Key Points
Analysis of coding genomes of FL tumor subpopulations reveals striking clonal diversity at diagnosis and progression.
Within a hierarchy of somatic evolution of FL coding genomes, many recurrent mutations are subclonal at diagnosis.
Abstract
Follicular lymphoma (FL) is currently incurable using conventional chemotherapy or immunotherapy regimes, compelling new strategies. Advances in high-throughput sequencing technologies that can reveal oncogenic pathways have stimulated interest in tailoring therapies toward actionable somatic mutations. However, for mutation-directed therapies to be most effective, the mutations must be uniformly present in evolved tumor cells as well as in the self-renewing tumor-cell precursors. Here, we show striking intratumoral clonal diversity within FL tumors in the representation of mutations in the majority of genes as revealed by whole exome sequencing of subpopulations. This diversity captures a clonal hierarchy, resolved using immunoglobulin somatic mutations and IGH-BCL2 translocations as a frame of reference and by comparing diagnosis and relapse tumor pairs, allowing us to distinguish early versus late genetic eventsduring lymphomagenesis. We provide evidence that IGH-BCL2 translocations and CREBBP mutations are early events, whereas MLL2 and TNFRSF14 mutations probably represent late events during disease evolution. These observations provide insight into which of the genetic lesions represent suitable candidates for targeted therapies.
Introduction
Follicular lymphoma (FL) is a common form of non-Hodgkin lymphoma (NHL) arising from mature B cells. FL tumor cells share identical immunoglobulin (Ig) gene rearrangements, indicating that the transforming founder mutation(s) occurs subsequent to VDJ recombination. These cells express markers of mature B-lineage including surface-Ig and CD19, and germinal center B-cell markers such as LMO2, CD10, and BCL6.1 The B-cell marker CD20, which is also the target of the anti-lymphoma therapy Rituximab,2,3 is expressed to a variable degree on FL tumors.4,5 CD20 levels can predict primary responsiveness of diverse lymphomas to Rituximab,6 and changes in CD20 expression contribute to Rituximab resistance and relapse.7,8 For instance, CD20 expression levels are associated with survival in patients with aggressive lymphomas treated with or without Rituximab.9 These studies each found CD20 to be variably expressed on cells within the same tumor, suggesting that this may be a marker of underlying clonal diversity.
The genetic hallmark of FL is the t(14;18)(q32;q21) translocation that places the antiapoptotic BCL2 oncogene under control of the Ig heavy-chain enhancer.10 This lesion is present in > 90% of FL cases,11 but is also detectable in the majority of older healthy adults suggesting that it is not sufficient to induce clinical disease.12 Recently, mutation of a histone methyltransferase gene, MLL2, was identified in 89% of FL cases, indicating that this may also be a founder mutation.13 Genes encoding other histone-modifying enzymes, such as CREBBP,14 are also mutated in this disease and they provide potentially attractive therapeutic targets.15 However, for gene mutation-directed therapy to eradicate disease, that mutation must be uniformly present in evolved tumor cells as well as all self-renewing tumor-cell precursors.16 To date, most mutations in NHL have been reported on a presence/absence basis with little attempt to evaluate their subclonal representation. We therefore aimed to investigate the degree of intratumoral genetic diversity within FL to evaluate the plausibility of gene mutation-directed therapies in this disease. We sorted tumor cell subpopulations based on their expression of CD20 and interrogated their mutational spectra using exome sequencing. This not only identified significant stratification of somatic mutations within tumor cell subpopulations within the same physical site, but also identified 2 contrasting patterns of disease relapse.
Methods
Patient samples
FL tumor specimens were acquired as part of the Stanford University Lymphoma Program Project and cryopreserved. We used 8 tumor samples acquired at diagnosis before any therapy, and 2 matching relapse samples acquired after combination chemotherapy without Rituximab (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Of these relapses, 1 was from a patient who responded well to therapy and was minimally treated before obtaining the relapse specimen (LPJ128), and the other from a patient who responded poorly to therapy and was heavily treated before obtaining the relapse specimen (LPJ041). All specimens were obtained with informed consent in accordance with the Declaration of Helsinki and this study was approved by Stanford University's Administrative Panels on Human Subjects in Medical Research.
Fluorescence activated cell sorting and DNA/RNA preparation
Cryopreserved samples were thawed, washed twice in PBS, and stained with anti-CD5(PECy7), anti-CD20(FITC), and either anti-IgM(APC), anti-CD10(APC) or anti-CD19(APC), depending on which marker combination was previously found in each individual case to show the least variability of expression on tumor B cells.4 All antibodies were from BD Bioscience. Samples were gated according to forward scatter and side scatter to identify lymphocytes and singlets. Tumor infiltrating CD5+ T cells were sorted and used for patient-matched normal DNA. Tumoral B cells were identified by their coexpression of CD20 and either IgM (if matching the tumor isotype), CD10 or CD19. Tumor cell subpopulations were sorted according to their CD20 expression: in all cases, the CD20hi subpopulation consisted of the top one-third of CD20-expressing tumor B cells, and the CD20int subpopulation consisted of the middle one-third of CD20-expressing tumor B cells. Fluorescence activated cell sorting was carried out on a BD FACS Aria II instrument (BD Bioscience). After sorting, cells were immediately pelleted and total RNA and DNA were extracted using the AllPrep system (QIAGEN) according to the manufacturer's instructions. For gene expression microarray analysis methodology (accession no. GSE38816), see supplemental Methods. Tumor purity was measured by quantitative assessment of t(14;18)(q32;q21)17 in 5 cases with major break region breakpoints (supplemental Methods). Using this approach, we found the low CD20-expressing cells to be enriched for nonmalignant B cells, and therefore excluded them from this study.
