enhance the expression of acceptor substrate (P antigen) for Fs synthase. Primers are listed in supplemental Table 2. All constructs were verified by DNA sequencing.
MEG-01 cells were grown in RPMI1640 supplemented with 10% fetal bovine serum (FBS). Transient transfection of 3 × 106 MEG-01 cells with 10 μg of GBGT1-allele–containing vector and 10 μg of B3GALNT1 vector, or 20 μg of empty vector as a negative control, was performed by electroporation at 280 V and 960 μF with GenePulser (Bio-Rad). The MEG-01 cells were resuspended in RPMI1640 supplemented with 10% FBS and grown for 48 hours before flow cytometric analysis of Fs expression. Experiments were run in triplicate (duplicate for mock-treated cells) and repeated 3 times.
Flow cytometry and serology
Flow cytometry was performed with MAb anti-Fs, donor plasma, and eluate made from Apae-RBCs adsorbed with group B plasma. Strongly Apae-hemagglutinating samples were selected by crossmatching. Fresh sera from these donors were used for hemolysin tests12 against native and papainised RBC (Apae/A1/B/O). For details, see supplemental Table 1 and supplemental Methods.
Three-dimensional modeling
A molecular model of human Fs synthase was derived by threading onto a human ABO glycosyltransferase crystal structure, PDB entry 2RJ7 (supplemental Methods).
Escherichia coli hemagglutination
A common K12 E coli strain, HB101, unmodified or containing either papG (HB101.AD110) or prsG (HB101.JFK102) operons was used for hemagglutination experiments (supplemental Methods). RBCs of various ABO/Apae phenotypes were used, including group O RBCs modified with synthetic glycolipid function-spacer-lipid (FSL) constructs, so-called kodecytes.21
Results
ABO testing of Apae propositi
The original serologic work13 was confirmed and extended with additional ABO reagents. Of 15 anti-A reagents, 3 of 4 polyclonal but only 1 MAb reagent (clone A003) reacted with Apae-RBCs, albeit with weak reactivity. Of 8 anti-A,B reagents, all 3 polyclonal and 1 MAb blend reagent were weakly positive. By ABO genotyping, both Apae individuals showed homozygosity for 261delG, the common group O polymorphism.22,23 These data argued strongly against Apae as a conventional A subgroup in the ABO system.
Glycolipid analysis
Based on the hypothesis that another glycolipid than A antigen may be responsible for the Apae phenotype, total neutral glycolipids from Apae#1 and Apae#2 RBC unit lysates were analyzed. Open-column fractionation and pooling resulted in 5 heterogeneous fractions containing glycolipids with > 4 sugars from each donor.
Thin layer chromatography-enzyme immunoassay.
Total glycolipids (Figure 1) and open column fractions (Figure 2) were analyzed by TLC and immunostaining with a variety of reagents. Staining of the heterogeneous open-column fractions of the 2 Apae individuals was very similar (Figure 2), and expected enhancement in reactivity of minor bands compared with the total glycolipid samples was seen. Total Apae glycolipids reacted strongly with Helix pomatia in the 5-sugar region consistent with Fs glycolipids (Figures 1B and 2B), with the CM65:35 fraction being of primary interest (Figure 2B lanes 5-14). Staining of total glycolipids with anti-A and anti-Fs MAbs (Figure 1C-D), and column fractions (Figure 2C) were concordant with the identity of Fs.
Thin layer chromatography staining of total Apae glycolipids. (A) Nomenclature and structures of selected glycolipids discussed in this paper. (B) Helix pomatia lectin staining of the Apae total glycolipid samples and blood group A1, A2, O, and B controls plus Fs glycolipid control. TLC migratory positions of known blood group glycolipids are identified in the margin. (C) Staining with MAb anti-Fs. (D) Staining with a representative broadly reactive MAb anti-A (clone 11H5). (E) Unidentified bands recognized by staining with MAb anti-A (003). (F) Staining with Lewis (anti-Lea, anti-Leb) and secretory A antigen glycolipid profiles (anti-A type 1, anti-ALeb). In all panels A1, A2, O, and B control samples are total erythrocyte glycolipids whereas Fs is a purified glycolipid fraction. Lewis phenotypes are expressed as red cell phenotypes [where low level Lea glycolipid is expected in the Le(a−b+) phenotype]. Because of minor migration differences between assays, direct comparison of absolute band positions should be viewed with caution. Black lines indicate where migration controls and unloaded lanes have been removed from the image. All lanes are from the same TLC plate and have not been repositioned.
Thin layer chromatography staining of total Apae glycolipids. (A) Nomenclature and structures of selected glycolipids discussed in this paper. (B) Helix pomatia lectin staining of the Apae total glycolipid samples and blood group A1, A2, O, and B controls plus Fs glycolipid control. TLC migratory positions of known blood group glycolipids are identified in the margin. (C) Staining with MAb anti-Fs. (D) Staining with a representative broadly reactive MAb anti-A (clone 11H5). (E) Unidentified bands recognized by staining with MAb anti-A (003). (F) Staining with Lewis (anti-Lea, anti-Leb) and secretory A antigen glycolipid profiles (anti-A type 1, anti-ALeb). In all panels A1, A2, O, and B control samples are total erythrocyte glycolipids whereas Fs is a purified glycolipid fraction. Lewis phenotypes are expressed as red cell phenotypes [where low level Lea glycolipid is expected in the Le(a−b+) phenotype]. Because of minor migration differences between assays, direct comparison of absolute band positions should be viewed with caution. Black lines indicate where migration controls and unloaded lanes have been removed from the image. All lanes are from the same TLC plate and have not been repositioned.
