Key Points
Human and mouse macrophages share partially conserved gene and protein expression programs in the resting or M2 activated state.
TGM2 is a novel M2 marker consistently induced in human and mouse M2 macrophages.
Abstract
The molecular repertoire of macrophages in health and disease can provide novel biomarkers for diagnosis, prognosis, and treatment. Th2-IL-4–activated macrophages (M2) have been associated with important diseases in mice, yet no specific markers are available for their detection in human tissues. Although mouse models are widely used for macrophage research, translation to the human can be problematic and the human macrophage system remains poorly described. In the present study, we analyzed and compared the transcriptome and proteome of human and murine macrophages under resting conditions (M0) and after IL-4 activation (M2). We provide a resource for tools enabling macrophage detection in human tissues by identifying a set of 87 macrophage-related genes. Furthermore, we extend current understanding of M2 activation in different species and identify Transglutaminase 2 as a conserved M2 marker that is highly expressed by human macrophages and monocytes in the prototypic Th2 pathology asthma.
Introduction
Macrophages exhibit beneficial or detrimental roles in disease, depending on their activation status and the pathology nature. Of particular interest is the alternative (M2) activation pathway induced by the Th2 cytokines IL-4 and IL-13 that leads to de novo expression of genes and selectively altered functions implicated in the modulation of inflammation, repair, and unexpected functions such as glucose homeostasis, thermogenesis, learning, and memory.1-6 This type of activation may be beneficial in classic inflammatory settings but deleterious in allergic and fibrotic diseases. Despite emerging knowledge from mouse studies, there is an unmet need for human M2 macrophage markers, which will be useful to study M2 macrophage contribution to disease pathogenesis and resolution and as potential biomarkers for diagnosis and treatment in diseases such as asthma and fibrosis. Prototypic mouse M2 markers are not applicable because there are no human homologs of particular genes (eg, Ym1 and Fizz1) or the gene is not regulated by these cytokines in human macrophages (eg, Arginase 1).7,8 In addition, the models used to study human macrophages are limited and overall there is poor access to ex vivo human material.
To define the macrophage contents useful for understanding the cell nature, predicting functions, and providing a repository of selective biomarkers for human and mouse studies, in the present study, we used microarray and proteomics to compare transcripts and proteins of resting and M2 human and mouse macrophages. The study included various in vitro and ex vivo macrophage models to establish multispecies and multimodel gene and protein signatures. We report on a range of macrophage expressed genes that make it possible to characterize these cells in human tissues in situ and identify Transglutaminase 2 (TGM2) as a uniquely functional conserved M2 marker in both species.
Methods
Reagents
Lung biopsies and blood from asthmatic patients and healthy donors were used for this study. The studies were carried out and approved locally at the medical ethics committee of the University Medical Center Groningen and the Oxford Center for Respiratory Medicine. Informed consent was obtained from all subjects in accordance with the Declaration of Helsinki.
Antibodies were purchased from Serotec and chemicals were from Sigma-Aldrich unless otherwise specified. Recombinant human and murine GM-CSF, M-CSF, and IL-4 were from PeproTech; RPMI 1640, l-glutamine, and penicillin/streptomycin were from PAA; OPTIMEM was from Invitrogen; X-VIVO 10 media were from Lonza; and FCS was from HyClone or PAA. Bacteriological 6-well plastic plates were from Greiner Bio-one; 6-well tissue-culture plastic plates were from Costar and Falcon; and Permanox 35-mm dishes were from Nunc.
Human macrophages
Human monocytes (98% CD14+, 13% CD16+) were obtained from healthy donor buffy coats by 2-step gradient centrifugation followed by an additional step using the Monocyte Isolation Kit II (Miltenyi Biotec).9
Human M-CSF–derived macrophages were obtained by culturing monocytes in FCS-coated dishes for 7 days in RPMI 1640 medium with l-glutamine, 20% FCS, and 100 ng/mL of M-CSF at a density of 1.5 × 105/cm2. A variant of this model used medium with 10% FCS and 50 ng/mL of M-CSF.
Autologous serum macrophages were obtained by culturing monocytes in tissue-culture plates for 7 days in X-VIVO 10 with 1% autologous serum at a density of 1.5 × 105/cm2. Off-the-clot serum from a pool of donors was used for maturation after decomplementation (56°C for 30 minutes).
Human GM-CSF–derived macrophages were obtained by culturing monocytes in tissue-culture plates for 7 days in RPMI 1640 medium with l-glutamine, 10% FCS, and 25 ng/mL of GM-CSF at a density of 1.5 × 105/cm2.
Mouse macrophages
Mouse macrophages (C57BL/6) were plated in 6-well plates at a density of 1-2 × 105/cm2. For BM-derived macrophages (BMMΦ), femora and tibiae of hind legs were flushed with PBS and cells were resuspended in RPMI 1640 medium with l-glutamine, 10% FCS, 100 units/mL of penicillin, and 100 mg/mL of streptomycin supplemented with 15% (vol/vol) L-cell–conditioned medium, and cultured for 7 days on bacteriologic plastic. BMMΦ were detached using PBS containing 10mM EDTA, washed, and resuspended in RPMI 1640 medium, 5% heat-inactivated FCS, 100 units/mL of penicillin, and 100 mg/mL of streptomycin.
Thioglycollate-elicited peritoneal macrophages (ThioMΦ) were isolated by peritoneal lavage 4 days after IP injection of 4% thioglycollate-broth. Biogel-elicited peritoneal macrophages (BioMΦ) were isolated from mice injected 4 days before with 2% wt/vol Biogel in PBS.
Transcriptional profiles study and data mining
RNA was isolated using the RNeasy Mini Kit (QIAGEN). Microarray raw data, RMA for Affymetrix, and background-subtracted data for Illumina were normalized by Quantiles in R Bioconductor. All datasets have been deposited in Geoprofiles: SuperSeries GSE35495. For translational purposes, we focused on 9006 transcripts detected by all microarray platforms and in both species. For the expression study, the average expression was calculated for each gene in each dataset and then ranked. After ranking, genes that ranked in the first 1000 were selected for all datasets. Using different microarray platforms with different detection probes compensates the quality detection bias intrinsic to microarray design. For alternative activation, differences in gene expression were assessed using R-Bioconductor, with Student t test and false discovery rate correction where possible (for single samples only the fold was considered). Genes with a false discovery rate ≤ 0.05 and fold change ≥ 1.5 were considered differentially expressed. The consensus of the IL-4 transcriptome includes genes regulated in at least 60% of the models. Pathway overrepresentation and novelty were assessed with IPA Ingenuity.
To investigate macrophage signature overrepresentation in other species, the following Gene Expression Omnibus (GEO) dataset series were used: rat peritoneal macrophages, series GSE24780 (samples GSM610350 to GSM610353), and chicken macrophage HD11 cells, series GSE23881 samples GSM610350 to GSM610353). Signature overrepresentation in major blood lymphocyte populations, CD8 and CD4 T cells, B cells, and CD56dim natural killer cells was done using Series GSE6887 (samples GSM158512 to GSM158516, GSM158532-36, GSM158543 to GSM158548, and GSM158613 to GSM158619). For every sample, the average ranking was established across replicates and the highest 500 transcripts were interrogated against the 87 macrophage signature IPA pathways template.
Proteomic analysis
For proteomic analysis, we cultured human M-CSF and autologous serum–derived macrophages or mouse BMMΦ and ThioMΦ on tissue culture plastic with 20 ng/mL of species-specific recombinant IL-4 for 48 hours. Cells were lysed in 20mM Tris, 140mM NaCl, 1mM EDTA, and 1% NP40, and samples centrifuged for 10 minutes at 10 000 rpm to remove nuclei. Four human and 4 mouse samples were prepared for ITRAQ proteomic analysis (Applied Biosystems). Each human sample included 300 μg of macrophage lysate formed by mixing 100 μg of protein lysates from 3 independent blood donors. Mouse samples included 300 μg of protein and were each derived from a pool of at least 3 mice. Samples were precipitated with cold acetone. After resuspension, 100 μg of sample was trypsinized overnight. Comparative analysis was done with ITRAQ and absolute quantitation using SINQ (supplemental Methods, see the Supplemental Materials link at the top of the article). Statistical processing was done with TMEV.10 Differences were assessed by ratio analysis.
Statistical analysis
For comparison of data from asthma patients and healthy controls, a 2-tailed unpaired t test with Welch correction was used. Statistical analysis was performed using Prism software (GraphPad) unless otherwise specified.
Results
Identification of conserved signatures in human and mouse macrophages
To establish molecular signatures shared by human and mouse macrophages, we used 2 experimental strategies: whole-genome microarray and proteomics analysis. We included validated human and mouse macrophage models representing diverse ex vivo and in vitro differentiation methods (Figure 1A and Table 1). Our knowledge about macrophages is biased toward specific markers; therefore, to understand the main components of the macrophage cellular machinery, we focused on all highly expressed genes. We wished to obtain an overview of molecules needed in mass, which may suggest new macrophage functions. The genes detected by microarrays were ranked according to their expression levels and the highest-ranked 1000 (ie, the most highly expressed) were compared across samples. Comparing the human datasets, we identified nearly 500 transcripts that were consistently detected as being highly expressed in different donors and models; a similar number was detected for the mouse. Between the human and mouse lists, we found an overlap of 231 genes (Figure 1B).