Exome sequencing and analysis
Standard shotgun-sequencing libraries were created according to Illumina protocol with the use of a highly accurate 4-plex indexing strategy targeting both ends of each amplicon allowing ultrasensitive detection of rare alleles.18 Exome libraries were created using SeqCap EZ Human Exome Library v2.0 targeting 36Mb of the human genome (Nimblegen). Libraries were normalized and combined (4-plex) before sequencing at a final concentration of 8pM. Paired-end sequencing (100 bp) was performed on a HiSeq 2000 sequencer with real time image analysis and base calling (Illumina), and de-indexed.18 Somatic nucleotide variants (SNVs) were called in tumor B cells with reference to patient-matched T cells using the Genome Analysis Toolkit (GATK)19 as previously described,20,21 with additional filtering criteria that allowed a false-negative rate < 4%. For detailed methodology, refer to supplemental Methods. Variants differing between subpopulations at 33% were classified as being discordantly represented. This threshold was empirically determined to allow 99% specificity for discordant representation, as determined by the representation of germ-line single nucleotide polymorphisms (SNPs) between intratumoral subpopulations (supplemental Figure 1). DNA copy number was called in B-cell populations with reference to matched T cells using VarScan 222 after normalization of total sample read depths per sample. Enrichment of mutated genes within functional categories was tested using the ToppGene suite.23
IgHV region analysis
Immunoglobulin heavy chain variable (IgHV) regions were amplified by PCR using an IGH gene clonality assay (Invivoscribe), according to the manufacturer's instructions. The resulting PCR product was cloned using a Zero Blunt PCR Cloning kit (Invitrogen), and Sanger sequenced using the M13 primer. Sequences are available through GenBank, accessions JX413134-JX413252 and JX413253-JX413349. IGH sequences were analyzed using IgBlast (http://www.ncbi.nlm.nih.gov/projects/igblast/), IMGT/V-Quest,24 and IgTree software25 to create immunoglobulin hierarchies.
Results
Sorting and whole-exome sequencing of tumor subpopulation from FL
Immunophenotyping of a large cohort of FL tumor specimens identified CD20 as a heterogeneously expressed cell-surface marker in FL4,5 (supplemental Figure 2). We therefore used it as a marker to separate tumor cell subpopulations from 8 diagnostic FL tumors. Using fluorescence activated cell sorting, we purified 2 subpopulations of tumor B cells based on their expression of CD20 (CD20int or CD20hi), as well as intratumoral T cells from FL tumor specimens of untreated patients (supplemental Figure 3). Cells with low expression of CD20 were found to be enriched for B cells lacking IGH-BCL2 translocation and the tumoral VDJ rearrangement and were therefore excluded from this study. We also sorted total tumor B cells from 2 matched relapse specimens. Using a quantitative real-time PCR (qPCR) assay for the IGH-BCL2 translocation, we found the average tumor purity to be 81.3% in the CD20-sorted subpopulations (supplemental Figure 4). Our exome-sequencing strategy enabled an average of 86.6% of each exome to be covered at ≥ 20 × depth (supplemental Figure 5), allowing us to confidently call somatic variants in each tumor exome with reference to patient-matched normal DNA. Assessment of germ-line single nucleotide polymorphisms between tumor subpopulations showed no subpopulation bias in allelic frequencies, suggesting that there was no significant PCR dropout or jackpot effects during library preparation, exome enrichment, or sequencing (supplemental Figure 1).
A dominance of subclones in FL inferred from somatic mutation allele frequencies
We sequenced whole-exomes from subpopulations of B cells and tumor infiltrating T cells from 8 diagnostic FL tumors. CD20int and CD20hi subpopulations were interrogated with reference to patient-matched normal cells (T cells). Transcriptome profiling data confirmed that T-cell fractions were of a high purity (supplemental Figure 6), allowing accurate calling of coding somatic nucleotide variants (cSNVs) in contrast to germ-line polymorphisms. We identified 877 cSNVs encoding missense and nonsense mutations in 569 unique genes within our 18 exomes (supplemental Table 2). These mutated genes were significantly enriched for those involved in chromosome and chromatin organization (FDR q < 0.001), and harboring bromodomains (FDR q = 0.0013). Of these, many were previously described in FL and/or diffuse large B-cell lymphoma (DLBCL),14,26-28 including mutation of CREBBP in 75% of our cases, MLL2 in 50% of cases (including 2 MLL2 mutations in 1 case), and TNFRSF14 (HVEM) and BCL2 in 25% of cases. We also identified novel recurrent somatic mutations in genes, such as ERBB2 (HER2), IKZF2 (HELIOS), and CD40, not previously described in any hematologic malignancy. Mutations in all aforementioned genes were validated by Sanger sequencing.