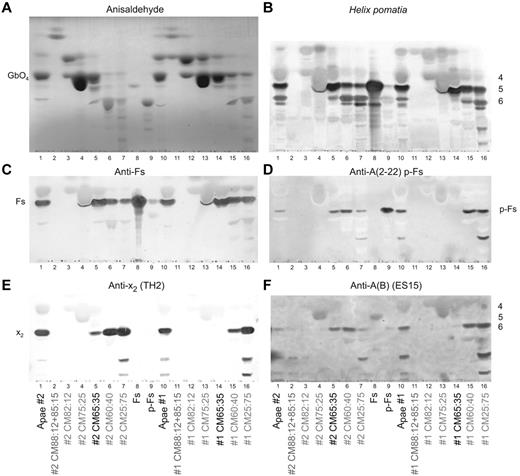
Thin layer chromatography staining of open column fractions. (A) Anisaldehyde semiquantitative chemical staining. (B) Helix pomatia lectin staining (with approximate sugar sizes indicated on the right of the plate). (C) MAb anti-Fs staining. (D) p-Fs crossreactive MAb anti-A 2-22 (clone AY209) staining. (E) Anti-x2 (clone TH2) staining. (F) p-Fs and Fs crossreactive MAb anti-A(B) (ES-15) staining. In all panels, total glycolipids are indicated as Apae#1, Apae#2, whereas open column fractions are identified by their elution solvent (with the fraction used for structural analysis indicated in black). Fs and p-Fs are purified reference glycolipids. Because of minor migration differences between assays, direct comparison of absolute band positions should be viewed with caution.
Thin layer chromatography staining of open column fractions. (A) Anisaldehyde semiquantitative chemical staining. (B) Helix pomatia lectin staining (with approximate sugar sizes indicated on the right of the plate). (C) MAb anti-Fs staining. (D) p-Fs crossreactive MAb anti-A 2-22 (clone AY209) staining. (E) Anti-x2 (clone TH2) staining. (F) p-Fs and Fs crossreactive MAb anti-A(B) (ES-15) staining. In all panels, total glycolipids are indicated as Apae#1, Apae#2, whereas open column fractions are identified by their elution solvent (with the fraction used for structural analysis indicated in black). Fs and p-Fs are purified reference glycolipids. Because of minor migration differences between assays, direct comparison of absolute band positions should be viewed with caution.
Present on the H pomatia plate (Figure 1B) with a migration similar to A-6-2, were also faint bands in both Apae samples and B and O controls. The migration and staining of these bands were consistent with p-Fs24 and/or x225 (Figure 2D-E) or A-6-2. Staining with an anti-A (MAb 2-22, Figure 2D) with known activity against p-Fs14 found activity in fractions CM65:35, CM60:40 and CM25:75 of the Apae#2 and fractions CM60:40 and CM25:75 of the Apae#1. Staining with another p-Fs-reactive MAb (ES-15) was also consistent with p-Fs in both Apae samples (Figure 2F). Staining with an x2-reactive MAb found only trace activity from x2 in fraction CM65:35 of Apae#2, although it was clearly present in all other later eluting fractions of both Apae samples (Figure 2E).
The nonstaining of Apae total glycolipids and concentrated fractions with broadly reactive anti-A reagents (Figure 1D, supplemental Figure 1) was indicative of the absence of A glycolipids. The bands seen with anti-A (A003, Figure 1E) in Apae#1 were not because of either Fs or A and remain unresolved yet consistent with the weak hemagglutination results observed with A003. Staining with 7 other anti-A MAbs (supplemental Table 1, supplemental Figure 1) was unable to detect any bands in the Apae samples. These results strongly suggest that the aberrant bands seen with some anti-A reagents are not because of A antigens. Finally, staining with Lewis MAbs established that both individuals are Le(b+) (Figure 1F), and, therefore, if blood group A they would be expected to have A-7-1 (ALeb) glycolipids,26,27 which were not found.
Overall both Apae samples could be demonstrated to have staining consistent with the presence of Fs, p-Fs, x2 but absence of A structures.
Structural analysis.
The LC-ESI/MS-MS and 1H-NMR results of both Apae samples were essentially the same. Only Apae#2 results are shown as it had less globoside content (Figure 2A). Released oligosaccharides from fraction CM65:35 were analyzed by LC-ESI/MS-MS.18 Despite the identical molecular masses of p-Fs and Fs (m/z 909 corresponding to an oligosaccharide with 2 HexNAc and 3 Hex) the reference pentasaccharides of Fs eluted as 2 peaks at 10.1-10.4 minutes retention time (Figure 3Aiii), whereas those of p-Fs eluted at 9.8-10.2 minutes (Figure 3Aiv). These elution time differences were used for differentiating Fs from p-Fs by LC-ESI/MS-MS. Fraction CM65:35 of both Apae samples had m/z 909 peaks at 10.0 and 10.4 minutes, corresponding with Fs (eg Apae#2, Figure 3Aii), whereas no peaks at 9.8-10.2 minutes were present. The MS-MS spectrum of the ion at m/z 909 (Figure 3Av) produced a series of C-type fragment ions (C2 at m/z 423, C3 at m/z 585, and C4 at m/z 747) consistent with a pentasaccharide with HexNAc-HexNAc-Hex-Hex-Hex sequence (Figure 3B). The fragmentation pattern obtained was identical to both reference Fs/p-Fs pentaglycosylceramides (Figure 3Avii-viii) but could be differentiated on the basis of retention time (Figure 3Aiii-iv). In addition, the Apae#2 CM65:35 fraction had a m/z 909 oligosaccharide eluting at 14.0-14.2 minutes (Figure 3Aii), with MS-MS spectrum (Figure 3Avi), consistent with a HexNAc-Hex-HexNAc-Hex-Hex sequence as in x2 (Figure 3B). Dominant ions in the base sample could be assigned to globoside (P/GbO4), neolactotetraosylceramide (nLc4), and the H-5-2 pentaglycosylceramide, respectively (Figure 3Ai).
Structural analysis of Apae glycolipids. Structural signatures indicative for the identification of Fs are indicated in red font. (A) LC-ESI/MS and MS-MS analysis of the oligosaccharides derived from fraction CM65:35 of Apae#2 glycolipids (#2) and Fs and p-Fs references. (Ai) Base peak chromatogram from LC-ESI/MS, representing the main oligosaccharides in fraction CM65:35 of Apae#2. (Aii) Mass chromatogram of oligosaccharides with m/z 909 (indicative for Fs, p-Fs, and x2) in the same fraction as in panel Ai shows peaks at retention time 10.1 and10.4, corresponding to Fs. (Aiii) Reference Fs m/z 909 mass chromatogram, showing the diagnostic retention time at 10.1 and10.4. (Aiv) Reference p-Fs m/z 909 mass chromatogram with retention time 9.8 and 10.2. (Av) MS-MS spectrum of the ion at m/z 909; see (Aii) retention time 10.4 minutes. (Avi) MS-MS spectrum of m/z 909 (retention time 14.0 minutes) indicating a x2 oligosaccharide. (Avii) Reference Fs MS-MS spectrum of m/z 909 (retention time 10.4 minutes). (Aviii) Reference p-Fs MS-MS spectrum of m/z 909 (retention time 9.8 minutes). (B) Interpretation MS-MS fragmentation formulas. (C) Low-field part of the 600-MHz proton NMR spectrum shown on top of the DQF-COSY spectrum, corresponding to the H1/H2 connectivities of the signature resonances for the structures present in the Apae#2 CM65:35 fraction. The anomeric resonances from the 2 terminal residues of the dominating globoside structure (GbO4) have been truncated at a height corresponding to approximately 20% of the total amplitude. In the COSY spectrum, connectivities belonging to expected structures (nLc4, P1, H-5-2, GbO5) have been outlined by black ellipses. H1/H2 connectivities of the 3 terminal residues (GalNAcα3GalNAcβ3Galα4−) of Fs are indicated with red ellipses. Also shown by blue circles is the absence in this fraction of the 3 terminal residues of p-Fs (GalNAcβ3GalNAcβ3Galα4−). (D) Reference NMR spectra of Fs and p-Fs glycolipids isolated from chicken and human erythrocytes, respectively.