Human and mouse macrophage expression profiles are highly conserved at the mRNA and protein levels. (A) In this study, we included variations of common human macrophage (hMΦ) models: M-CSF– or autologous serum (AS)–differentiated macrophages cultured on tissue culture plastic (TCP) or bacteriological plastic (BP). For murine macrophages (mMΦ), we included BM-derived macrophages (BMMΦ) and 2 primary macrophage populations isolated ex vivo: peritoneal macrophages elicited by injection of Biogel (BioMΦ) or inflammatory macrophages elicited by injection of thioglycollate (ThioMΦ); macrophages were cultured on different surfaces (TCP, BP, or Permanox [PP]). (B) comparing basal transcriptomes of all human macrophage culture models, we identified 489 transcripts consistently detected in the first 1000 expression-ranked genes. A total of 459 transcripts were detected in common for all murine culture models; 231 transcripts, approximately 50% of each species consensus list, were shared by human and mouse macrophages. (C) Using proteomics, we investigated the protein content of 4 macrophage models: human macrophages differentiated in the presence of M-CSF or AS and murine BMMΦ and ThioMΦ. We detected 977 and 1038 proteins in human and mouse macrophages, respectively. Of these, 513 were detected in macrophages of both species. (D) Overlap between the conserved mRNA and protein signatures; 231 highly expressed genes and 513 detected proteins, respectively, amounted to 87 genes.
Human and mouse macrophage expression profiles are highly conserved at the mRNA and protein levels. (A) In this study, we included variations of common human macrophage (hMΦ) models: M-CSF– or autologous serum (AS)–differentiated macrophages cultured on tissue culture plastic (TCP) or bacteriological plastic (BP). For murine macrophages (mMΦ), we included BM-derived macrophages (BMMΦ) and 2 primary macrophage populations isolated ex vivo: peritoneal macrophages elicited by injection of Biogel (BioMΦ) or inflammatory macrophages elicited by injection of thioglycollate (ThioMΦ); macrophages were cultured on different surfaces (TCP, BP, or Permanox [PP]). (B) comparing basal transcriptomes of all human macrophage culture models, we identified 489 transcripts consistently detected in the first 1000 expression-ranked genes. A total of 459 transcripts were detected in common for all murine culture models; 231 transcripts, approximately 50% of each species consensus list, were shared by human and mouse macrophages. (C) Using proteomics, we investigated the protein content of 4 macrophage models: human macrophages differentiated in the presence of M-CSF or AS and murine BMMΦ and ThioMΦ. We detected 977 and 1038 proteins in human and mouse macrophages, respectively. Of these, 513 were detected in macrophages of both species. (D) Overlap between the conserved mRNA and protein signatures; 231 highly expressed genes and 513 detected proteins, respectively, amounted to 87 genes.
List of macrophage models used in the study
| Dataset . | Maturation stimulus/dose duration . | Activation stimulus/dose duration . |
|---|---|---|
| Human dataset 1 (hMΦ1) | M-CSF 100 ng/mL/7 d | IL-4 20 ng/mL/18 h |
| Human dataset 2 (hMΦ2) | 1% Autologous serum/7 d | IL-4 20 ng/mL/24 h |
| Human dataset 3 (hMΦ3) | 1% Autologous serum/7 d | IL-4 10 ng/mL/48 h |
| Human dataset 4 (hMΦ4) | 10% Autologous serum/7 d | IL-4 20 ng/mL/7 d |
| Human dataset 5 (hMΦ5) | 10% Autologous serum/7 d | 50 ng/mL of IL-4 or 50 U/mL of IFN-γ + 12.5 ng/mL of TNF-α or 50 ng/mL of M-CSF or 50 ng/mL of IL-10/5 d |
| Mouse dataset mBMMΦ | L-cell–conditioned medium (M-CSF)/7 d | IL-4 20 ng/mL/18 h |
| Mouse datasets ThioMΦ1, ThioMΦ2, and BioMΦ* | Thioglycollate or Biogel/4 d | mIL-4/X63Ag8-653 supernatant/48 h |
| Mouse dataset (ThioMΦ3) | Thioglycollate/4 d | IL-4 10 ng/mL/48 h |
| Dataset . | Maturation stimulus/dose duration . | Activation stimulus/dose duration . |
|---|---|---|
| Human dataset 1 (hMΦ1) | M-CSF 100 ng/mL/7 d | IL-4 20 ng/mL/18 h |
| Human dataset 2 (hMΦ2) | 1% Autologous serum/7 d | IL-4 20 ng/mL/24 h |
| Human dataset 3 (hMΦ3) | 1% Autologous serum/7 d | IL-4 10 ng/mL/48 h |
| Human dataset 4 (hMΦ4) | 10% Autologous serum/7 d | IL-4 20 ng/mL/7 d |
| Human dataset 5 (hMΦ5) | 10% Autologous serum/7 d | 50 ng/mL of IL-4 or 50 U/mL of IFN-γ + 12.5 ng/mL of TNF-α or 50 ng/mL of M-CSF or 50 ng/mL of IL-10/5 d |
| Mouse dataset mBMMΦ | L-cell–conditioned medium (M-CSF)/7 d | IL-4 20 ng/mL/18 h |
| Mouse datasets ThioMΦ1, ThioMΦ2, and BioMΦ* | Thioglycollate or Biogel/4 d | mIL-4/X63Ag8-653 supernatant/48 h |
| Mouse dataset (ThioMΦ3) | Thioglycollate/4 d | IL-4 10 ng/mL/48 h |
All assays were done in biological triplicates except that for mouse datasets ThioMΦ1, ThioMΦ2, and BioMΦ, which was performed in single using pooled RNA.
The best antibodies with which to detect macrophages are directed against membrane and cytosolic proteins with more value for histology than secreted proteins. Therefore, we determined protein levels in macrophages using unfractionated cytosol and membrane protein samples and detected approximately 1000 proteins that were highly expressed in human or mouse macrophages; 513 proteins were detected in macrophages of both species (Figure 1C). The overlap between the conserved 231 genes and 513 proteins amounted to 87 genes (Figure 1D and supplemental Table 1). The proteomics study strengthens the predictions from microarray but per se are not amenable to intra-array comparisons.
Approximately 50% of the 87 macrophage signature genes have been detected in macrophages or macrophage cell lines before, but few of these genes represent established macrophage markers. This validates our selection strategy, but also highlights the novelty of our findings. A total of 43 of 87 genes have not been associated with human or mouse macrophages so far (supplemental Table 1). Previously unknown macrophage signature genes suggesting functional properties include glutathione S-transferase ω 1 (GSTO1) and glutathione peroxidase (GPX1), both of which are associated with redox balance. Examples of established functionally relevant genes in the signature are Annexin A1 (ANXA1), Galectin-3 (LGALS3), peroxiredoxin (PRDX1), S100A4, and integrin β2 (ITGB2, CD18), which are involved in classic macrophage tasks, as discussed later. The extended signature also comprises ubiquitously expressed but established macrophage genes with more basic roles, including ribosomal and cytoskeletal proteins such as actin β (ACTB), ribosomal protein S20 (RPS20), ribosomal protein L27 (RPL27), and mitochondrial protein ubiquinol-cytochrome c reductase core protein (UQCRC1; supplemental Table 2).
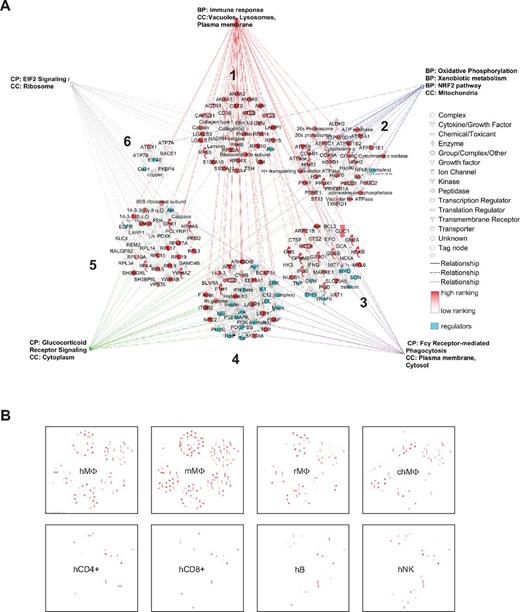
The 87-gene signature is composed of 6 separate expression networks forming a “visual” macrophage landscape (Figure 2A and supplemental Table 1). Overrepresentation analysis of functional and canonical pathways (supplemental Table 1) links them to functions including chemotaxis, lipid storage, immune recognition, and metabolism of xenobiotics. Interestingly, the prototypic macrophage markers CD68, CSF1 receptor (CSFR1), and PU.1 (SFPI1), are not part of our selection, showing that they do not belong to the most highly expressed macrophage genes. The signature defined herein will therefore help to focus on highly expressed molecules, improve our understanding of macrophage biology, and potentially aid the discovery of neglected functions.