The distribution of mutation frequencies within individual cases showed patterns that were indicative of subclonal representation of the majority of cSNVs (Figure 1A). In 6 of 8 tumors, the majority of cSNVs show less than a 20% frequency, probably corresponding to their heterozygous representation in a fraction of tumor cells. Notably, only a minority of cSNVs resided within motifs for activation induced cytidine deaminase (AICDA; supplemental Table 2), and AICDA-mediated somatic mutations in immunoglobulin (Ig) genes showed a contrasting distribution of allelic frequencies than those in non-Ig genes (supplemental Figure 7), suggesting that this clonal diversity is not primarily driven by AICDA activity. To test whether non-tumor B cells may contribute a subset of these low-frequency mutations, we evaluated the frequency of these mutations compared with the estimated frequency of non-tumor B-cell clones. Using VDJ clonotyping of 2 cases with particularly high mutational burden, and which possessed several mutations with low mutation frequency, we found that no single non-tumor clonotype existed at high enough frequency to account for the observed variant fraction (supplemental Figure 8). This indicates that variants detected here are contributed by tumor cell subclones that make a variable proportion of the total tumor cell pool. Further, the low allelic frequencies for cSNVs are not simply an artifact of low tumor purity, as high tumor representation (> 80% on average) was confirmed by quantitative assessment of the IGH-BCL2 translocation. In line with these observations, correcting variant frequency for tumor purity within the 5 cases for which it was known resulted in the variant frequencies forming clusters indicative of their probable representation within the tumor cell pool (Figure 1B). Only 3.5% (14/401) of variants were present at allelic frequencies that may correspond to their homozygous representation in all tumor cells. Interestingly, this included 4/4 CREBBP mutations, and 1/4 MLL2 mutations within this cohort. Another 19.5% (78/401) of variants formed a cluster indicative of uniformly heterozygous representation within all tumor cells, including the remaining 3/4 MLL2 mutations within this cohort. However, the majority of mutations were present at allelic frequencies reflecting subclonal heterozygosity (74.1%, 297/401), with the remaining 3.0% (12/401) of variants inferred to be subclonally homozygous (Figure 1B). These distributions are not the result of allelic bias during amplification, as these patterns were not observed for germ-line polymorphisms (supplemental Figure 1). These results therefore suggest that the majority of cSNVs present within FL tumors are in minor tumor subclones.
Variant frequency distributions reflect pervasive subclonal representation of most somatic coding mutations in FL. (A) Kernel density plots (shaded gray) depict the distribution of somatic nucleotide variant frequencies in each of 8 patients. For each case, peaks can be seen corresponding to clusters of variants with different frequencies. For 6 of 8 cases, the major peak resides less than 20% frequency, indicating that the majority of mutations are present only in a minor subclone within the total tumor cell pool. Red vertical bars represent conservative estimates of tumor purity defined by IGH-BCL2 translocation qPCR assay (supplemental Figure 2). Vertical black bars in LPJ103 and LPJ108 represent homozygous CREBBP mutations that correlate with tumor purity. (B) Variant frequencies in 5 cases were corrected for tumor purity measured by IGH-BCL2 translocation and plotted against depth of sequencing for the given nucleotide. Two clear clusters can be seen, corresponding to mutations that are probably heterozygous in the entire tumor pool (uniformly heterozygous) or within a subclone (subclonally heterozygous). The remaining points probably represent mutations that are homozygous within the entire tumor pool (uniformly homozygous) or within a minor clone (subclonally homozygous). Notably, 3 of 4 MLL2 mutations (pink) fall within uniformly heterozygous cluster and the remaining mutation within the uniformly homozygous cluster. All 4 CREBBP mutations (blue) fall within the uniformly homozygous cluster.
Variant frequency distributions reflect pervasive subclonal representation of most somatic coding mutations in FL. (A) Kernel density plots (shaded gray) depict the distribution of somatic nucleotide variant frequencies in each of 8 patients. For each case, peaks can be seen corresponding to clusters of variants with different frequencies. For 6 of 8 cases, the major peak resides less than 20% frequency, indicating that the majority of mutations are present only in a minor subclone within the total tumor cell pool. Red vertical bars represent conservative estimates of tumor purity defined by IGH-BCL2 translocation qPCR assay (supplemental Figure 2). Vertical black bars in LPJ103 and LPJ108 represent homozygous CREBBP mutations that correlate with tumor purity. (B) Variant frequencies in 5 cases were corrected for tumor purity measured by IGH-BCL2 translocation and plotted against depth of sequencing for the given nucleotide. Two clear clusters can be seen, corresponding to mutations that are probably heterozygous in the entire tumor pool (uniformly heterozygous) or within a subclone (subclonally heterozygous). The remaining points probably represent mutations that are homozygous within the entire tumor pool (uniformly homozygous) or within a minor clone (subclonally homozygous). Notably, 3 of 4 MLL2 mutations (pink) fall within uniformly heterozygous cluster and the remaining mutation within the uniformly homozygous cluster. All 4 CREBBP mutations (blue) fall within the uniformly homozygous cluster.