Structural analysis of Apae glycolipids. Structural signatures indicative for the identification of Fs are indicated in red font. (A) LC-ESI/MS and MS-MS analysis of the oligosaccharides derived from fraction CM65:35 of Apae#2 glycolipids (#2) and Fs and p-Fs references. (Ai) Base peak chromatogram from LC-ESI/MS, representing the main oligosaccharides in fraction CM65:35 of Apae#2. (Aii) Mass chromatogram of oligosaccharides with m/z 909 (indicative for Fs, p-Fs, and x2) in the same fraction as in panel Ai shows peaks at retention time 10.1 and10.4, corresponding to Fs. (Aiii) Reference Fs m/z 909 mass chromatogram, showing the diagnostic retention time at 10.1 and10.4. (Aiv) Reference p-Fs m/z 909 mass chromatogram with retention time 9.8 and 10.2. (Av) MS-MS spectrum of the ion at m/z 909; see (Aii) retention time 10.4 minutes. (Avi) MS-MS spectrum of m/z 909 (retention time 14.0 minutes) indicating a x2 oligosaccharide. (Avii) Reference Fs MS-MS spectrum of m/z 909 (retention time 10.4 minutes). (Aviii) Reference p-Fs MS-MS spectrum of m/z 909 (retention time 9.8 minutes). (B) Interpretation MS-MS fragmentation formulas. (C) Low-field part of the 600-MHz proton NMR spectrum shown on top of the DQF-COSY spectrum, corresponding to the H1/H2 connectivities of the signature resonances for the structures present in the Apae#2 CM65:35 fraction. The anomeric resonances from the 2 terminal residues of the dominating globoside structure (GbO4) have been truncated at a height corresponding to approximately 20% of the total amplitude. In the COSY spectrum, connectivities belonging to expected structures (nLc4, P1, H-5-2, GbO5) have been outlined by black ellipses. H1/H2 connectivities of the 3 terminal residues (GalNAcα3GalNAcβ3Galα4−) of Fs are indicated with red ellipses. Also shown by blue circles is the absence in this fraction of the 3 terminal residues of p-Fs (GalNAcβ3GalNAcβ3Galα4−). (D) Reference NMR spectra of Fs and p-Fs glycolipids isolated from chicken and human erythrocytes, respectively.
The 1D-1H-NMR spectra (results not shown) were complex and 2D-DQF-COSY spectra was required to determine the presence of Fs and differentiate it from p-Fs pentaglycosylceramides. The most abundant glycolipids present in Apae#2 fraction CM65:35 (Figure 3C) were GbO4, followed by nLc4, H-5-2, P1, and GbO5. The COSY spectrum (Figure 3C) identified Fs pentaglycosylceramide (as indicated by the H1/H2 connectivities outlined by red ellipses in Figure 3C) as a minor glycolipid. With respect to connectivities and reference glycolipids, identification of Fs was unambiguous. Equally clear in this fraction was the absence of p-Fs pentaglycosylceramide (essentially excluded by fractionation) and indicated by blue circles in Figure 3C. The same structures were also identified in fraction CM65:35 of Apae#1. Importantly, there was no evidence for any structures carrying blood group A determinants. Sugar sequences and NMR resonances are given in supplemental Table 3.
Extended serologic analysis
Positive reactions were obtained by flow cytometry using MAb anti-Fs against ovine, canine and Apae-RBCs, whereas other human RBCs were negative (Figure 4A). Approximately 6% of 278 random blood donor plasmas agglutinated Apae-RBCs by the indirect antiglobulin test (anti-IgG/-C3d) and ∼ 23% caused direct IgM agglutination. Selected plasmas reacting strongly with Apae-RBCs were further investigated (Figure 4B). By adsorption of one such plasma onto Apae#2-RBCs and testing the eluted antibodies against various RBCs, reactivity against both Apae and nonprimate mammal RBCs but not other ABO groups was noted (Figure 4C-D), thus strongly suggesting that the antibody eluted is not crossreactive anti-A but indeed anti-Fs.
Flow cytometric and serologic testing of the 2 Apae individuals. The y-axis displays the number of cells on a linear scale whereas the x-axis shows the fluorescence intensity on a logarithmic scale. Flow cytometry line color codes: Sheep RBCs (green), dog RBCs (purple), Apae #1 (red), Apae #2 (blue) group A1 (light blue), and group O RBCs (black). (A) MAb anti-Fs with goat anti-rat IgM labeled with phycoerythrin as secondary antibody. Sheep RBCs were used as a strong positive control and human RBCs of various ABO groups as negative controls (only A1 and O are shown for clarity). (B) Reactivity of plasma antibodies from a group B donor against Apae RBCs and visualized with rabbit anti-human IgM labeled with FITC as secondary antibody. Group O RBCs were used as a negative control. Similar reactions could be obtained with O plasma (not shown). Because of the expected, strong agglutination of group A RBCs, that is the positive control, it was not possible to run the A RBCs on the flow cytometer hence they are not included in the histogram. (C) Reactivity of an eluate prepared from group B plasma antibodies adsorbed to and eluted from Apae#2 RBCs and visualized with rabbit anti-human IgM FITC secondary antibody. Group A1 and O RBCs were included as negative controls. (D) Gel column hemagglutination test between the eluate [prepared from Apae#2 cells, also used in (C)] against RBCs of various phenotypes. Negative reactions were observed with the common human blood groups (A1, A2 and O), whereas positive reactions were seen with RBCs from the Apae#1 individual and both dog and sheep RBCs known to express Fs antigen. (E-F) The hemolysin test was performed with 2 strongly Apae-reactive sera. Lysis patterns for group O and B serum are displayed in panels E and F, respectively. Results for RBCs of Apae and 3 common ABO phenotypes (A1, B, and O) are shown. ABO-compatible and ABO-incompatible combinations serve as negative and positive controls, respectively. Incubations were performed with both native (gray) and papain-treated (black) RBCs. Diagrams show hemoglobin levels (g/L) in all the tubes tested, whereas the photos show the results with papainised cells.