A consistent macrophage signature characterizes macrophages of different species and highlights conserved functions. (A) The set of 87 consistently detected genes identifiable in human and mouse macrophages at the mRNA and protein levels form 6 functionally and expression-related networks (numbered 1-6) according to the Ingenuity Knowledge Base. Expression ranking is pseudocolored in red; regulators are shown in turquoise. BP indicates biologic process; CC, cellular component; and CP, canonical pathway. Many of the genes are immune response-related, such as CD36, S100A10, S100A11, LGALS1, and LGALS3. The lysosomal cathepsins and cystatins, LAMP1, HEXA, and other catabolic enzymes are also included in this category. Furthermore, the categories oxidative phosphorylation (connected to energy pathways) and xenobiotic metabolism are overrepresented. These genes form part of the NRF2 gene targets important for redox balance. Further overrepresented categories include glucocorticoid signaling, phagocytosis, and EIF pathways; all pathways are listed in supplemental Table 1. (B) The consistent macrophage signature as a whole can be used to distinguish macrophages from other cell types. Overlap of the 500 most highly expressed genes in unrelated human GM-CSF macrophage samples (hMΦ), mouse macrophage samples (mMΦ), and even rat and chicken macrophage samples (rMΦ and chMΦ) shows a clear overrepresentation of the signature. In contrast, the consistent signature is scarcely represented when overlaid with the 500 most highly expressed genes in human, CD4 and CD8 T cells (hCD4+ and hCD8+), B cells (hB), or NK cells (hNK).
A consistent macrophage signature characterizes macrophages of different species and highlights conserved functions. (A) The set of 87 consistently detected genes identifiable in human and mouse macrophages at the mRNA and protein levels form 6 functionally and expression-related networks (numbered 1-6) according to the Ingenuity Knowledge Base. Expression ranking is pseudocolored in red; regulators are shown in turquoise. BP indicates biologic process; CC, cellular component; and CP, canonical pathway. Many of the genes are immune response-related, such as CD36, S100A10, S100A11, LGALS1, and LGALS3. The lysosomal cathepsins and cystatins, LAMP1, HEXA, and other catabolic enzymes are also included in this category. Furthermore, the categories oxidative phosphorylation (connected to energy pathways) and xenobiotic metabolism are overrepresented. These genes form part of the NRF2 gene targets important for redox balance. Further overrepresented categories include glucocorticoid signaling, phagocytosis, and EIF pathways; all pathways are listed in supplemental Table 1. (B) The consistent macrophage signature as a whole can be used to distinguish macrophages from other cell types. Overlap of the 500 most highly expressed genes in unrelated human GM-CSF macrophage samples (hMΦ), mouse macrophage samples (mMΦ), and even rat and chicken macrophage samples (rMΦ and chMΦ) shows a clear overrepresentation of the signature. In contrast, the consistent signature is scarcely represented when overlaid with the 500 most highly expressed genes in human, CD4 and CD8 T cells (hCD4+ and hCD8+), B cells (hB), or NK cells (hNK).
The 6 networks forming the macrophage signature suggest coregulation by specific transcription factors. We analyzed 381 possible promoter sequences from the 87 genes to find that ETS, NR2F, and HOMF Genomatix-defined transcription factor families had binding sites in all sequences. ETS1, HNF4A (NR2F family), and SP1 (ETS family) were also predicted with IPA Knowledge Database (supplemental Table 1). Of the 11 members of the ETS family, ELF2 and, as expected, PU.1 appeared to be specifically expressed in human tissue macrophages (human protein atlas).11
We found substantial overlap when we compared the macrophage “landscape” with macrophage samples not included in the analysis (eg, human monocyte derived GM-CSF macrophages or chicken and rat macrophages; Figure 2B). In CD4+/CD8+ T cells, B cells, or NK cells, the overlap was less marked (Figure 2B). In addition to the information gained from the analysis of pathways overrepresented in the 87-gene signature, more details were obtained for human and mouse macrophages when we analyzed species-specific signatures. For example, the 489 genes consistently detected as being highly expressed at the mRNA level in human samples organized into 25 gene-expression networks, 238 canonical pathways, and 1186 associated biologic functions and toxicological pathways (supplemental Table 2).
Microarray and proteomics predict the protein repertoire of human tissue macrophages and validate the signature concept
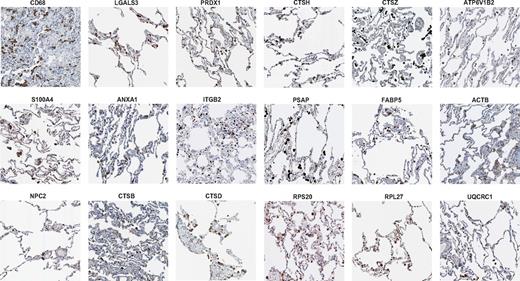
To determine whether the conserved 87-gene macrophage signature could predict the content of human tissue macrophages, we analyzed their expression concentrating on the lung as an organ with clearly detectable macrophages, as shown by the expression pattern of CD68 as an established immunohistochemical macrophage marker11 (Figure 3). CD68 itself was not contained in the conserved signature and therefore does not belong to the most highly expressed genes in macrophages. This is a useful reminder that results are highly dependent on methods. Many of the highly expressed genes with direct functional relevance, either previously known or newly associated with macrophages, were indeed expressed in human alveolar macrophages (eg, LGALS3, PRDX1, S100A4, ANXA1, ITGB2, and NPC2; Figure 3). Other molecules confirmed in lung macrophages included the lysosomal compartment proteins, cathepsins (CTSB, CTSD, CTSH, and CTSZ), and the ATPase, H+ transporting, lysosomal V1 subunit B2 (ATP6V1B2). Additional human macrophage signature genes expressed included prosaposin (PSAP), fatty acid binding protein 5 (FABP5), ACTB, RPS20, RPL27, and UQCRC1 (Figure 3 and supplemental Tables 2 and 3). The lung is amenable to the study of macrophages without colocalization because of their clear distribution; however, more focused studies should be performed to understand the expression of these genes in tissue cells, particularly in the mononuclear phagocyte system. For most of the genes mentioned, macrophage-specific localization could be confirmed in other macrophage-rich tissues such as the gut, spleen, and liver.11 In conclusion, analysis of tissue expression shows that we have generated a useful resource of novel markers for tissue macrophage detection that complements conventional markers. Single markers do not define the macrophage compared with a signature network of multiple functional genes.
The consistent macrophage signature predicts in situ macrophage-specific expression in human tissues. The gene signature of resting macrophages detected by proteomics and microarray ranking can be detected in lung-resident tissue macrophages. Shown are selected immunohistochemical stainings from human lung tissue using antibodies against CD68, Galectin-3 (LGALS3), Peroxiredoxin (PPRDX1), S100A4, Annexin A1 (ANXA1), ITGB2 (Integrin β 2, CD18), Niemann Pick type C2 (NPC2), cathepsins (CTSB, CTSD, CTSH, CTSZ), ATPase, H+ transporting, lysosomal V1 subunit B2 (ATP6V1B2), Prosaposin (PSAP), fatty acid binding protein 5 (FABP5), actin β (ACTB), ribosomal protein S20 (RPS20), ribosomal protein L27 (RPL27), and the mitochondrial protein ubiquinol-cytochrome c reductase core protein (UQCRC1). Data were generated with the Human Protein Atlas and staining was selected only if the antibodies had supportive scores for immunohistochemistry.
The consistent macrophage signature predicts in situ macrophage-specific expression in human tissues. The gene signature of resting macrophages detected by proteomics and microarray ranking can be detected in lung-resident tissue macrophages. Shown are selected immunohistochemical stainings from human lung tissue using antibodies against CD68, Galectin-3 (LGALS3), Peroxiredoxin (PPRDX1), S100A4, Annexin A1 (ANXA1), ITGB2 (Integrin β 2, CD18), Niemann Pick type C2 (NPC2), cathepsins (CTSB, CTSD, CTSH, CTSZ), ATPase, H+ transporting, lysosomal V1 subunit B2 (ATP6V1B2), Prosaposin (PSAP), fatty acid binding protein 5 (FABP5), actin β (ACTB), ribosomal protein S20 (RPS20), ribosomal protein L27 (RPL27), and the mitochondrial protein ubiquinol-cytochrome c reductase core protein (UQCRC1). Data were generated with the Human Protein Atlas and staining was selected only if the antibodies had supportive scores for immunohistochemistry.
Identification of conserved gene and protein signatures in human and mouse alternatively activated macrophages
In addition to novel markers for human tissue macrophage analysis, there is a great need for markers that detect and characterize alternatively activated (M2) macrophages in human disease. Therefore, in the present study, we investigated changes in gene and protein expression induced by the Th2 cytokine IL-4 in human and murine macrophages (Figure 4) using the different macrophage culture models described above (Figure 1A). IL-13, another important Th2 cytokine that targets in part the IL-4 receptor, was not included in this study.