Tumor cell subpopulations are genetically divergent
To further evaluate the subclonal representation of cSNVs within FL tumors, we compared the variant frequencies of CD20int and CD20hi tumor cell subpopulations. Consensus clustering of variant allelic frequency profiles showed that subpopulations from the same tumor were invariably more similar to each other than to subpopulations from other tumors (supplemental Figure 9). This is probably driven by the shared set of founder mutations that are unique to each tumor, and present in all tumor cells before clonal divergence. Nonetheless, only a minority of mutations (mean = 27.2%, range = 3.8%-63.6%) were present at comparable frequencies within each subpopulation (Figure 2). Interestingly, the magnitude of variant frequency discordance between CD20int and CD20hi subpopulations increased proportionally with the total number of mutations detected in each case (supplemental Figure 10). This suggests that tumors with either more active acquisition of mutations or longer evolutionary history have greater clonal diversity.
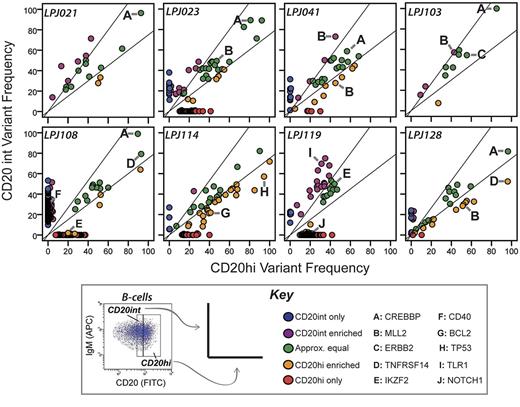
Intratumoral subpopulations are genetically divergent. Variant allele frequencies are compared between the CD20int (y-axis) and CD20hi (x-axis) intratumoral subpopulations. In most cases, only a minority of mutations are represented at approximately equal frequencies in each subpopulation (green dots). Many mutations comparably are enriched in either the CD20int (violet dots) or CD20hi (orange dots) subpopulation, or detected only in the CD20int (blue dots) or CD20hi (red dots) subpopulation. Mutations in selected genes of biologic interest are highlighted by letters A through J. Whereas CREBBP mutations (A) always appear in approximately equal frequencies in tumor cell subpopulations, MLL2 mutations are relatively enriched in 1 subpopulation in 3 of 4 tumors. Diagonal lines represent the threshold for 33% enrichment of variants in 1 subpopulation with reference to the alternate subpopulation (y = 1.33x and x = 1.33y), which allows 99% specificity for true subpopulation bias (supplemental Figure 1).
Intratumoral subpopulations are genetically divergent. Variant allele frequencies are compared between the CD20int (y-axis) and CD20hi (x-axis) intratumoral subpopulations. In most cases, only a minority of mutations are represented at approximately equal frequencies in each subpopulation (green dots). Many mutations comparably are enriched in either the CD20int (violet dots) or CD20hi (orange dots) subpopulation, or detected only in the CD20int (blue dots) or CD20hi (red dots) subpopulation. Mutations in selected genes of biologic interest are highlighted by letters A through J. Whereas CREBBP mutations (A) always appear in approximately equal frequencies in tumor cell subpopulations, MLL2 mutations are relatively enriched in 1 subpopulation in 3 of 4 tumors. Diagonal lines represent the threshold for 33% enrichment of variants in 1 subpopulation with reference to the alternate subpopulation (y = 1.33x and x = 1.33y), which allows 99% specificity for true subpopulation bias (supplemental Figure 1).
In all tumors, there were sets of mutations that had allele frequencies in one subpopulation that were > 1.33-fold higher than that in the alternate subpopulation (Figure 2). This included mutations in genes, such as TP53 and BCL2, and 4/5 MLL2 mutations. Interestingly, of the 2 MLL2 mutations identified in patient LPJ041, one is enriched in the CD20int subpopulation and the other enriched in the CD20hi subpopulation. In contrast, all mutations in CREBBP were consistently represented between tumor cell subpopulations. Increasing CREBBP mutation frequency was also associated with decreasing abundance of BCL6 target genes29 (supplemental Figure 11), suggesting that CREBBP mutations alter BCL6 activity in FL, as previously described in DLBCL.14
In addition to the biased allelic representation of many mutations relatively enriched within either the CD20int or CD20hi fractions, we identified sets of mutations exclusive to one subpopulation (ie, < 1% of reads of the alternate subpopulation). This observation was not biased by relative tumor purity between these subpopulations, because different sets of mutations were enriched in different directions within each tumor, and correcting for tumor purity did not alter this observation (supplemental Figure 12). In addition, we found no such subpopulation diversity in common FL DNA copy number abnormalities identified in this data (supplemental Figure 13), and differences in DNA copy number state at the site of cSNVs did not appear to be the cause of broad differences in cSNV frequency between intratumoral subpopulations (supplemental Figure 14). Together, these data provide evidence that variants within recurrently mutated genes are infrequently uniformly represented within the tumor cell population (Figure 3). The non-uniform representation of variants probably indicates that they are late events during FL disease genesis.
Subclonal representation of recurrent somatic mutations. Genes containing cSNVs in 2 or more diagnostic specimens within this study exhibit different patterns of representation between CD20int and CD20hi subpopulations. The top panel shows the recurrence frequency of mutations within each gene in our cohort. The bottom panel shows the fraction of those mutations that are represented approximately equally (≤ 33% deviation; green) between each subpopulation, those that show skewed representation (> 33% deviation; purple), and those that are identified exclusively in one subpopulation and not the other (gray). It can be seen that cSNVs in the majority of genes show skewed or unequal representation between tumor subpopulations, and CREBBP mutations are the only variants that are equally represented between subpopulations in all instances.