Flow cytometric and serologic testing of the 2 Apae individuals. The y-axis displays the number of cells on a linear scale whereas the x-axis shows the fluorescence intensity on a logarithmic scale. Flow cytometry line color codes: Sheep RBCs (green), dog RBCs (purple), Apae #1 (red), Apae #2 (blue) group A1 (light blue), and group O RBCs (black). (A) MAb anti-Fs with goat anti-rat IgM labeled with phycoerythrin as secondary antibody. Sheep RBCs were used as a strong positive control and human RBCs of various ABO groups as negative controls (only A1 and O are shown for clarity). (B) Reactivity of plasma antibodies from a group B donor against Apae RBCs and visualized with rabbit anti-human IgM labeled with FITC as secondary antibody. Group O RBCs were used as a negative control. Similar reactions could be obtained with O plasma (not shown). Because of the expected, strong agglutination of group A RBCs, that is the positive control, it was not possible to run the A RBCs on the flow cytometer hence they are not included in the histogram. (C) Reactivity of an eluate prepared from group B plasma antibodies adsorbed to and eluted from Apae#2 RBCs and visualized with rabbit anti-human IgM FITC secondary antibody. Group A1 and O RBCs were included as negative controls. (D) Gel column hemagglutination test between the eluate [prepared from Apae#2 cells, also used in (C)] against RBCs of various phenotypes. Negative reactions were observed with the common human blood groups (A1, A2 and O), whereas positive reactions were seen with RBCs from the Apae#1 individual and both dog and sheep RBCs known to express Fs antigen. (E-F) The hemolysin test was performed with 2 strongly Apae-reactive sera. Lysis patterns for group O and B serum are displayed in panels E and F, respectively. Results for RBCs of Apae and 3 common ABO phenotypes (A1, B, and O) are shown. ABO-compatible and ABO-incompatible combinations serve as negative and positive controls, respectively. Incubations were performed with both native (gray) and papain-treated (black) RBCs. Diagrams show hemoglobin levels (g/L) in all the tubes tested, whereas the photos show the results with papainised cells.
Freshly drawn group B and O sera from normal (non-Apae) donors were incubated with native and papainised Apae and control RBCs. Hemolysis was observed with Apae and ABO-incompatible RBCs (Figure 4E-F) and quantified. These in vitro results indicate that anti-Fs can activate complement and may have the potential to cause intravascular lysis of Fs-positive RBCs.
Genetic analysis
The sequence of GBGT1.
Based on the finding that Apae RBCs carry Fs antigens, exons 1-7 of the GBGT1 gene were sequenced in 11 individuals because this supposed human pseudogene is homologous to mammal Fs synthase genes.28 No deviations from the GenBank consensus sequence were detected in 9 non-Apae individuals, whereas in both Apae#1 and Apae#2, heterozygosity for a previously unreported single nucleotide polymorphism (SNP), 887G > A, that results in Arg296Gln was identified (supplemental Figure 2). In addition, Apae#2 was homozygous for a previously noted SNP, 58C > T (Leu20Phe), which shows that Apae has arisen from heterozygosity for different alleles in the 2 families, with or without 58C > T but including 887G > A. Apae#1 also had another SNP in exon 7, 363C > A, predicting a premature stop codon following residue 120 (supplemental Figure 2). Cloning and allele-specific amplification with subsequent sequencing verified that 363C > A and 887G > A are located on different alleles.
Nine non-Apae individuals with different ABO phenotypes conformed to consensus, although 58C > T and 363C > A were detected in 5 and 1, respectively, of the 18 examined alleles. The prevalences of GBGT1-SNPs are given in supplemental Table 4.
Exon 7 of GBGT1 in 6 different non-human primates was also sequenced, verifying that all had the human consensus 887G, as opposed to non-primates.15,28,29
PCR-ASP assays were designed to screen saliva DNA from members of the 2 Apae families, and random blood donors for 363C > A and 887G > A (supplemental Figure 2, supplemental Table 4). The Apae versus non-Apae phenotypes defined in the 1987 paper13 were concordant with the presence of 887G > A (supplemental Figures 3-4).
Transcript analysis.
It is unknown whether Fs synthase is expressed during hematopoiesis, especially in erythroid cells, although GBGT1-mRNA has been detected in other tissues.15
We measured transcript levels in buffy coats from 4 random blood samples of different ABO groups (A1/A2/B/O) and the 2 Apae blood samples. Transcripts were readily detected in mRNA preparations from all samples but no apparent quantitative differences were noted, suggesting that lack of Fs is not because of absence of mRNA. Transcript levels were ∼ 4 × higher in erythroid cultured cells from bone marrow (supplemental Figure 5).
Transfection of MEG-01.
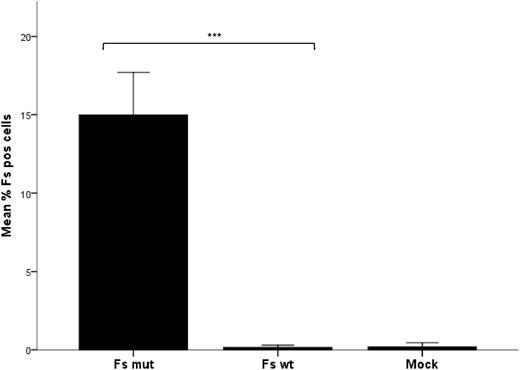
Based on the hypothesis that 887G > A in GBGT1 induces Fs synthesis in Apae individuals, a Fs-negative megakaryoblastic cell line, MEG-01, was cotransfected with the genes encoding P synthase (B3GALNT1) and 2 allelic forms of Fs synthase (GBGT1). Mutant (887A) but not wild-type (887G) GBGT1 resulted in a statistically significant increase in Fs-positive cells (Figure 5). This indicates that 887G > A (Arg296Gln) changes the inactive human Fs synthase to become enzymatically active.