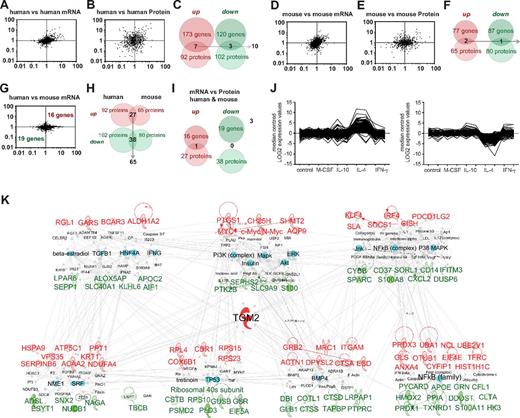
Identification of conserved M2 alternative activation markers in human and mouse. Fold regulation of M2 versus M0 macrophages plotted for each dataset. (A) Pairwise comparisons of human samples follow an inclined ellipse contour, with many genes up- or down-regulated by IL-4 in common. (B) Comparison of proteomics data shows a more spread distribution. (C) Genes up-regulated or down-regulated by IL-4 in all human samples and coincidences between microarray and proteomics. (D-E) Pairwise comparisons of mouse samples. The plots for gene (D) and protein (E) expression analysis follow similar patterns as human samples. (F) Microarrays and proteomics of mouse samples. Genes consistently up-regulated or down-regulated by IL-4 and coincidences between microarray and proteomics are shown. (G) Scatter plots of comparisons of human and mouse microarray profiles show star-like rather than inclined ellipse-like contours. The coincidences of genes regulated in macrophages from both species consisted of 35 genes. (H) With proteomics, the coincidences amounted to 65 proteins. (I) Comparing the regulation of transcripts and proteins, only one gene was consistently induced by IL-4: TGM2. (J) K means clustering of genes modulated by IL-4 in human macrophages in macrophages stimulated with M-CSF, IL-10, and IFN-γ. (K) Alternative activation profile conserved in the human and the mouse organized into 3 and 4 gene and protein expression networks (top and bottom, respectively). At the center of the networks is the highly connected TGM2. The networks comprise key gene-expression regulators and signaling mediators. Expression ranking is pseudocolored in red for up-regulated and green for down-regulated genes and is influenced by key regulators of macrophage activation (selected in turquoise; the rest are in white). Label color: red indicates; green, down-regulated; and black, regulator. Label size is for highlighting purposes.
Identification of conserved M2 alternative activation markers in human and mouse. Fold regulation of M2 versus M0 macrophages plotted for each dataset. (A) Pairwise comparisons of human samples follow an inclined ellipse contour, with many genes up- or down-regulated by IL-4 in common. (B) Comparison of proteomics data shows a more spread distribution. (C) Genes up-regulated or down-regulated by IL-4 in all human samples and coincidences between microarray and proteomics. (D-E) Pairwise comparisons of mouse samples. The plots for gene (D) and protein (E) expression analysis follow similar patterns as human samples. (F) Microarrays and proteomics of mouse samples. Genes consistently up-regulated or down-regulated by IL-4 and coincidences between microarray and proteomics are shown. (G) Scatter plots of comparisons of human and mouse microarray profiles show star-like rather than inclined ellipse-like contours. The coincidences of genes regulated in macrophages from both species consisted of 35 genes. (H) With proteomics, the coincidences amounted to 65 proteins. (I) Comparing the regulation of transcripts and proteins, only one gene was consistently induced by IL-4: TGM2. (J) K means clustering of genes modulated by IL-4 in human macrophages in macrophages stimulated with M-CSF, IL-10, and IFN-γ. (K) Alternative activation profile conserved in the human and the mouse organized into 3 and 4 gene and protein expression networks (top and bottom, respectively). At the center of the networks is the highly connected TGM2. The networks comprise key gene-expression regulators and signaling mediators. Expression ranking is pseudocolored in red for up-regulated and green for down-regulated genes and is influenced by key regulators of macrophage activation (selected in turquoise; the rest are in white). Label color: red indicates; green, down-regulated; and black, regulator. Label size is for highlighting purposes.
When we analyzed the IL-4–regulated genes and proteins in macrophages from both species, it became clear that the macrophage response to IL-4 is more variable across models than the signatures identified in resting macrophages showing that M2 programs are sensitive to culture conditions. However, we were able to detect genes and proteins consistently regulated by IL-4 in human (Figure 4A-C) or mouse (Figure 4D-F) macrophages using the consensus that the markers were regulated in at least 60% of the samples. With this consensus, we identified 35 genes and 65 proteins (99 in total) regulated in the macrophages of both species (Figure 4G-H, Tables 2 and 3, and supplemental Table 3), but within this list, we found only 1 molecule that was regulated at both the mRNA and protein levels (Figure 4I). A detailed literature search performed with IPA ingenuity showed that approximately half of these genes/proteins have never been reported to be regulated by IL-4 or expressed in macrophages, whereas 18 genes/proteins have been and include several described alternative activation markers (Tables 2 and 3 and supplemental Table 3). We found few coincidences between the mRNA and protein levels in alternative activation (Figure 4C,F,I), mainly because of the fact that with proteomics we can only study highly abundant proteins, which restricts the chances for overlap with the microarray data, but also suggests that the cytokine modulates expression at different levels and that the main mRNA changes occur in genes with medium to low expression not detectable by proteomics.
List of genes regulated by IL-4 in both human and mouse macrophages as detected with microarrays
| Symbol . | EntrezGene name . | Fold change . | Reported in M2 macrophages . |
|---|---|---|---|
| KLF4 | Kruppel-like factor 4 (gut) | 5.25 | Liao12 |
| PTGS1 | Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 3.93 | Rehli25 |
| AQP9 | Aquaporin 9 | 3.69 | |
| IRF4 | Interferon regulatory factor 4 | 3.39 | El Chartouni14 |
| PDCD1LG2 | programmed cell death 1 ligand 2 | 3.49 | Huber17 |
| CISH | cytokine inducible SH2-containing protein | 3.09 | Rehli25 |
| CH25H | cholesterol 25-hydroxylase | 2.91 | |
| SOCS1 | suppressor of cytokine signaling 1 | 2.78 | Whyte13 |
| TGM2 | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | 2.59 | |
| RGL1 | ral guanine nucleotide dissociation stimulator-like 1 | 2.28 | |
| BCAR3 | breast cancer anti-estrogen resistance 3 | 2.01 | |
| GARS | glycyl-tRNA synthetase | 1.97 | |
| ALDH1A2 | aldehyde dehydrogenase 1 family, member A2 | 1.86 | |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | 1.73 | Pello19 |
| SLA | Src-like-adaptor | 1.69 | |
| SHMT2 | serine hydroxymethyltransferase 2 (mitochondrial) | 1.64 | |
| CD37 | CD37 molecule | −1.52 | |
| IFITM3 | interferon induced transmembrane protein 3 | −1.59 | |
| SLC9A9 | solute carrier family 9 (sodium/hydrogen exchanger), member 9 | −1.60 | |
| CXCL2 | chemokine (C-X-C motif) ligand 2 | −1.76 | Rehli25 |
| SPARC | secreted protein, acidic, cysteine-rich (osteonectin) | −1.79 | |
| KLHL6 | kelch-like 6 (Drosophila) | −1.80 | |
| APOC2 | apolipoprotein C-II | −1.80 | |
| DUSP6 | dual specificity phosphatase 6 | −1.81 | |
| SEPHS2 | selenophosphate synthetase 2 | −1.81 | |
| ALOX5AP | arachidonate 5-lipoxygenase-activating protein | −1.86 | Chaitidis47 |
| PTK2B | PTK2B protein tyrosine kinase 2 beta | −1.87 | |
| SORL1 | sortilin-related receptor, L(DLR class) A repeats containing | −1.94 | |
| S100A8 | S100 calcium binding protein A8 | −2.09 | Lugering48 |
| SLC40A1 | solute carrier family 40 (iron-regulated transporter), member 1 | −2.12 | |
| LPAR6 | lysophosphatidic acid receptor 6 | −2.23 | |
| AIF1 | allograft inflammatory factor 1 | −2.26 | |
| SEPP1 | selenoprotein P, plasma, 1 | −2.27 | |
| CD14 | CD14 molecule | −2.30 | Landmann49 |
| CYBB | cytochrome b-245, beta polypeptide | −2.32 | Balce50 |
| Symbol . | EntrezGene name . | Fold change . | Reported in M2 macrophages . |
|---|---|---|---|
| KLF4 | Kruppel-like factor 4 (gut) | 5.25 | Liao12 |
| PTGS1 | Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 3.93 | Rehli25 |
| AQP9 | Aquaporin 9 | 3.69 | |
| IRF4 | Interferon regulatory factor 4 | 3.39 | El Chartouni14 |
| PDCD1LG2 | programmed cell death 1 ligand 2 | 3.49 | Huber17 |
| CISH | cytokine inducible SH2-containing protein | 3.09 | Rehli25 |
| CH25H | cholesterol 25-hydroxylase | 2.91 | |
| SOCS1 | suppressor of cytokine signaling 1 | 2.78 | Whyte13 |
| TGM2 | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | 2.59 | |
| RGL1 | ral guanine nucleotide dissociation stimulator-like 1 | 2.28 | |
| BCAR3 | breast cancer anti-estrogen resistance 3 | 2.01 | |
| GARS | glycyl-tRNA synthetase | 1.97 | |
| ALDH1A2 | aldehyde dehydrogenase 1 family, member A2 | 1.86 | |
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | 1.73 | Pello19 |
| SLA | Src-like-adaptor | 1.69 | |
| SHMT2 | serine hydroxymethyltransferase 2 (mitochondrial) | 1.64 | |
| CD37 | CD37 molecule | −1.52 | |
| IFITM3 | interferon induced transmembrane protein 3 | −1.59 | |
| SLC9A9 | solute carrier family 9 (sodium/hydrogen exchanger), member 9 | −1.60 | |
| CXCL2 | chemokine (C-X-C motif) ligand 2 | −1.76 | Rehli25 |
| SPARC | secreted protein, acidic, cysteine-rich (osteonectin) | −1.79 | |
| KLHL6 | kelch-like 6 (Drosophila) | −1.80 | |
| APOC2 | apolipoprotein C-II | −1.80 | |
| DUSP6 | dual specificity phosphatase 6 | −1.81 | |
| SEPHS2 | selenophosphate synthetase 2 | −1.81 | |
| ALOX5AP | arachidonate 5-lipoxygenase-activating protein | −1.86 | Chaitidis47 |
| PTK2B | PTK2B protein tyrosine kinase 2 beta | −1.87 | |
| SORL1 | sortilin-related receptor, L(DLR class) A repeats containing | −1.94 | |
| S100A8 | S100 calcium binding protein A8 | −2.09 | Lugering48 |
| SLC40A1 | solute carrier family 40 (iron-regulated transporter), member 1 | −2.12 | |
| LPAR6 | lysophosphatidic acid receptor 6 | −2.23 | |
| AIF1 | allograft inflammatory factor 1 | −2.26 | |
| SEPP1 | selenoprotein P, plasma, 1 | −2.27 | |
| CD14 | CD14 molecule | −2.30 | Landmann49 |
| CYBB | cytochrome b-245, beta polypeptide | −2.32 | Balce50 |
Included markers were regulated in at least 60% of the samples and the fold change represents the average of the samples that were scored as changed.