Subclonal representation of recurrent somatic mutations. Genes containing cSNVs in 2 or more diagnostic specimens within this study exhibit different patterns of representation between CD20int and CD20hi subpopulations. The top panel shows the recurrence frequency of mutations within each gene in our cohort. The bottom panel shows the fraction of those mutations that are represented approximately equally (≤ 33% deviation; green) between each subpopulation, those that show skewed representation (> 33% deviation; purple), and those that are identified exclusively in one subpopulation and not the other (gray). It can be seen that cSNVs in the majority of genes show skewed or unequal representation between tumor subpopulations, and CREBBP mutations are the only variants that are equally represented between subpopulations in all instances.
Contrasting patterns of disease relapse
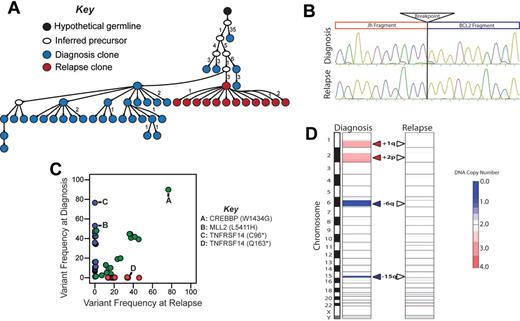
We next evaluated the relationship between clonal diversity at diagnosis to paired tumors at first disease relapse in 2 patients without clinical or histologic evidence of transformation to aggressive lymphoma. Importantly, the paired tumors shared identical Ig heavy-chain VDJ rearrangements and t(14;18)(q32;q21) breakpoints (Figures 4–5), demonstrating that they are clonally related. However, IgHV sequencing suggested different patterns of relapse of these cases. In LPJ041, 40% of clones from the diagnostic specimen shared an identical IgHV sequence (Figure 4A). This exact IgHV sequence represented 74% of clones from the relapse specimen, and was the clone most closely related to the predicted germ-line sequence within this specimen. In LPJ128, IgHV clones from the diagnostic and relapse specimens segregated to different branches of the hierarchy, with the majority of diagnostic and relapse clones being separated by 4 nucleotide substitutions and linked by an inferred precursor that was not detected (Figure 5A).
Relapse emanating from a progressed tumor clone in LPJ041. (A) Molecular cloning of productively rearranged Ig heavy-chain genes show a clonal relationship between the diagnosis and relapse specimen in LPJ041. Fifty-five unique clones were sequenced from the diagnostic specimen (blue), and 43 from the relapse specimen (red). Forty percent of the clones from the diagnostic specimen and 74% of the clones from the relapse specimen shared identical IgHV sequences (indicated by red/blue split). This was the closest sequence to the germ-line identified in the relapse specimen, indicating that the relapse emanated from an evolved tumor clone. Each arrowed line represents a single nucleotide substitution, with numbers representing additional substitutions. (B) Sanger sequencing showed identical t(14;18)(q32;q21) breakpoints in the diagnostic and relapse specimen for LPJ041. Four clones were sequenced from each of the diagnostic and relapse specimens and the breakpoint established by alignment of the sequences with the human genome consensus. (C) Comparison of variant frequencies in diagnostic and relapse specimens for LPJ041 showed the maintenance of the majority of mutations in the relapse that were detected within the diagnostic specimen (green), including 2 mutations in MLL2 that were variably represented in diagnostic subpopulations and a mutation in CREBBP that was uniformly represented at diagnosis. The relapse sample lost a subset of mutations that were identified in the diagnostic specimen (blue), and acquired other mutation that were not detected in the diagnostic specimen (red), including a premature stop codon in TNFAIP3. All highlighted mutations have been validated by Sanger sequencing. (D) Comparison of DNA copy number alterations in diagnostic and relapse specimens of LPJ041 show detectable DNA copy number gains (red) and losses (blue) that are maintained from diagnosis to relapse. The relapse acquires an additional major abnormality, loss of chromosome X.
Relapse emanating from a progressed tumor clone in LPJ041. (A) Molecular cloning of productively rearranged Ig heavy-chain genes show a clonal relationship between the diagnosis and relapse specimen in LPJ041. Fifty-five unique clones were sequenced from the diagnostic specimen (blue), and 43 from the relapse specimen (red). Forty percent of the clones from the diagnostic specimen and 74% of the clones from the relapse specimen shared identical IgHV sequences (indicated by red/blue split). This was the closest sequence to the germ-line identified in the relapse specimen, indicating that the relapse emanated from an evolved tumor clone. Each arrowed line represents a single nucleotide substitution, with numbers representing additional substitutions. (B) Sanger sequencing showed identical t(14;18)(q32;q21) breakpoints in the diagnostic and relapse specimen for LPJ041. Four clones were sequenced from each of the diagnostic and relapse specimens and the breakpoint established by alignment of the sequences with the human genome consensus. (C) Comparison of variant frequencies in diagnostic and relapse specimens for LPJ041 showed the maintenance of the majority of mutations in the relapse that were detected within the diagnostic specimen (green), including 2 mutations in MLL2 that were variably represented in diagnostic subpopulations and a mutation in CREBBP that was uniformly represented at diagnosis. The relapse sample lost a subset of mutations that were identified in the diagnostic specimen (blue), and acquired other mutation that were not detected in the diagnostic specimen (red), including a premature stop codon in TNFAIP3. All highlighted mutations have been validated by Sanger sequencing. (D) Comparison of DNA copy number alterations in diagnostic and relapse specimens of LPJ041 show detectable DNA copy number gains (red) and losses (blue) that are maintained from diagnosis to relapse. The relapse acquires an additional major abnormality, loss of chromosome X.