Expression of Fs antigen in transfected MEG-01 measured by flow cytometry. The bar graph shows the mean percentage of Fs-positive cells after 3 independent transfection experiments, each performed in triplicate with the open reading frame of B3GALNT1 and either GBGT1 887A (Fs mut) or 887G (Fs WT). As a negative (background) control included in all 3 experiments, cells were transfected with the empty vector without insert (mock) and run in duplicate. The mean percentage of Fs-positive cells among the viable transfected (GFP-positive) cells is shown. Non-viable cells were disqualified by exclusion gating of cells positive for 7AAD. Error bars depict the standard error of the mean (SEM). The difference in Fs antigen expression after transfection was calculated to be statistically significant for the Fs mutant compared with the Fs wild-type cells and mock-treated cells according to the independent Kruskal-Wallis test. The significance level for the difference between wild-type and mutant is shown by the asterisks above the bars. Data were considered statistically significant with respect to the following criteria: *P < .05, **P < .01, and ***P < .001.
Expression of Fs antigen in transfected MEG-01 measured by flow cytometry. The bar graph shows the mean percentage of Fs-positive cells after 3 independent transfection experiments, each performed in triplicate with the open reading frame of B3GALNT1 and either GBGT1 887A (Fs mut) or 887G (Fs WT). As a negative (background) control included in all 3 experiments, cells were transfected with the empty vector without insert (mock) and run in duplicate. The mean percentage of Fs-positive cells among the viable transfected (GFP-positive) cells is shown. Non-viable cells were disqualified by exclusion gating of cells positive for 7AAD. Error bars depict the standard error of the mean (SEM). The difference in Fs antigen expression after transfection was calculated to be statistically significant for the Fs mutant compared with the Fs wild-type cells and mock-treated cells according to the independent Kruskal-Wallis test. The significance level for the difference between wild-type and mutant is shown by the asterisks above the bars. Data were considered statistically significant with respect to the following criteria: *P < .05, **P < .01, and ***P < .001.
Human Fs synthase model
The close homology (45% amino-acid identity) between ABO and GBGT1 permitted creation of a 3-dimensional model of the Fs-synthesizing enzyme based on a crystal structure of ABO transferase.30 The modeled structure of the putative human Fs synthase was generated by threading onto a human group B glycosyltransferase (GTB) variant (PDB entry 2RJ7). The model also shows an overall similarity to the human group A glycosyltransferase (GTA) structure (PDB entry 1LZI), a 3-α-N-acetylgalactosaminyltransferase similar to Fs synthase (Figure 6A). Moreover, the catalytically important His301 in GTA maps to the same position as Arg296 in Fs synthase. UDP-Galactose (Gal) co-crystallized with the GTB structure can be easily superimposed into the binding pocket of modeled Fs synthase. In sharp contrast to His301 in GTA, none of the preferred side chain rotamers of Arg296 in the Fs synthase model seems well suited to create the critical hydrogen bond to the O6 of the donor-sugar moiety. Furthermore, assuming that UDP-N-acetylgalactosamine (GalNAc) binds in a similar way to Fs synthase as UDP-Gal binds to GTB, the GalNAc can be easily accommodated in the binding pocket (Figure 6B). One reason for this is that the corresponding position of Met-266 in GTB (important for UDP-Gal specificity by sterically preventing binding of UDP-GalNAc)30 is glycine in Fs synthase (Gly261), which leaves ample space for GalNAc. The Apae-associated Gln296 mutant is more probable to form a hydrogen bond with O6 of the donor substrate (Figure 6C), which could explain why the Gln296 enzyme gets activated.
Superimposition of a model human Fs synthase threaded onto human ABO glycosyltransferase. This GTB (a GTB variant, PDB entry 2RJ7) contains UDP-Gal and an antigen acceptor derivative in the binding site. Furthermore, this enzyme structure has adopted a closed conformation where an internal loop (residue 180-200) and the C-terminus (residue 345-354) have folded over the active site. The threading and minimization was done using Moe and resulted in a model highly similar to the GTB structure. (A) The GTA structure (orange; PDB entry 1LZI) has been superimposed onto the human Fs synthase model (green), although this GTA structure has a disordered internal loop and C-terminus. UDP-Gal (gray) from the GTB mutant (PDB entry 2RJ7) was also superimposed onto the figure to highlight the probable location of the donor substrate. The catalytically important His301 in GTA/GTB is shown as sticks and potentially forms a hydrogen bond to the O6 position on the Gal of the UDP-Gal (yellow dashed line). In the human Fs synthase model the Arg296, which corresponds to and overlaps His301 in ABO transferase, is shown as sticks and has adopted a rotamer side chain conformation away from the Gal. In almost any other conformation, the Arg296 side chain would be clashing with either nearby residues in the enzyme itself or with the donor. The H-antigen acceptor from the GTB structure is shown as black sticks and the manganese ion as a purple sphere. (B) Close-up of the active site from panel A. A GalNAc molecule from the HIC-Up database (yellow) is superimposed onto the Gal without introducing any clashes to the molecule. Assuming the UDP-GalNAc binds in a similar way to the Fs synthase as UDP-Gal does to GTB, the acetamido group can be easily accommodated. One reason for this is that at the corresponding position of Met-266 in GTB (known to be important for UDP-Gal specificity by sterically preventing binding of UDP-GalNAc) there is a glycine in Fs synthase which leaves ample space for the acetamido group. Color scheme as described. (C) The threaded structure of the modeled human Fs synthase. Close-up of the active site showing the position of the Arg296 (green) versus the Gln296 mutant (light-brown) as it was modeled in Moe with the residue replacement introduced. It is evident that residue Gln296 in the mutant is more probable to form a hydrogen bond with O6 of the Gal which could explain why the enzyme goes from inactive to active when the mutant is introduced.