List of proteins regulated by IL-4 in both human and mouse macrophages as detected with proteomics
| Symbol . | EntrezGene name . | Fold change . | Reported in M2 macrophages . |
|---|---|---|---|
| MRC1 | mannose receptor, C type 1 | 10.76 | Stein15 |
| NDUFA4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa | 8.08 | |
| CBR1 | carbonyl reductase 1 | 5.87 | |
| KRT1 | keratin 1 | 5.86 | |
| EIF4E | eukaryotic translation initiation factor 4E | 4.34 | |
| SERPINB6 | serpin peptidase inhibitor, clade B (ovalbumin), member 6 | 4.09 | |
| ATP5C1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 3.82 | |
| TFRC | transferrin receptor (p90, CD71) | 3.62 | Weiss16 |
| DPYSL2 | dihydropyrimidinase-like 2 | 3.40 | |
| OTUB1 | OTU domain, ubiquitin aldehyde binding 1 | 2.81 | |
| ACAA2 | acetyl-CoA acyltransferase 2 | 2.71 | |
| RPS23 | ribosomal protein S23 | 2.67 | |
| ESD | esterase D | 2.56 | |
| CYFIP1 | cytoplasmic FMR1 interacting protein 1 | 2.36 | |
| PPT1 | palmitoyl-protein thioesterase 1 | 2.32 | |
| GRN | granulin | 2.29 | |
| ANXA4 | annexin A4 | 2.27 | |
| RPL4 | ribosomal protein L4 | 2.19 | |
| HSPA9 | heat shock 70kDa protein 9 (mortalin) | 2.12 | |
| ACTN1 | actinin, alpha 1 | 1.96 | |
| NCL | nucleolin | 1.95 | |
| UBA1 | ubiquitin-like modifier activating enzyme 1 | 1.95 | |
| RPS15 | ribosomal protein S15 | 1.89 | |
| VPS35 | vacuolar protein sorting 35 homolog (S. cerevisiae) | 1.86 | |
| UBE2V1 | ubiquitin-conjugating enzyme E2 variant 1 | 1.68 | |
| ITGAM | integrin, alpha M (complement component 3 receptor 3 subunit) | 1.65 | |
| DDOST | dolichyl-diphosphooligosaccharide–protein glycosyltransferase | −1.58 | |
| HMOX2 | heme oxygenase (decycling) 2 | −1.63 | |
| COTL1 | coactosin-like 1 (Dictyostelium) | −1.64 | |
| PTPRC | protein tyrosine phosphatase, receptor type, C | −1.65 | |
| GUSB | glucuronidase, beta | −1.67 | |
| GRB2 | growth factor receptor-bound protein 2 | −1.71 | |
| PRDX1 | peroxiredoxin 1 | −1.71 | |
| COX6B1 | cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) | −1.73 | |
| CTSA | cathepsin A | −1.74 | |
| CLTA | clathrin, light chain A | −1.75 | |
| CTSS | cathepsin S | −1.75 | Balce50 (up-regulated) |
| EIF5A | eukaryotic translation initiation factor 5A | −1.75 | |
| CFL1 | cofilin 1 (non-muscle) | −1.77 | |
| GLB1 | galactosidase, beta 1 | −1.78 | |
| PPIA | peptidylprolyl isomerase A (cyclophilin A) | −1.79 | |
| DBI | diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) | −1.90 | |
| S100A11 | S100 calcium binding protein A11 | −1.95 | |
| PRDX3 | peroxiredoxin 3 | −1.96 | |
| CSTB | cystatin B (stefin B) | −2.03 | |
| GSR | glutathione reductase | −2.03 | |
| TBCB | tubulin folding cofactor B | −2.04 | |
| PYCARD | PYD and CARD domain containing | −2.17 | |
| TXNRD1 | thioredoxin reductase 1 | −2.24 | |
| ESYT1 | extended synaptotagmin-like protein 1 | −2.35 | |
| RPS10 | ribosomal protein S10 | −2.47 | |
| SNX2 | sorting nexin 2 | −2.49 | |
| TAPBP | TAP binding protein (tapasin) | −2.61 | |
| CTSD | cathepsin D | −2.71 | |
| GLS | glutaminase | −2.73 | |
| LRPAP1 | low density lipoprotein receptor-related protein associated protein 1 | −2.81 | |
| PLD3 | phospholipase D family, member 3 | −2.84 | |
| APOE | apolipoprotein E | −2.92 | |
| HIST1H1C | histone cluster 1, H1c | −3.28 | |
| HK3 | hexokinase 3 (white cell) | −3.28 | |
| NUCB1 | nucleobindin 1 | −3.39 | |
| PSMD2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | −3.68 | |
| NAGA | N-acetylgalactosaminidase, alpha- | −4.35 | |
| ADSL | adenylosuccinate lyase | −5.84 |
| Symbol . | EntrezGene name . | Fold change . | Reported in M2 macrophages . |
|---|---|---|---|
| MRC1 | mannose receptor, C type 1 | 10.76 | Stein15 |
| NDUFA4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa | 8.08 | |
| CBR1 | carbonyl reductase 1 | 5.87 | |
| KRT1 | keratin 1 | 5.86 | |
| EIF4E | eukaryotic translation initiation factor 4E | 4.34 | |
| SERPINB6 | serpin peptidase inhibitor, clade B (ovalbumin), member 6 | 4.09 | |
| ATP5C1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 3.82 | |
| TFRC | transferrin receptor (p90, CD71) | 3.62 | Weiss16 |
| DPYSL2 | dihydropyrimidinase-like 2 | 3.40 | |
| OTUB1 | OTU domain, ubiquitin aldehyde binding 1 | 2.81 | |
| ACAA2 | acetyl-CoA acyltransferase 2 | 2.71 | |
| RPS23 | ribosomal protein S23 | 2.67 | |
| ESD | esterase D | 2.56 | |
| CYFIP1 | cytoplasmic FMR1 interacting protein 1 | 2.36 | |
| PPT1 | palmitoyl-protein thioesterase 1 | 2.32 | |
| GRN | granulin | 2.29 | |
| ANXA4 | annexin A4 | 2.27 | |
| RPL4 | ribosomal protein L4 | 2.19 | |
| HSPA9 | heat shock 70kDa protein 9 (mortalin) | 2.12 | |
| ACTN1 | actinin, alpha 1 | 1.96 | |
| NCL | nucleolin | 1.95 | |
| UBA1 | ubiquitin-like modifier activating enzyme 1 | 1.95 | |
| RPS15 | ribosomal protein S15 | 1.89 | |
| VPS35 | vacuolar protein sorting 35 homolog (S. cerevisiae) | 1.86 | |
| UBE2V1 | ubiquitin-conjugating enzyme E2 variant 1 | 1.68 | |
| ITGAM | integrin, alpha M (complement component 3 receptor 3 subunit) | 1.65 | |
| DDOST | dolichyl-diphosphooligosaccharide–protein glycosyltransferase | −1.58 | |
| HMOX2 | heme oxygenase (decycling) 2 | −1.63 | |
| COTL1 | coactosin-like 1 (Dictyostelium) | −1.64 | |
| PTPRC | protein tyrosine phosphatase, receptor type, C | −1.65 | |
| GUSB | glucuronidase, beta | −1.67 | |
| GRB2 | growth factor receptor-bound protein 2 | −1.71 | |
| PRDX1 | peroxiredoxin 1 | −1.71 | |
| COX6B1 | cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) | −1.73 | |
| CTSA | cathepsin A | −1.74 | |
| CLTA | clathrin, light chain A | −1.75 | |
| CTSS | cathepsin S | −1.75 | Balce50 (up-regulated) |
| EIF5A | eukaryotic translation initiation factor 5A | −1.75 | |
| CFL1 | cofilin 1 (non-muscle) | −1.77 | |
| GLB1 | galactosidase, beta 1 | −1.78 | |
| PPIA | peptidylprolyl isomerase A (cyclophilin A) | −1.79 | |
| DBI | diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) | −1.90 | |
| S100A11 | S100 calcium binding protein A11 | −1.95 | |
| PRDX3 | peroxiredoxin 3 | −1.96 | |
| CSTB | cystatin B (stefin B) | −2.03 | |
| GSR | glutathione reductase | −2.03 | |
| TBCB | tubulin folding cofactor B | −2.04 | |
| PYCARD | PYD and CARD domain containing | −2.17 | |
| TXNRD1 | thioredoxin reductase 1 | −2.24 | |
| ESYT1 | extended synaptotagmin-like protein 1 | −2.35 | |
| RPS10 | ribosomal protein S10 | −2.47 | |
| SNX2 | sorting nexin 2 | −2.49 | |
| TAPBP | TAP binding protein (tapasin) | −2.61 | |
| CTSD | cathepsin D | −2.71 | |
| GLS | glutaminase | −2.73 | |
| LRPAP1 | low density lipoprotein receptor-related protein associated protein 1 | −2.81 | |
| PLD3 | phospholipase D family, member 3 | −2.84 | |
| APOE | apolipoprotein E | −2.92 | |
| HIST1H1C | histone cluster 1, H1c | −3.28 | |
| HK3 | hexokinase 3 (white cell) | −3.28 | |