Relapse emanating from an early precursor clone in LPJ128. (A) Molecular cloning of productively rearranged Ig heavy-chain genes showed identical VDJ breakpoints for the diagnostic and relapse specimens of LPJ128, indicating clonal origin. Seventy-four clones were sequenced from the diagnostic specimen (blue), and 43 from the relapse (red). The sequences obtained from the relapse specimen clones were unique from those obtained from the diagnostic specimen clones, and indicated that the relapse emanated from an early tumor precursor clone. Each arrowed line represents a single nucleotide substitution, with numbers representing additional substitutions. (B) Sanger sequencing showed identical t(14;18)(q32;q21) breakpoints in the diagnostic and relapse specimen for LPJ128, indicating that this is an early event. Four clones were sequenced from each of the diagnostic and relapse specimens and the breakpoint established by alignment of the sequences with the human genome consensus. (C) Comparison of variant frequencies in diagnostic and relapse specimens for LPJ128 showed mutations that the relapse specimen loses the majority of mutations in the relapse that were detected in the diagnostic specimen (blue), including mutations in MLL2 and TNFRSF14 that were variably represented in diagnostic subpopulations. A subset of mutations that were uniformly represented in the diagnostic specimen, including that in CREBBP, were maintained at relapse (green). The relapse also acquired additional mutations (red), including a unique TNFRSF14 mutation after loss of the mutation in the diagnostic specimen. All highlighted mutations were validated by Sanger sequencing. (D) Comparison of DNA copy number alterations in diagnostic and relapse specimens of LPJ128 show detectable DNA copy number gains (red) and losses (blue) that are all lost from diagnosis to relapse.
Relapse emanating from an early precursor clone in LPJ128. (A) Molecular cloning of productively rearranged Ig heavy-chain genes showed identical VDJ breakpoints for the diagnostic and relapse specimens of LPJ128, indicating clonal origin. Seventy-four clones were sequenced from the diagnostic specimen (blue), and 43 from the relapse (red). The sequences obtained from the relapse specimen clones were unique from those obtained from the diagnostic specimen clones, and indicated that the relapse emanated from an early tumor precursor clone. Each arrowed line represents a single nucleotide substitution, with numbers representing additional substitutions. (B) Sanger sequencing showed identical t(14;18)(q32;q21) breakpoints in the diagnostic and relapse specimen for LPJ128, indicating that this is an early event. Four clones were sequenced from each of the diagnostic and relapse specimens and the breakpoint established by alignment of the sequences with the human genome consensus. (C) Comparison of variant frequencies in diagnostic and relapse specimens for LPJ128 showed mutations that the relapse specimen loses the majority of mutations in the relapse that were detected in the diagnostic specimen (blue), including mutations in MLL2 and TNFRSF14 that were variably represented in diagnostic subpopulations. A subset of mutations that were uniformly represented in the diagnostic specimen, including that in CREBBP, were maintained at relapse (green). The relapse also acquired additional mutations (red), including a unique TNFRSF14 mutation after loss of the mutation in the diagnostic specimen. All highlighted mutations were validated by Sanger sequencing. (D) Comparison of DNA copy number alterations in diagnostic and relapse specimens of LPJ128 show detectable DNA copy number gains (red) and losses (blue) that are all lost from diagnosis to relapse.
We also identified mirroring patterns of clonal evolution within cSNVs in these cases using exome sequencing. In LPJ041, the relapse sample maintained the majority of mutations identified in the diagnostic specimen (39/49) and acquired an additional 36 mutations, including 1 in TNFAIP3 (Figure 4C). This pattern was also conserved within the context of DNA copy number abnormalities, where the relapse specimen maintained all DNA copy number alterations identified in the diagnostic specimen and acquired 1 additional alteration, loss of chromosome X (Figure 4D). These patterns of maintenance of the majority of somatic alterations suggest that the clone of origin for the relapse in LPJ041 was probably a progressed malignant clone present in the diagnostic specimen that possessed founder, driver, and accelerator mutations such as t(14;18)(q32;q21), CREBBP mutation, and MLL2 mutations, respectively.
In contrast, in patient LPJ128, tumor at first relapse showed absence of the majority (22/39) of mutations identified in the diagnostic specimen, including mutations in MLL2 and TNFRSF14 (Figure 5C). Interestingly, the relapse also showed presence of an additional 16 mutations, including a unique premature stop codon in TNFRSF14. This pattern was also seen at the level of DNA copy number abnormalities, where 4/4 DNA copy number alterations observed in the diagnostic specimen were absent in the relapse (Figure 5D). These patterns of somatic alterations are consistent with the clone of origin for the relapse in LPJ128 being an early malignant precursor possessing founder and driver mutations, such as t(14;18)(q32;q21) and CREBBP mutation, respectively, but lacking accelerator mutations, such as those in MLL2 and TNFRSF14.