Superimposition of a model human Fs synthase threaded onto human ABO glycosyltransferase. This GTB (a GTB variant, PDB entry 2RJ7) contains UDP-Gal and an antigen acceptor derivative in the binding site. Furthermore, this enzyme structure has adopted a closed conformation where an internal loop (residue 180-200) and the C-terminus (residue 345-354) have folded over the active site. The threading and minimization was done using Moe and resulted in a model highly similar to the GTB structure. (A) The GTA structure (orange; PDB entry 1LZI) has been superimposed onto the human Fs synthase model (green), although this GTA structure has a disordered internal loop and C-terminus. UDP-Gal (gray) from the GTB mutant (PDB entry 2RJ7) was also superimposed onto the figure to highlight the probable location of the donor substrate. The catalytically important His301 in GTA/GTB is shown as sticks and potentially forms a hydrogen bond to the O6 position on the Gal of the UDP-Gal (yellow dashed line). In the human Fs synthase model the Arg296, which corresponds to and overlaps His301 in ABO transferase, is shown as sticks and has adopted a rotamer side chain conformation away from the Gal. In almost any other conformation, the Arg296 side chain would be clashing with either nearby residues in the enzyme itself or with the donor. The H-antigen acceptor from the GTB structure is shown as black sticks and the manganese ion as a purple sphere. (B) Close-up of the active site from panel A. A GalNAc molecule from the HIC-Up database (yellow) is superimposed onto the Gal without introducing any clashes to the molecule. Assuming the UDP-GalNAc binds in a similar way to the Fs synthase as UDP-Gal does to GTB, the acetamido group can be easily accommodated. One reason for this is that at the corresponding position of Met-266 in GTB (known to be important for UDP-Gal specificity by sterically preventing binding of UDP-GalNAc) there is a glycine in Fs synthase which leaves ample space for the acetamido group. Color scheme as described. (C) The threaded structure of the modeled human Fs synthase. Close-up of the active site showing the position of the Arg296 (green) versus the Gln296 mutant (light-brown) as it was modeled in Moe with the residue replacement introduced. It is evident that residue Gln296 in the mutant is more probable to form a hydrogen bond with O6 of the Gal which could explain why the enzyme goes from inactive to active when the mutant is introduced.
E coli hemagglutination
P-fimbriae are well-known virulence factors of uropathogenic E coli.31 The papG adhesin recognizes the terminal Galα4Gal moiety of Pk and P1 blood group glycolipids but also the internal Galα4Gal of P (globoside) antigen. The prsG adhesin recognizes Galα4Gal-containing glycolipids less well but preferentially binds to the Fs structure terminating in GalNAcα3GalNAc.32 The papG adhesin readily agglutinates all common human RBCs, although the prsG adhesin may crossreact weakly with A1 (but not A2/B/O) RBCs but binds strongly to sheep and dog RBCs which are Fs-positive. We tested the ability of E coli strain HB101, unmodified or expressing either the papG (HB101.AD110) or prsG (HB101.JFK102) adhesin to agglutinate RBCs. Table 1 shows that neither the parent (HB101) nor the prsG-expressing (HB101.JFK102) strain agglutinates A1/A2/O RBCs under these conditions, whereas the papG-expressing (HB101.AD110) strain agglutinates all tested RBCs as expected. Only Apae-RBCs, sheep RBCs, and human group O RBCs containing synthetic FSL-Forssman constructs (Fs kodecytes), but not FSL-B (B kodecytes), were readily agglutinated by prsG-expressing (HB101.JFK102) bacteria.
Hemagglutinating ability of E coli displaying different fimbriae
| Strain . | Reactions against RBCs . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| A1 . | A2 . | O . | O . | Apae#1 . | Apae#2 . | Sheep . | Fs kodecyte* . | B kodecyte* . | |
| HB101† | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HB101.AD110‡ | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ | ++ |
| HB101.JFK102§ | 0 | 0 | 0 | 0 | ++ | ++ | +++ | ++ | 0 |
| Strain . | Reactions against RBCs . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| A1 . | A2 . | O . | O . | Apae#1 . | Apae#2 . | Sheep . | Fs kodecyte* . | B kodecyte* . | |
| HB101† | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HB101.AD110‡ | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ | ++ |
| HB101.JFK102§ | 0 | 0 | 0 | 0 | ++ | ++ | +++ | ++ | 0 |
Grading according to standard blood bank practice, where ++++ is the strongest, +++, ++, and + are gradually weaker but positive reactions, and 0 is negative.
Group O human RBCs modified with synthetic Fs and B FSL glycolipid-like KODE constructs.
Parental strain.
Expressing papG adhesin.
Expressing prsG adhesin.
Discussion
We report biochemical, serologic, and genetic data that show conclusively for the first time the expression of Fs antigen on human RBCs. Fs is better known for its expression on animal RBCs,33 and has been implicated as a target for a variety of microorganisms and toxins.34-37 The discovery that humans can also express Fs on their RBCs was a consequence of the immunochemical evaluation of glycolipids from individuals with the so-called ABO subgroup, Apae,13 the terminology of which now needs to be revised.
Before 2012, there were 30 blood-group systems.38 Six of these are carbohydrate-based and their genes code for glycosyltransferases. To define a new blood-group antigen, the International Society of Blood Transfusion (ISBT) requires that it be shown independent of all other blood-group antigens, be expressed on RBCs, and is inheritable. Furthermore, at least 1 individual with the corresponding antibody because of lack of the antigen, must be known (in this case naturally occurring anti-Fs is already known to exist).37,39,40 We have proven the unambiguous existence of structurally defined Fs glycolipids on the surface of RBCs from 2 unrelated Apae individuals and have excluded Fs in RBCs from others. Related and potentially crossreactive glycolipids, such as p-Fs, x2 and blood-group A antigens, were excluded as causative. In addition to the phenotypic characterization, we linked this phenotype to a SNP in GBGT1, the human Fs synthase gene. The inheritance of Arg296Gln was associated with the Fs-positive phenotype in Apae families. As our results satisfy the ISBT criteria, we proposed to designate this new system FORS, in honor of Prof John Forssman who a century ago discovered antibodies to this heterophile antigen by injecting extracts from guinea pig tissues into rabbits whose immune sera hemolyzed sheep RBCs.6 In July 2012, Fs was recognized as a blood-group antigen by ISBT in the newly formed 31st blood-group system given the name FORS.