| NUCB1 | nucleobindin 1 | −3.39 | |
| PSMD2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | −3.68 | |
| NAGA | N-acetylgalactosaminidase, alpha- | −4.35 | |
| ADSL | adenylosuccinate lyase | −5.84 |
Included markers were regulated in at least 60% of the samples and the fold change represents the average of the samples that were scored as changed.
With the consensus used, only one gene was detected as being regulated by IL-4 at both the protein and mRNA levels in the majority of culture models and species (Figure 4I). We analyzed the specificity of M2 markers by comparing at the whole-genome level human IL-4–treated macrophages with macrophages treated with the Th1 cytokines IFN-γ and TNF, the maturation related cytokine M-CSF, and the deactivating cytokine IL-10. This analysis showed great specificity for the majority of our markers and the IL-4 profile in general (Figure 4J). We hypothesize that this may be true at protein level as well. The 35 genes and 65 proteins consistently regulated in both species can be organized into networks, 3 for mRNA and 4 for proteins (Figure 4K). Overrepresented pathways detected include transmigration of cells, proliferation of T lymphocytes, homeostasis of iron ions, and efflux of phospholipids (supplemental Table 3). The lists include genes previously associated with alternative activation, including signaling modulators such as Kruppel-like factor 4 (KLF4),12 suppressor of cytokine signaling 1 (SOCS1),13 IFN regulatory factor 4 (IRF4),14 and membrane molecules such as the mannose receptor (MRC1),15 the transferrin receptor (TFRC),16 and programmed cell death 1 ligand 2 (PDCD1LG2).17 The networks also contain potential novel markers, for example, ANXA4 and TGM2. TGM2 represents the only molecule with regulation detected by both approaches in both species using the criteria chosen. Based on the networks, we identified known regulators of M2 activation such as PI3K, AKT, and NF-κB1,18 (Figure 4K). In parallel with known regulators, potential new regulators also appeared, including signaling molecules and soluble factors such as BMP4, TGFB1, insulin, and IFNs (Figure 4K). Overrepresented transcription factor analysis detected the well-known STAT6 and also HNF4A, STAT3, HOXA10, PPARA, and MYC, described recently as a major regulator of IL-4 profiles19 (Figure 4K and supplemental Table 3).
For the M2 signature, a more detailed view of the human or mouse macrophage phenotype can be achieved by taking into consideration species-specific genes. With an emphasis on the human system, we investigated the 293 genes consistently regulated in all human samples (173 up-regulated and 120 down-regulated) to find 22 overrepresented networks, 298 canonical pathways, and 1246 biologic and toxicological pathways (supplemental Table 4). The canonical pathway G-protein coupled receptor signaling had the greatest number of genes affected by M2 activation. At the protein level, we investigated the 194 proteins consistently regulated (92 up-regulated and 102 down-regulated) to find 12 overrepresented networks and 160 canonical pathways. At the protein level, the major functional networks seem to be controlled by MYC, STAT6, and PPARD according to IPA analysis. Among the networks, lipid metabolism is outstanding, with down-regulation of genes related to metabolism/accumulation of triacylglycerol and cholesterol such as apolipoprotein C1, E, and H, as well as up-regulation of transporters such as fatty acid binding protein 4 (FABP4) and ANXA11.
TGM2 is a novel functionally relevant alternative activation gene
Among several genes detected as being conserved at either the mRNA or protein level in both species, the only gene regulated by IL-4 in the majority of conditions that was detectable with microarrays and proteomics was TGM2 (Figure 4I,K). TGM2 is an enzyme associated with broad biologic functions, including cross-linking of cellular proteins, phagocytosis of apoptotic cells, and extracellular matrix generation.20 This molecule is not only expressed in macrophages but also in epithelial cells. To confirm IL-4–mediated up-regulation of TGM2 protein levels in vitro in different human and murine macrophage culture models, we performed Western blot analysis and compared TGM2 expression with the expression of other proteins detected as being up-regulated after M2 activation according to our proteomics analysis (Figure 5A-B). We included MRC1 as M2 marker previously described in the human and mouse, TFRC as a marker previously described in mouse M2 macrophages,16 and the potential novel marker ANXA4.
Conserved macrophage alternative activation gene and protein signatures reveal TGM2 as the most stable marker. (A) Up-regulation of TGM2 protein by M2 activation can be confirmed by Western blot in different human macrophage culture models (M-CSF, GM-CSF, and AS). Analysis of Mannose receptor (MRC1) and Actin expression were used as alternative activation and loading controls, respectively. In addition, expression of transferrin receptor (TFRC) and annexin A4 (ANXA4) was determined. (B) In mouse macrophage protein samples, IL-4–induced Tgm2 up-regulation can be confirmed in thioglycollate- (mThioMΦ) and Biogel (mBioMΦ)–elicited peritoneal macrophages and BM-derived macrophages (mBMMΦ). Mrc1, Tfrc, Anxa4, and Actin expression was analyzed in parallel. (C-D) Time-course analysis of TGM2/Tgm2 in human AS macrophages (C) and mouse ThioMΦ (D). (E-F) Transglutaminase activity was determined in human AS macrophages (E; n = 3) and mouse ThioMΦ (F; n = 4). *P < 0.05. Shown are means ± SEM. Transglutaminase activity was normalized to the amount of protein and to the unstimulated control.
Conserved macrophage alternative activation gene and protein signatures reveal TGM2 as the most stable marker. (A) Up-regulation of TGM2 protein by M2 activation can be confirmed by Western blot in different human macrophage culture models (M-CSF, GM-CSF, and AS). Analysis of Mannose receptor (MRC1) and Actin expression were used as alternative activation and loading controls, respectively. In addition, expression of transferrin receptor (TFRC) and annexin A4 (ANXA4) was determined. (B) In mouse macrophage protein samples, IL-4–induced Tgm2 up-regulation can be confirmed in thioglycollate- (mThioMΦ) and Biogel (mBioMΦ)–elicited peritoneal macrophages and BM-derived macrophages (mBMMΦ). Mrc1, Tfrc, Anxa4, and Actin expression was analyzed in parallel. (C-D) Time-course analysis of TGM2/Tgm2 in human AS macrophages (C) and mouse ThioMΦ (D). (E-F) Transglutaminase activity was determined in human AS macrophages (E; n = 3) and mouse ThioMΦ (F; n = 4). *P < 0.05. Shown are means ± SEM. Transglutaminase activity was normalized to the amount of protein and to the unstimulated control.