Discussion
Several recent studies have identified recurrent somatic mutations in several genes in FL using high next generation sequencing.13,14,28 Together, these important studies genetically characterized bulk tumor populations from 13 patients with FL, but did not dissect the subclonal heterogeneity of somatic events at diagnosis with relationship to relapse, or constant features of tumor clones including tumoral VDJ and IGH-BCL2 translocations. Here, we show that intratumoral subpopulations in FL have diverse genetic profiles. Using a heterogeneously expressed marker, CD20, we sorted subpopulations of FL tumor cells and interrogated them with gene expression microarrays and whole-exome sequencing. Using this approach, we identified patterns of mutational diversity that could not be inferred by analysis of bulk tumor specimens. This allowed us to model the genetic evolution of FL, and thereby provide insight into the potential for gene-directed therapy in this disease.
CD20 is an important molecule in FL because it is a target of several monoclonal antibody therapies including Rituximab. Separation of tumor subpopulations using CD20 allowed us to characterize their clearly divergent mutation profiles. However, it is not clear as to whether CD20 is unique in this respect. It is possible that the tumor cell pool in FL is so heterogeneous that subpopulations with various mutation profiles could potentially be isolated using a variably expressed markers other than CD20. Importantly, low CD20 expression has been associated with inferior outcome in lymphoma, independent of whether patients were treated with or without Rituximab.9 However, we found no genes containing cSNVs that were overrepresented within the lower CD20-expressing subpopulation in multiple cases, suggesting that this association with poor outcome is not the result of underlying genetic differences.
By exome sequencing tumor subpopulations from diagnostic FL tumors with reference to their matched germ-line sequence, we identified a large number of cSNVs. The majority of these cSNVs were present at frequencies that indicated their subclonal representation. This was confirmed by evaluating their respective frequencies in subpopulations from the same tumor, which showed that the majority of cSNVs were discordantly represented between each population, particularly in cases with a large mutational burden. Importantly, the clonal representation of cSNVs could not be accurately predicted by assessing their location within either the distribution or clustering of variant frequencies from the bulk tumor alone, as previously used.30-32 However, by separately evaluating tumor subpopulations, we were able to identify numerous mutations that were variably represented among subpopulations, as well as a subset that were lost in progression from diagnosis to relapse, highlighting them as probable late events during clonal evolution. Conversely, we were also able to identify mutations that were present at frequencies consistent with their uniform representation among the entire tumor cell pool, that did not vary in frequency between tumor cell subpopulations, and that were maintained between diagnosis and relapse, highlighting these as probable early events during lymphomagenesis.
Based on our exome sequencing results, we propose a model for FL tumorigenesis (Figure 6, supplemental Figures 15-16): (1) A primary founder mutation is acquired to a premalignant tumor cell precursor and elongates the life of a clone sufficiently for it to acquire secondary mutations. (2) Secondary driver mutation(s) may then be acquired resulting in an early malignant clone that may or may not release the cell from reliance on the founder mutation. (3) Tertiary mutations acquired subsequently to this may either act as accelerator or passenger mutations. Accelerator mutations would provide a selective advantage to a progressed malignant subclone, result in progressive outgrowth of that clone compared with competing clones, and cause clinical disease. Passenger mutations would have no selective advantage and accumulate only if present in the same clone as an accelerator mutation.
A model of genetic evolution of FL. A normal B-cell undergoes immunoglobulin recombination, followed by acquisition of the t(14;18)(q32;q21) founder IGH-BCL2 translocation yielding a premalignant tumor cell precursor, detectable in the majority of older adults in the absence of tumors.16 This precursor acquires 1 or more driver mutations, such as in CREBBP, yielding an early malignant clone. Accelerator mutations are then acquired, such as those in MLL2 and/or TNFRSF14, resulting in a progressed malignant clone with a selective advantage that yields clinical disease. Relapses may originate from either an early malignant clone as in patient LPJ128 (Figure 5), and therefore possess only founder and driver mutations, or from a progressed malignant clone as in patient LPJ041 (Figure 4), and therefore possess a full repertoire of founder, driver and accelerator mutations.
A model of genetic evolution of FL. A normal B-cell undergoes immunoglobulin recombination, followed by acquisition of the t(14;18)(q32;q21) founder IGH-BCL2 translocation yielding a premalignant tumor cell precursor, detectable in the majority of older adults in the absence of tumors.16 This precursor acquires 1 or more driver mutations, such as in CREBBP, yielding an early malignant clone. Accelerator mutations are then acquired, such as those in MLL2 and/or TNFRSF14, resulting in a progressed malignant clone with a selective advantage that yields clinical disease. Relapses may originate from either an early malignant clone as in patient LPJ128 (Figure 5), and therefore possess only founder and driver mutations, or from a progressed malignant clone as in patient LPJ041 (Figure 4), and therefore possess a full repertoire of founder, driver and accelerator mutations.
Mutations biased in frequency between tumor cell subpopulations in this study are late genetic events and therefore probably represent accelerator or passenger mutations. Because we only sampled a single temporal point along the evolutionary continuum for 6 of our tumors, it is impossible to establish whether mutations that are represented equally within these subpopulations and at frequencies that suggest their uniform representation in the tumor cell pool of these samples are drivers, accelerators or passengers. However, using our diagnosis/relapse pairs we show 2 contrasting patterns of disease relapse and, from these, can provide some inference into the role of various genetic alterations. Because both diagnosis and relapse pairs have the same t(14;18(q32;q21) breakpoint, but this translocation is not sufficient to induce disease,12 we assume IGH-BCL2 translocation is the primary event in lymphomagenesis and highlight it as a candidate founder mutation in FL. The high frequency and uniform representation of CREBBP mutations between subpopulations, as well their maintained presence between diagnosis and relapses, highlights it as secondary genetic event and a candidate driver mutation in these cases. In contrast, the variable representation of MLL2 and TNFRSF14 mutations between subpopulations within diagnostic specimens, and their loss at relapse in LPJ128, indicates that these are tertiary genetic events and highlights them as candidate accelerator mutations in these cases. We have provided multiple lines of evidence to support this order of events within our cohort; however, these observations should be tested in a larger population. Furthermore, the only way to definitively establish a temporal hierarchy of genetic events is to genotype tumor clones at the single-cell level.
CREBBP encodes a histone acetyltransferase (HAT) that also has a role in acetylation of BCL6 and p53. Mutations in this gene have been described in 32% of FL cases13 and linked with loss of HAT activity, associated attenuation of CREBBP-mediated BCL6 acetylation, and a resulting increase in activity of the BCL6 oncogene.14 Notably, 6/6 CREBBP mutations identified here were located within the HAT domain, and were associated with altered expression of BCL6 target genes, providing a mechanism for a CREBBP-mediated lymphomagenic role in FL. This is particularly notable because the uniform representation of CREBBP mutations highlight this as a potential therapeutic target in FL that may be potentially actionable via targeting of the aberrantly active BCL6 oncogene.33
MLL2 encodes a histone methyltransferase that was previously found to be mutated in 89% of FL cases.14 The high frequency of MLL2 mutations has led to some speculation that it may be a possible secondary requisite alteration, in tandem with t(14;18)(q32;q21), for FL lymphomagenesis.13 However, we found that 3/4 of these mutations showed unequal representation between these subpopulations. In 1 case that possessed 2 MLL2 mutations, LPJ041, 1 mutation was enriched in the CD20int subpopulation and the other mutation in the CD20hi subpopulation. In another case, LPJ128, a SET domain mutation of MLL2 was enriched in the CD20hi subpopulation and was not present in the relapse in this patient, indicating that MLL2 mutation occurred as a late event in a subclone that was more evolved than that from which the relapse originated. Our data therefore suggest that MLL2 mutation is a secondary event in FL lymphomagenesis that may act as an accelerator or passenger mutation rather than a central driver mutation. However, a more detailed functional analysis of these candidate genes is required to definitively define their capacity to act as driver or accelerator mutations.
Together, these observations demonstrate substantial clonal diversity in the coding genome of FL. Surprisingly, we observed subclonal distribution of mutations for several genes which are known to be recurrently mutated in FL and on that basis previously thought to be early drivers of pathogenesis. The intratumoral heterogeneity seen within the tumors studied here confirms IGH-BCL2 translocation as an early event during FL tumorigenesis, but provides the first evidence for MLL2 arising later in disease evolution. This disparity is particularly notable, because both lesions are found in a similarly high fraction of patients with FL (circa 90%). Indeed, the frequency of mutation within a given gene across 441 patients with mature B-cell malignancies studied by high throughput sequencing (FL = 21, BL = 29, MM = 38, DLBCL = 157, CLL = 196) correlated poorly with clonal dominance within individual FL tumors in this study (supplemental Figure 17). For instance, despite being less prevalent in patients with FL, CREBBP mutations were consistently represented between tumor subpopulations and between diagnosis and relapse.
Nevertheless, the majority of mutations identified in this study were subclonal and therefore not ideal candidates for therapeutic intervention. These observations highlight the need for closer examination of the clonal representation of mutations before their therapeutic targeting. The framework described here provides important insight into lymphomagenesis and is a key step in prioritization of candidates for gene mutation-directed therapies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge the contribution of the Stanford Shared FACS Facility, and Dr Ramit Mehr for sharing IgTree software.
This work was supported by grants from the USPHS/NIH (P01 CA034233, U54 CA149145, R01 CA151748), The Leukemia and Lymphoma Society (Specialized Center of Research Program Award), The Albert and Mary Yu Gift Fund, Evelyn Leung Gift Fund, and Lymphoma Research Foundation (AAA). R.L. is an American Cancer Society Clinical Research Professor. M.R.G. and A.A.A. are Special Fellows of Leukemia and Lymphoma Society. A.A.A. is a Doris Duke Charitable Foundation Clinical Investigator.
National Institutes of Health
Authorship
Contribution: M.R.G. designed and performed research, analyzed data, and wrote the paper; A.J.G. analyzed data; R.V.N. analyzed data; S.K. performed research; J.M.I. designed and performed research; C.L.L. analyzed data; I.K. designed research; E.S.H. performed research; J.H.M. performed research; H.J. designed research; S.K.P. analyzed data; and R.L. and A.A.A. analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ash A. Alizadeh or Ronald Levy, Division of Oncology, Dept of Medicine, Stanford University, Stanford, CA 94305; e-mail: arasha@stanford.edu or levy@stanford.edu.