Previous studies have deemed the Fs synthase inactive because of mutations in its catalytic domain encoded by exon 7 of GBGT1.15 The human protein differs from the canine enzyme by 58 amino-acids but can be reactivated by adding the canine C-terminus to the human N-terminus.15 The consensus non-primate sequence corresponding to human residue 296 is Gln, whereas all primate and human (excluding Apae) sequences code for Arg. GBGT1 has been considered a human pseudogene, together with other homologous genes except ABO in the GT6 superfamily.28 Interestingly, a recent study noted that GBGT1 has not decayed as much as other related pseudogenes, and was therefore predicted to have retained some unknown function.29 Although speculative, the apparently inactive human consensus form of GBGT1-encoded enzyme (with Arg296) may have as yet unrecognized enzymatic activity. In contrast, the Gln296 variant described here has become effective at synthesizing Fs, in analogy with the enzyme in many non-primate mammals. The Apae-associated substitution was not found in 256 blood donors but its frequency in other populations around the world remains to be established. In addition to the 3 originally reported families (2 of which were studied here and surprisingly found to have 2 different 887G > A-containing alleles), we noted another 887G > A-positive individual (phenotype unknown) in a database (supplemental Table 4).
To establish if Arg296Gln directly causes Fs synthesis, we expressed the wild-type and mutant forms in a hematopoietic cell line. After optimization of the experiment, including cotransfection of B3GALNT1 (P synthase) to increase the number of precursors/acceptors for Fs synthase, a striking increase in Fs-positive cells (from background level) was noted with the Apae construct. A possible mechanism behind the reactivation of the enzyme was suggested by the Fs synthase model based on the crystal structure of the homologous GTA/GTB.30
GBGT1 is transcribed in several human tissues but does not appear to result in Fs antigen expression.15 However, Fs antigen may be expressed in carcinomas41,42 and has been suggested to constitute the so-called incompatible A antigen found in non–group-A individuals with cancer.42 We demonstrated here for the first time that GBGT1-mRNA is readily detectable in human blood and erythroid bone marrow cultures at levels similar to, if not higher than, ABO-mRNA.43 This indicates that Fs synthase is probably expressed in erythroid progenitors and thus able to make Fs on RBCs if mutated Arg296Gln.
The carbohydrate histo-blood group systems in man are ancient and the biologic pressures that created and maintained them are still largely unknown.44 The generally accepted belief is that these polymorphisms arose as a consequence of selective pressure exerted by microorganisms.8,45 We investigated the possible biologic consequences of Fs expression on human cells and found that uropathogenic E coli containing a PrsG-encoding plasmid bound only to Apae (and animal) cells. Thus, by acquiring an animal-associated GBGT1 polymorphism, Apae individuals express an antigen which protects against shiga toxin 1 (Stx1)34 but risks making them more susceptible to Stx2e, a common variant of Stx2 in porcine feces, recently reported to bind Fs.37 E coli strains capable of infecting the urinary tracts of canine and other Fs-expressing mammals adhere to uroepithelial cells via Fs-binding PrsG adhesins.32,35 Expression of canine Fs synthase induces binding of canine E coli strains to human and monkey epithelial cell lines.15 As suggested by hemagglutination, Apae individuals could potentially be more prone to certain toxins and pathogens. However, it remains to be established whether Fs is also present in non-erythroid tissues and secretions of Apae individuals, where it may constitute a biologically more relevant target. From these observations, we predict that Fs glycolipids will be expressed outside the hematopoietic compartment when Fs synthase is activated by Arg296Gln, on the basis that other globoseries glycolipids, such as Pk and P (the precursor of Fs), are expressed in uroepithelial cells. Furthermore, GBGT1 is transcribed in most tissues investigated.15
This report provides an explanation for the enigmatic Apae phenotype. Because Apae is unrelated to the ABO system and its gene, the ISBT accepted that this old terminology should be abolished and replaced by FORS, a new system based on our findings. Accordingly, the (former) Apae phenotype was redefined as FORS1-antigen positive and independent of ABO group, that is, not comparable with weak A subgroups. We identified a missense SNP that according to transfection data offers a plausible mechanism of reactivation of Fs-synthase activity, and allows for genetic screening of the phenotype. Even if our hemolysin results are limited in scope and mainly show an effect on papainized RBCs, it can be questioned if FORS1-positive individuals should be accepted as blood/organ donors. It cannot be excluded that anti-Fs, such as anti-A or anti-P, can be hemolytic in certain donor-recipient combinations and thereby constitute a threat to patient safety. If FORS1-positive units were to be transfused, we recommend that they are not given after electronic (type-and-screen) crossmatch but require a negative reaction between patient plasma and donor RBCs. However, it should be emphasized that the clinical consequences of naturally occurring anti-Fs are still unknown and require further studies. Taken together, these findings provide the genetic and structural bases for the new histo-blood group system FORS with possible implications for both transfusion and transplantation medicine as well as trans-species microbial susceptibility.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Fs glycolipid was a kind gift from Prof Göran Larson, and NMR-verified by Dr Jonas Nilsson at the Department of Clinical Chemistry and Transfusion Medicine, The Sahlgrenska Academy, Gothenburg. Prof Michael Breimer, Department of Surgery, Institute of Clinical Sciences, and Prof Bo Samuelsson, Department of Clinical Chemistry and Transfusion Medicine, The Sahlgrenska Academy Gothenburg, are thanked for their support of this project. Dr Britt Thuresson, Department of Laboratory Medicine, Lund University, is thanked for advice on technical assistance with real-time PCR and helpful suggestions concerning cell culture and transfection. DNA samples from 6 primate species were a kind gift from Prof Ulf Arnason, Lund University, Sweden. Dr Maj-Lis Svensson and Prof Catharina Svanborg, Department of Laboratory Medicine, Lund University, are thanked for help with the selection of bacterial strains. Dr Magnus Jöud at the Department of Laboratory Medicine, Lund University, is thanked for help with database mining. Joyce Poole at the International Blood Group Reference Laboratory in Bristol, United Kingdom, is thanked for sending Apae samples for ABO genotyping more than a decade ago, thereby bringing them to our attention. Finally, the participation of the Apae donors and their families is gratefully acknowledged.
This work was supported by the Swedish Research Council/Medicine (grants 14251 to M.L.O., 12628 to S.T., and 11612 to Michael Breimer), Claes Högman SAGMAN stipendium (Baxter Medical AB) to L.S., governmental ALF research grants to Lund University Hospital and Labmedicin Skåne (M.L.O.), the Medical Faculty at Lund University (M.L.O.) and the Skåne County Council's Research and Development foundation, Sweden (M.L.O.), and the Swedish Cancer Foundation (S.T.). The use of the LTQ linear quadrupole ion trap mass spectrometer (obtained by a grant from the Swedish Research Council, grant 342-2004-4434 to Prof Gunnar Hansson, Gothenburg), and the Varian 600-MHz machine at the Swedish NMR Center, Hasselblad Laboratory, University of Gothenburg, are gratefully acknowledged.