We found that TGM2 was indeed modulated in all samples (Figure 5A-B; full Western blot images including molecular weight standards are shown in supplemental Figure 1). Interestingly, the expression of TGM2 in murine BM-derived macrophages (BMMΦ) appeared to be dependent on the cell culture conditions (supplemental Figure 2). Although the known M2 marker MRC1 was also up-regulated after IL-4 stimulation in all culture models (Figure 5A-B), the other markers tested were more restricted: TFRC was up-regulated in all mouse macrophage models (Figure 5B),but in only one human macrophage model, and was actually down-regulated in M2-activated GM-CSF–derived human macrophages (Figure 5A). ANXA4 was not up-regulated in GM-CSF–derived human macrophages and thioglycollate-elicited peritoneal mouse macrophages (Figure 5A-B). We conclude from our analysis that TGM2 represents a universal M2 marker. Time-course analysis showed that its expression in macrophages increases over time and is most prominent after 48 hours of IL-4 stimulation (Figure 5C-D). When we quantified transglutaminase activity in macrophage cell lysates, we also found that M2 activation leads to significantly increased transglutaminase activity in human and mouse macrophages (Figure 5E-F).
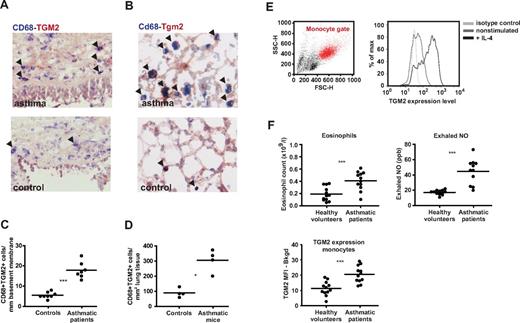
We undertook an analysis of TGM2 expression in vivo in asthma, a chronic allergic Th2 pathology characterized by high levels of IL-4 and IL-13. In a mouse model, M2 macrophages have been shown to contribute to allergic lung inflammation.21 We first analyzed TGM2 and MRC1 expression in human macrophages from bronchial biopsies (Figure 6A,C). The human biopsies used in this study were previously characterized as being rich in M2 macrophages in an independent study using MRC1 as a marker.22 Our staining with TGM2, MRC1, and CD68 corroborated the previous results and showed abundant double-positive cells in asthmatic samples compared with healthy donor biopsies (Figure 6A MRC1 and data not shown). TGM2 was not restricted to CD68+ cells, but was expressed more broadly. We observed a similar pattern in lungs isolated from mice exposed to house dust mite (HDM) to induce allergic airway inflammation; TGM2 was expressed diffusely with strong staining in CD68+ macrophages (Figure 6B). TGM2 can therefore serve as an M2 marker when used in combination with macrophage-specific antibodies. To analyze this further, we quantified the number of TGM2/CD68 double-positive cells in lung sections from human asthmatic patients and healthy controls (Figure 6C), as well as from mice exposed to HDM, compared with control mice (Figure 6D). In both cases, we detected a statistically significant increase in the number of TGM2+ macrophages at the basement membrane (Figure 6C-D).
TGM2 is a conserved M2 marker that is highly expressed in asthma. (A-B) TGM2 and CD68 expression was analyzed in bronchial biopsies of asthma patients (A) and lung sections of mice with asthma (B). Shown are representative sections stained for CD68 (blue) and TGM2 (red). Arrowheads indicate double-positive cells. (C) Airway wall tissue of asthmatic patients contains more TGM2+ macrophages than that of healthy controls. ***P < .0001. (D) Lung tissue of mice with asthma contains more TGM2+ macrophages than that of control mice. *P = .0286 by 2-tailed unpaired t test. (E) TGM2 is also expressed in monocytes as detected by FACS. Monocytes were gated according to their forward/side scatter (red); monocytes were left untreated or stimulated with IL-4. (F) Asthmatic patients showed significantly increased eosinophil counts (***P < .0001) and exhaled NO levels (***P = .0003). In blood samples of these patients, TGM2 levels in circulating monocytes were significantly higher in asthmatic patients than in healthy controls (***P = .0002 by 2-tailed unpaired t test with Welch correction). TGM2 MFI − bkgd = mean fluorescence intensity (MFI) detected with a primary polyclonal antibody subtracted by the MFI detected with control Igs. Eos indicates eosinophil counts.
TGM2 is a conserved M2 marker that is highly expressed in asthma. (A-B) TGM2 and CD68 expression was analyzed in bronchial biopsies of asthma patients (A) and lung sections of mice with asthma (B). Shown are representative sections stained for CD68 (blue) and TGM2 (red). Arrowheads indicate double-positive cells. (C) Airway wall tissue of asthmatic patients contains more TGM2+ macrophages than that of healthy controls. ***P < .0001. (D) Lung tissue of mice with asthma contains more TGM2+ macrophages than that of control mice. *P = .0286 by 2-tailed unpaired t test. (E) TGM2 is also expressed in monocytes as detected by FACS. Monocytes were gated according to their forward/side scatter (red); monocytes were left untreated or stimulated with IL-4. (F) Asthmatic patients showed significantly increased eosinophil counts (***P < .0001) and exhaled NO levels (***P = .0003). In blood samples of these patients, TGM2 levels in circulating monocytes were significantly higher in asthmatic patients than in healthy controls (***P = .0002 by 2-tailed unpaired t test with Welch correction). TGM2 MFI − bkgd = mean fluorescence intensity (MFI) detected with a primary polyclonal antibody subtracted by the MFI detected with control Igs. Eos indicates eosinophil counts.
The in situ study of such biomarkers in human tissue macrophages is limited and invasive compared with blood monocytes. It is unclear if local M2 activation observed in tissue macrophages can be recapitulated in monocytes because the conventional M2 marker MRC1 is restricted to the mature macrophage forms. In the present study, we used FACS analysis to determine whether monocytes expressed TGM2 and whether this was regulated by IL-4. Up-regulation of TGM2 by IL-4 was not only detected in human circulating monocytes, but also in total blood and within the PBMC fraction (Figure 6E). To establish whether, like their tissue counterparts, monocytes could demonstrate alternative activation in vivo, we examined TGM2 expression in monocytes from both healthy controls and asthma patients. The asthma patients were confirmed to exhibit significantly increased eosinophil counts and exhaled NO levels compared with healthy controls (Figure 6F). TGM2 levels were significantly increased in monocytes from asthma patients and a significant correlation was detected for monocytic TGM2 expression, eosinophil counts, and exhaled NO levels (not shown). We conclude that TGM2 is a potential biomarker of M2 activation and asthma in circulating blood, where conventional M2 markers cannot be applied.
Discussion
In the present study, we investigated gene and protein repertoires in human and mouse macrophages under resting conditions and on stimulation with the Th2 cytokine IL-4. Because existing studies on macrophage gene expression in response to INF-γ and lipopolysaccharide (M1) or IL-4 (M2) activation are based on selected macrophage culture models9,17,23-25 and there are only limited data available in the human system, we included several prototypic human and mouse macrophage models in our analysis and focused on IL-4 effects. Our study was designed to understand common markers in routinely used models for M2 activation. The study design may mask coincidences that could be found between mouse and humans if all parameters were kept constant. Despite stressing the differences between the models, our results support a great degree of conservation of molecular macrophage signatures among different species and models. The conservation in resting macrophages exceeded that found in response to M2 activation, which appears to be tuned by additional factors, as reflected in our culture models by cellular source, maturation stimulus, culture surface, and other factors. Accordingly, care has to be taken when comparing results generated using different experimental protocols. We hypothesize that the variations detected in vitro systems will be verified in vivo because of different tissue environments and degrees of inflammation.
Using in vitro and ex vivo models, we have defined herein a set of molecules highly expressed in resting macrophages from the human and mouse, representing a consistent 87-gene macrophage molecular signature. The proposed signature is consistent with a revised definition of macrophages, which are heterogeneous cells that are difficult to define by single markers. The 87-gene selection is biased by our arbitrary cutoff, but identifies the best expressed transcripts and proteins and provides a more comprehensive view of the basic contents of the macrophage. The combined signature appears to be specific to macrophages and not lymphocytes, but its expression in other myeloid cells requires further studies focused on specificity and not a general repertoire. The list extends and gathers scattered information about macrophage molecules, only 50% of which have been associated with these cells in independent publications and many of these only with macrophage cell lines and not primary or tissue macrophages. With the current macrophage signature, a macrophage functional picture begins to emerge. The list includes, for example, ANXA1 and ANXA2, with Annexin 1 having potent anti-inflammatory effects26 and ANXA2 being proposed to promote inflammation.27 Both annexins appear to be involved in the phagocytosis of apoptotic cells28 ; also involved is Galectin-3 (LGALS3), a β-galactoside–binding lectin known to be abundantly expressed in macrophages.29 Galectin-3 plays a regulatory role in inflammation and fibrosis30 and is critically involved in alternative macrophage activation because of a positive feedback loop leading to sustained PI3K activation.31 Additional examples of identified markers with functional relevance include genes involved in redox balance. Peroxiredoxin 1 (PRDX1) is an antioxidant and molecular chaperone, but is also able to trigger TLR4 signaling and activation of NF-κB, resulting in the secretion of TNF-α and IL-6.32 Two important selenoproteins involved in cellular antioxidative defense systems and redox control, glutathione peroxidase (GPX1) and thioredoxin reductase (TXNRD1), are also present.33 GPX1, and selenoproteins in general, have recently been linked to the production of endogenous activators that mediate the PPARγ-dependent switch from the M1 to the M2 phenotype in the presence of IL-4.34 Another gene associated with redox-related functions is glutathione S-transferase ω1 (GSTO1). This protein is highly expressed in alveolar macrophages,35 and GSTO1 polymorphisms have been associated with increased atherosclerosis, macrophage infiltration, and inflammation.36 Further general functions overrepresented in the 87-gene signature are adhesion and chemotaxis, for example, integrin β 2 (ITGB2, CD18), which can mediate cell adhesion,37 and S100A10 and S100A4, which regulate chemotaxis.38,39 Because our 87-gene signature contains many genes with known specialized macrophage functions, we hypothesize that the analysis of other genes within the signature may reveal previously unknown macrophage functions and/or specializations.
In addition to the consistent macrophage signature defined for resting macrophages, we also identified herein genes and proteins consistently regulated in alternatively activated macrophages from both the human and mouse, including known and previously unknown molecules. The IL-4 profiles are very specific, as demonstrated by comparison with regulation of the respective genes by other prototypic cytokines of opposed nature, such as IFN-γ, or even if compared with stimuli inappropriately termed M2, such as M-CSF or IL-10. Our data may provide a better understanding of IL-4–induced alternative macrophage activation in humans and its overlap and differences with the widely used mouse macrophage models.
For human macrophages, we found that the G protein–coupled receptor signaling pathway had the greatest number of genes affected by M2 activation. Heterotrimeric G proteins are key players in transmembrane signaling coupling a multitude of receptors to enzymes, channel proteins, and other molecules. Our data show that in human macrophages, M2 activation leads to the up-regulation of several GPCRs, including cysteinyl-leukotriene receptor 1 (CYSLTR1), prostaglandin E receptor 2 (PTGER2), G protein–coupled receptor 35 (GPR35), and G protein–coupled receptor 183 (GPR183). CYSLTR1 has been implicated in asthma and was found previously to be associated with IL-4.40 The antagonist of human CYSLTR1, trade name Singulair, is currently in use for the treatment of asthma and others are being developed (eg, Zeneca ZD 3523 is in phase 3 clinical trials). GPCRs that were down-regulated by IL-4 include complement component 3a receptor 1 (C3AR1) and prostaglandin E receptor 4 (PTGER4), both previously shown to be down-regulated by Th2 cytokines.41,42
Many of the GPCRs modulated by M2 activation recognize eicosanoids or other lipid products that represent established mediators of human bronchial asthma.43 Consistent with the modulated receptor expression, our network analysis shows that IL-4 stimulation generally affects arachidonic acid metabolism and eicosanoid signaling pathways. Down-regulation of ALOX5, ALOX5AP, and GPX3 may lead to reduced levels of 5-HPETE, 5-HETE, 15(S)-HETE, and LTA4, whereas production of PGG2 and PGF2, but not PGE2, may be induced via up-regulation of PTGS, CBR1, and simultaneous down-regulation of PTGES2.
Although in the present study, we concentrated on TGM2, we propose a wider panel of genes to characterize human alternative activation. SOCS1, FCER2, and CCL17 have been previously confirmed, but many are newly associated with IL-4 and macrophages, including aldehyde dehydrogenase 1 A2 (ALDH1A2), receptor (G protein–coupled) activity modifying protein 1 (RAMP1), and guanylate cyclase activator 1A (GUCA1A).
The multifunctional enzyme TGM2 provides a relatively consistent activation biomarker for mouse and human M2 macrophages and monocytes. TGM2 has been implicated in functions that fit with the M2 macrophage role, such as matrix remodeling linked to wound healing, fibrosis, and metastasis.20,44 TGM2 can also activate TGF-β, inducing anti-inflammatory and profibrotic cytokines.20 From our analysis of TGM2 expression in macrophages and monocytes from asthma patients, it can be concluded that TGM2 is clearly expressed in human alternatively activated macrophages in vivo. Although TGM2 is regulated only by IL-4 of all cytokines considered in this study, other stimuli such as pathogens or TLRs require investigation. Until now, it was believed that M2 activation happens in situ and not systemically, and it has been demonstrated that M2 macrophages can arise from local proliferation within the affected tissue rather than from the recruitment of monocytes from the blood.45,46 TGM2 is potentially useful because it can be studied in circulating monocytes, making it possible to study the relationship between local and systemic alternative activation. Our studies show that a combination of markers such as TGM2, MRC1, and CD68 may help to clarify alternative activation heterogeneity in tissue. We hypothesize that TGM2 used in combination with other M2 markers may provide a tool to stratify asthmatic patients by FACS analysis of fresh blood or sputum.
In conclusion, in the present study, we have defined macrophage gene and protein signatures for human and mouse macrophages. The resting and alternative gene signatures provide candidates with which to detect the presence and activation of these cells in tissues and will help to uncover new functions and provide a platform for further studies in vitro and in situ.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Medical Research Council and the German Research Foundation. F.O.M. was supported by the British Heart Foundation (PG/09/076). B.N.M. and C.D. were supported by grant 3.2.10.056 from the Netherlands Asthma Foundation. M.F., M.L., and A.M. were supported by Italian Association for Cancer Research (AIRC), Regione Lombardia (LIIN project), and the European Community's Seventh Framework Programme [FP7-2007-2013] under grant agreement HEALTH-F4-2011-281608 (TIMER).
Authorship
Contribution: F.O.M. analyzed the data; F.O.M. and L.H. designed and performed the experiments and wrote the manuscript; R.M., A.V., B.N.M, C.D., and V.C.J. performed the experiments; B.T. performed the mass spectrometry; M.F., N.A.K., J.H., D.R.G., M.L., and A.M. supervised the research; A.C and L.P.H organized and collected the cohort of human asthmatic and healthy samples; N.H.T.H. collected the human lung samples; and S.G. supervised the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fernando O. Martinez, Sir William Dunn School of Pathology, University of Oxford, Oxford, United Kingdom, e-mail: fernando.martinezestrada@path.ox.ac.uk; Laura Helming, Institute for Medical Microbiology, Immunology and Hygiene, Technische Universität München, Munich, Germany; e-mail: laura.helming@mikrobio.med.tum.de; or Siamon Gordon, Sir William Dunn School of Pathology, University of Oxford, South Parks Road, Oxford OX1 3RE, United Kingdom; e-mail: siamon.gordon@path.ox.ac.uk.
References
Author notes
F.O.M. and L.H. contributed equally to this work.

![Figure 1. Human and mouse macrophage expression profiles are highly conserved at the mRNA and protein levels. (A) In this study, we included variations of common human macrophage (hMΦ) models: M-CSF– or autologous serum (AS)–differentiated macrophages cultured on tissue culture plastic (TCP) or bacteriological plastic (BP). For murine macrophages (mMΦ), we included BM-derived macrophages (BMMΦ) and 2 primary macrophage populations isolated ex vivo: peritoneal macrophages elicited by injection of Biogel (BioMΦ) or inflammatory macrophages elicited by injection of thioglycollate (ThioMΦ); macrophages were cultured on different surfaces (TCP, BP, or Permanox [PP]). (B) comparing basal transcriptomes of all human macrophage culture models, we identified 489 transcripts consistently detected in the first 1000 expression-ranked genes. A total of 459 transcripts were detected in common for all murine culture models; 231 transcripts, approximately 50% of each species consensus list, were shared by human and mouse macrophages. (C) Using proteomics, we investigated the protein content of 4 macrophage models: human macrophages differentiated in the presence of M-CSF or AS and murine BMMΦ and ThioMΦ. We detected 977 and 1038 proteins in human and mouse macrophages, respectively. Of these, 513 were detected in macrophages of both species. (D) Overlap between the conserved mRNA and protein signatures; 231 highly expressed genes and 513 detected proteins, respectively, amounted to 87 genes.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/9/10.1182_blood-2012-06-436212/4/m_zh89991302450001.jpeg?Expires=1765888767&Signature=iBF7Yo8Ck90SctjRuBgTU~H8Fc55b12z~z0FB8wLLDwi1tDmyZL-PD48C9N16uGby21Z8Q0g5MpX-JQfGWO6uWHyQKppMTBmOU2eFG0-bGQTKlWXUlP3eV2KmjDmhbE3jm9FKtrfNLV9WMbIxn2044uy2oXMrx9Yx50MKyuBn6Kp6WRxt2fLBNi3aFVGGFZHEFynmkhMkwom5aYjETI5yFXyVd9aEafJs2GJhrfaL3Y2lZNFLU-NF8u8WwdvccPUXmDCesOYkOkOkYQmdGXS57YZAKSrHlWHUquqpu0dXwnfOYPr0CHOJnCvOIwQ2eqtg3QUv~dg-pUo4zWDr43Rmw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)