Authorship
Contribution: A.H. and L.S. performed most experiments; R.S. provided and characterized samples; J.Å. and S.T. obtained and analyzed the structural data; J.R.S. designed and performed the bacterial binding experiments; R.J. made the molecular modeling experiments and figures; L.S., A.H., S.M.H., L.R., and M.L.O. designed the study and interpreted data; and L.S., A.H., S.M.H., and M.L.O. wrote the paper and the remaining coauthors reviewed, revised, and approved the paper.
Conflict-of-interest disclosure: The authors declare no competing financial intersts.
Correspondence: Martin L. Olsson, Division of Hematology and Transfusion Medicine, Dept of Laboratory Medicine, Lund University, Lund, Sweden SE-22185; e-mail: Martin_L.Olsson@med.lu.se; or Stephen M. Henry, AUT University, Auckland, New Zealand; e-mail: kiwi@aut.ac.nz.
References
Author notes
L.S. and A.K.H. contributed equally to this paper.
S.M.H. and M.L.O. contributed equally to this paper.

![Figure 1. Thin layer chromatography staining of total Apae glycolipids. (A) Nomenclature and structures of selected glycolipids discussed in this paper. (B) Helix pomatia lectin staining of the Apae total glycolipid samples and blood group A1, A2, O, and B controls plus Fs glycolipid control. TLC migratory positions of known blood group glycolipids are identified in the margin. (C) Staining with MAb anti-Fs. (D) Staining with a representative broadly reactive MAb anti-A (clone 11H5). (E) Unidentified bands recognized by staining with MAb anti-A (003). (F) Staining with Lewis (anti-Lea, anti-Leb) and secretory A antigen glycolipid profiles (anti-A type 1, anti-ALeb). In all panels A1, A2, O, and B control samples are total erythrocyte glycolipids whereas Fs is a purified glycolipid fraction. Lewis phenotypes are expressed as red cell phenotypes [where low level Lea glycolipid is expected in the Le(a−b+) phenotype]. Because of minor migration differences between assays, direct comparison of absolute band positions should be viewed with caution. Black lines indicate where migration controls and unloaded lanes have been removed from the image. All lanes are from the same TLC plate and have not been repositioned.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/8/10.1182_blood-2012-10-455055/4/m_zh89991302060001.jpeg?Expires=1763510528&Signature=fLESZ0yAIOyV2jmW46pwMC7UEAMJBoqHcJiJz-1k7VJFPFNs4jBszVdS5CQgMF5GJQwLBLUaEtAzdLEbohv3wLzesx~3jER0LNq-ZdGWe1CxjCG3DKsM3UZYbs~1jcDFh7oEiB6Z-shu1L2zhRvoa4LCm0XbvB3X6c~Mxo8jzl~7pZtIT7fXV7gQ~Oh5CrafgWhP~IlKDRKvpm098-U-H4uoC4LwfC22Pz3e6oWjLyyHgSesrip-GtJDFG8bhq-jAm4jX87A3WyNNu0cHwblBcgn4oU6ZetFPUHV43uD6krw1Q36TFaybXfhbrenVkg13GCT0pJOjKys2N-OyWEFNw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 4. Flow cytometric and serologic testing of the 2 Apae individuals. The y-axis displays the number of cells on a linear scale whereas the x-axis shows the fluorescence intensity on a logarithmic scale. Flow cytometry line color codes: Sheep RBCs (green), dog RBCs (purple), Apae #1 (red), Apae #2 (blue) group A1 (light blue), and group O RBCs (black). (A) MAb anti-Fs with goat anti-rat IgM labeled with phycoerythrin as secondary antibody. Sheep RBCs were used as a strong positive control and human RBCs of various ABO groups as negative controls (only A1 and O are shown for clarity). (B) Reactivity of plasma antibodies from a group B donor against Apae RBCs and visualized with rabbit anti-human IgM labeled with FITC as secondary antibody. Group O RBCs were used as a negative control. Similar reactions could be obtained with O plasma (not shown). Because of the expected, strong agglutination of group A RBCs, that is the positive control, it was not possible to run the A RBCs on the flow cytometer hence they are not included in the histogram. (C) Reactivity of an eluate prepared from group B plasma antibodies adsorbed to and eluted from Apae#2 RBCs and visualized with rabbit anti-human IgM FITC secondary antibody. Group A1 and O RBCs were included as negative controls. (D) Gel column hemagglutination test between the eluate [prepared from Apae#2 cells, also used in (C)] against RBCs of various phenotypes. Negative reactions were observed with the common human blood groups (A1, A2 and O), whereas positive reactions were seen with RBCs from the Apae#1 individual and both dog and sheep RBCs known to express Fs antigen. (E-F) The hemolysin test was performed with 2 strongly Apae-reactive sera. Lysis patterns for group O and B serum are displayed in panels E and F, respectively. Results for RBCs of Apae and 3 common ABO phenotypes (A1, B, and O) are shown. ABO-compatible and ABO-incompatible combinations serve as negative and positive controls, respectively. Incubations were performed with both native (gray) and papain-treated (black) RBCs. Diagrams show hemoglobin levels (g/L) in all the tubes tested, whereas the photos show the results with papainised cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/8/10.1182_blood-2012-10-455055/4/m_zh89991302060004.jpeg?Expires=1763510528&Signature=ougOmVgq~WbKz0faeJfXUaWc0ndsAUbO6LnvsatEVKLTv56SoMBAex4GMnupaRKYv527~sfNvi17kU35slV8VFy7Q4Z-Re7whY4FwP117zgxh95rErQ-RcnUxB0bcNS3CUeOn1Q9ovNgbtiWIPjXA8Co6~8~QkJS09aVoF9oAb5M3L3Y6rbE1U8eqWwz66mZD5zIIdpq7kQsLgmBEtwfQbbtm6hyAEiLnyJ5nQ~fPFHXXQtMRBtIU7LgZnak0NwqpssCBxAnm7AB6wVpMP-HAlW2mrkX2ynQ0dc0UMYW2DKTNngj6X651ljuxl47~Vt4JHrZr-Kl8Pi5fC~tBOAcPw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal