Key Points
Single-cell heterogeneity, rather than lineage reprogramming, explains the remarkable complexity and functional diversity of human Tregs.
Altered homeostasis of Treg subpopulations in patients developing acute graft-versus-host disease.
Abstract
Understanding the heterogeneity of human CD4+FOXP3+ regulatory T cells (Tregs) and their potential for lineage reprogramming is of critical importance for moving Treg therapy into the clinics. Using multiparameter single-cell analysis techniques, we explored the heterogeneity and functional diversity of human Tregs in healthy donors and in patients after allogeneic hematopoietic stem cell transplantation (alloHSCT). Human Tregs displayed a level of complexity similar to conventional CD4+ effector T cells with respect to the expression of transcription factors, homing receptors and inflammatory cytokines. Single-cell profiling of the rare Treg producing interleukin-17A or interferon-γ showed an overlap of gene expression signatures of Th17 or Th1 cells and of Tregs. To assess whether Treg homeostasis is affected by an inflammatory and lymphopenic environment, we characterized the Treg compartment in patients early after alloHSCT. This analysis suggested a marked depletion of Treg with a naive phenotype in patients developing acute graft-versus-host disease, compared with tolerant patients. However, single-cell profiling showed that CD4+FOXP3+ T cells maintain the Treg gene expression signature and Treg-suppressive activity was preserved. Our study establishes that heterogeneity at the single-cell level, rather than lineage reprogramming of CD4+FOXP3+ T cells, explains the remarkable complexity and functional diversity of human Tregs.
Introduction
CD4+FOXP3+ regulatory T cells (Tregs) are essential in maintaining peripheral self-tolerance and preventing inflammatory disease.1-4 The perspective of harnessing the potent immunosuppressive activity of Treg for clinical applications (such as prevention or treatment of graft-versus-host disease [GVHD] after allogeneic hematopoietic stem cell transplantation [alloHSCT], improved tolerance after solid organ transplantation and treatment of autoimmune diseases5 ) has sparked numerous studies that revealed that Tregs are not phenotypically or functionally homogeneous.6-9 Distinct human Treg subsets have been described by analyzing cell-surface or intracellular markers such as ICOS, CD45RA, Helios, HLA-DR, and chemokine receptors.10-16 It is unclear how the Treg subsets so defined relate to each other, and which subset may be more suitable for therapeutic applications.
A further concern comes from studies showing that Tregs may convert into CD4+ effector T cells with proinflammatory functions, in particular, in an inflammatory environment.17-23 However, the notion of Treg reprogramming remains controversial, with a study demonstrating a remarkable stability of Tregs even under highly inflammatory conditions.24 Genetic fate mapping identified a small subpopulation of Foxp3− conventional T cells (Tconvs) that transiently upregulates Foxp3 and could explain the emergence of “ex-Treg” cells.25
The role of Tregs in allogeneic bone marrow transplantation was initially examined in murine models of GVHD. Depletion of CD25+ cells from splenocytes increased GVHD severity and lethality after major histocompatibility complex–mismatched bone marrow transplantation, providing evidence that Tregs ameliorate the detrimental effects of alloreactive donor effector T cells in the host.26,27 Treg transplantation at high ratios could completely protect recipient mice from GVHD development, even when lethal doses of Tconv cells were administered.26-28 Clinical studies have demonstrated reduced CD4+FOXP3+ T-cell frequencies in the blood or the in mucosal tissues of patients with GVHD.29,30 Recently, the adoptive transfer of freshly isolated or in vitro–expanded Tregs was used in early clinical trials.31-34 However, the dynamics of CD4+FOXP3+ T cells and their heterogeneity early after alloHSCT, at the onset of acute GVHD (aGVHD), have never been studied because only minute numbers of cells are available for analyses. This analysis is important to understand the pathophysiology of the disease.35
We have used multiparameter single-cell analyses to explore the heterogeneity and functional diversity of human Tregs in healthy donors, and in patients developing aGVHD after alloHSCT. Our results show that the human Treg compartment displays a similar level of complexity and functional heterogeneity as conventional CD4+ effector T cells. Analysis of Treg homeostasis at the time of engraftment after alloHSCT revealed that overall frequency of CD4+FOXP3+ T cells, single-cell gene expression profiles, and suppressive activity of Tregs isolated from aGVHD patients were preserved. However, the Treg compartment in patients developing aGVHD showed a marked depletion of CD45RA+HLA-DR− “naive” Tregs, compared with tolerant patients.
Methods
Patients
This study was performed in compliance with the Declaration of Helsinki after approval by the ethics committee of the Hospital Saint Louis (Paris, France). Written informed consent was obtained from all participants. Patients’ characteristics are presented in supplemental Table 2 (available on the Blood website). Umbilical cord blood was collected from normal deliveries after maternal informed consent according to approved institutional guidelines (Assistance Publique–Hôpitaux de Paris Cord Blood Bank, Paris, France).
Cell isolation
Peripheral blood mononuclear cells (PBMCs) were isolated from adult peripheral buffy coats or from cord blood mononuclear cells by Ficoll gradient centrifugation. CD4+ T cells were isolated using anti-CD4 beads (Miltenyi Biotec). Cells were labeled with anti-CD4–Pacific Blue, anti-HLA-DR–PECy7 (BD Biosciences), anti-CD25–phycoerythrin, anti-CD45RA–fluorescein isothiocyanate, anti-CD127–allophycocyanin (Miltenyi Biotec) and sorted into CD4+CD25−CD45RA+ (naive Tconvs), CD4+CD25highCD127−CD45RA+HLADR− (“naive” Treg), CD4+CD25highCD127−CD45RA−HLADR− (“memory” Treg), and CD4+CD25highCD127−CD45RA−HLADR+ (“activated” Treg) populations using FACSAria II (BD Biosciences).
Flow cytometric analysis
Phenotypic analysis was performed on purified peripheral blood mononuclear cells, cord blood mononuclear cells, or CD4+ T cells. Data were analyzed with FlowJo software (Tree Star, Inc.) and statistical comparisons were performed with a 2-tailed nonparametric Mann-Whitney test (Prism).
Suppression assay
The suppressive capacity of Tregs toward responder cells in coculture was expressed as the relative inhibition of the percentage of carboxyfluorescein diacetate succinimidyl ester (CFSE)low cells [100 × (1 − % CFSElow Tconv cells in coculture/% CFSElow Tconv alone)].
Isolation of cytokine-secreting Tregs and Tconvs
Tconv and Treg subpopulations were sorted by fluorescence-activated cell sorting according to the expression of CD25, CD45RA, and HLA-DR. Each population was stimulated with staphylococcal enterotoxin B (1 µg/mL) in the presence of irradiated autologous CD14+ cells for 7 hours (antigen-presenting cell:T ratio, 3:1). Interleukin-17 (IL-17) and interferon-γ (IFN-γ) secreting cells were labeled using IFN-γ and IL-17 Secretion Assay Cell Enrichment and Detection kits (Miltenyi Biotec).
Single-cell gene expression analysis
Single cells were sorted into 96-well polymerase chain reaction (PCR) plates containing 9 µL of reverse transcriptase (RT) preamplification mix (CellsDirect One-Step qRT-PCR kit; Invitrogen) with a mix of a 0.2× concentration of pooled Thermo Scientific Solaris quantitative PCR (qPCR) gene expression assays (supplemental Table 3). Single-cell gene profiling was performed using BioMark 48.48 or 96.96 Dynamic Arrays (Fluidigm) as described in the manufacturer’s protocol.
The means of the cycle thresholds (Cts) from ACTB and B2M for all cells of the experiment and the SD were calculated, and cells with a mean Ct of ACTB and B2M exceeding twofold the SD were eliminated from the analysis.
Microarrays
Sorted Treg populations and Tconv (CD4+CD25−CD127dim/+CD45RA+HLA-DR−) from healthy donors were stimulated for 24 hours with anti-CD3/CD28 beads (Life Technologies) prior to extraction of RNA (RNeasy Micro kit; Qiagen). Gene expression profiling and data analysis were performed using Applied Biosystems 1700 microarray platform as previously described.36-38 Microarray data were deposited in the MACE database (http://www.ihes.fr/jsp/site/Portal.jsp) with the accession number 2312657974.
See supplemental Methods for additional methods.
Results
Heterogeneity of human CD4+FOXP3+ T cells
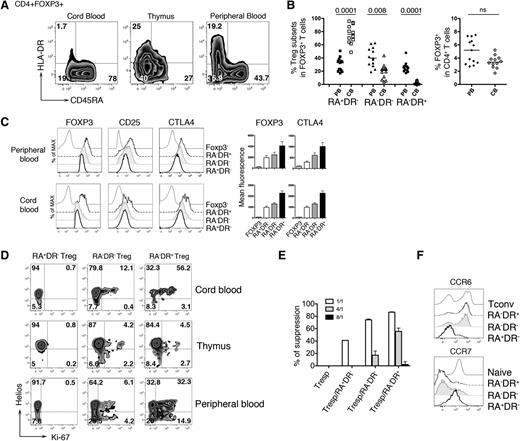
Phenotypic analysis of the CD4+FOXP3+ T-cell compartment revealed 3 distinct Treg populations (CD45RA+HLADR−, CD45RA−HLA-DR−, and CD45RA−HLA-DR+) present at different frequencies in peripheral blood, thymus, and cord blood. The majority of cord blood Treg exhibits a “naive” phenotype (RA+DR−), while higher percentages of RA−DR− and RA−DR+ Tregs are found in adult peripheral blood. The frequency of total CD4+FOXP3+ T cells is not significantly different in neonates and adults (Figure 1A-B). The Treg subpopulations vary for expression of Treg markers (Figure 1C), proliferation state, and expression of Helios, a transcription factor primarily expressed by Tregs of thymic origin15 (Figure 1D). In the thymus, the 3 Treg subpopulations show weak proliferative activity, while in cord blood and peripheral blood, Treg proliferation is observed, mainly in the RA−DR+ subpopulation. This subpopulation has also the highest suppressive activity in vitro, consistent with higher expression of molecules associated with Treg function (Figure 1C,E). We also observed that RA−DR+ Tregs, as well as RA−DR− Tregs, express CCR6 but not CCR7, suggesting homing to mucosal or inflamed tissues. In contrast, RA+DR− Tregs express high levels of CCR7 but lack CCR6, indicating that these cells may preferentially home to secondary lymphoid organs (Figure 1F).
HLA-DR and CD45RA expression delineates 3 regulatory T-cell subpopulations. (A) CD4+ T cells from cord blood, thymus, and adult peripheral blood of healthy individuals were analyzed by flow cytometry. Plots shown were gated on CD4+FOXP3+ T cells. After staining for extracellular antigens, cells were fixed, permeabilized, and intracellular staining of CTLA4, FOXP3, Helios, Ki-67 was performed using eBioscience FOXP3 Staining Buffer Set, following manufacturer instructions. (B) Plots show (left panel) the percentage of each Treg subpopulation in adult peripheral blood samples (n = 13) and cord blood samples (n = 12) and (right panel) the percentage of total FOXP3+ cells within CD4+ T cells. P values were calculated using a 2-tailed nonparametric Mann-Whitney test. (C) Expression of FOXP3, CD25, and CTLA4 were assessed in each Treg subpopulation by flow cytometry (left panel). Bar histograms represent the mean fluorescence intensity of FOXP3 and CTLA4 for adult peripheral blood (n = 13) and cord blood (n = 12). (D) Expression of Ki-67 and Helios in each Treg subpopulation was analyzed in cord blood, thymus, and adult peripheral blood of healthy individuals by flow cytometry. (E) Sorted CD4+CD25−CD45RA+ Tconvs (Tresp) were used as responder cells, stained with 1mM CFSE (CellTrace CFSE Cell Proliferation Kit; Life Technologies) and 104 CFSE-labeled responder CD4+CD25−CD45RA+ T cells were cultured alone or cocultured with 104 unlabeled Treg cells at different ratio Treg:Tresp (1:1; 1:4; 1:8) in triplicates. (Tresp:Treg, white bar, 1:1; gray bar, 4:1; black bar, 8:1) in the presence of CD3/CD28 beads. Proliferation of CFSE-labeled cells was assessed by flow cytometry after 84 to 90 hours of culture. Histograms represent the percentage of suppression of Tresp proliferation. (F) CCR6 and CCR7 expression on Treg subpopulations, naive CD4 T cells (Naive), and RA−DR− CD4 Tconvs (Tconv) were assessed by flow cytometry. Histograms are representative of 4 different healthy individuals. Tresp, T responder.
HLA-DR and CD45RA expression delineates 3 regulatory T-cell subpopulations. (A) CD4+ T cells from cord blood, thymus, and adult peripheral blood of healthy individuals were analyzed by flow cytometry. Plots shown were gated on CD4+FOXP3+ T cells. After staining for extracellular antigens, cells were fixed, permeabilized, and intracellular staining of CTLA4, FOXP3, Helios, Ki-67 was performed using eBioscience FOXP3 Staining Buffer Set, following manufacturer instructions. (B) Plots show (left panel) the percentage of each Treg subpopulation in adult peripheral blood samples (n = 13) and cord blood samples (n = 12) and (right panel) the percentage of total FOXP3+ cells within CD4+ T cells. P values were calculated using a 2-tailed nonparametric Mann-Whitney test. (C) Expression of FOXP3, CD25, and CTLA4 were assessed in each Treg subpopulation by flow cytometry (left panel). Bar histograms represent the mean fluorescence intensity of FOXP3 and CTLA4 for adult peripheral blood (n = 13) and cord blood (n = 12). (D) Expression of Ki-67 and Helios in each Treg subpopulation was analyzed in cord blood, thymus, and adult peripheral blood of healthy individuals by flow cytometry. (E) Sorted CD4+CD25−CD45RA+ Tconvs (Tresp) were used as responder cells, stained with 1mM CFSE (CellTrace CFSE Cell Proliferation Kit; Life Technologies) and 104 CFSE-labeled responder CD4+CD25−CD45RA+ T cells were cultured alone or cocultured with 104 unlabeled Treg cells at different ratio Treg:Tresp (1:1; 1:4; 1:8) in triplicates. (Tresp:Treg, white bar, 1:1; gray bar, 4:1; black bar, 8:1) in the presence of CD3/CD28 beads. Proliferation of CFSE-labeled cells was assessed by flow cytometry after 84 to 90 hours of culture. Histograms represent the percentage of suppression of Tresp proliferation. (F) CCR6 and CCR7 expression on Treg subpopulations, naive CD4 T cells (Naive), and RA−DR− CD4 Tconvs (Tconv) were assessed by flow cytometry. Histograms are representative of 4 different healthy individuals. Tresp, T responder.
To molecularly characterize each Treg subpopulation, we obtained specific gene expression profiles (supplemental Figure 1A; supplemental Table 1). Because Tregs need to be activated to exert immunosuppressive activity, sorted T-cell populations were stimulated for 24 hours with anti-CD3/CD28 antibodies prior to microarray analysis. This analysis also defined a common “Treg core signature” of 41 genes that is shared by the 3 Treg populations. Hierarchical clustering of samples based on the expression level of the core signature genes not only discriminates Tregs from naive CD4+ Tconvs, but also accurately separates the 3 Treg populations, indicating that the core signature genes are expressed at characteristically different levels in the 3 Treg subpopulations (supplemental Figure 1B).
Single-cell gene expression profiling reveals heterogeneity within Treg subpopulations
The expression patterns of Helios, CCR6, and CCR7 suggested that Tregs display marked heterogeneity also within the subpopulations defined by CD45RA and HLA-DR. To examine heterogeneity at the single-cell level within each subset, we selected genes from the Treg signatures (supplemental Table 1) and analyzed their expression in single T-cell receptor activated Tconvs and Tregs. Hierarchical clustering analysis of gene expression shows that the signatures defined at the population level segregate each Treg subpopulation at the single-cell level (Figure 2A) and reveals a strong heterogeneity of gene expression within each subpopulation. Conversely, expression of the housekeeping gene B2M fluctuated considerably less (Figure 2B). Consistent with the flow cytometry data (Figure 1C), expression of Treg cell markers FOXP3 and CTLA4 is higher in the RA−DR+ Treg subpopulation, and CCR6 expression is restricted to the RA−DR− and RA−DR+ Treg populations. These results demonstrate an as-yet unappreciated heterogeneity of the human Treg compartment at the single-cell level, reminiscent of the heterogeneity of effector CD4+ T-cell populations.
Single-cell gene expression profiling reveals heterogeneity within Treg subpopulations. The 3 Treg subpopulations were sorted from adult peripheral blood of healthy donors. Each population was stimulated with CD3/CD28 coated beads for 24 hours then sorted as single cells into 96-well plates. Expression of the indicated mRNA transcript was assessed by single-cell quantitative RT-PCR using microfluidic arrays (Fluidigm). (A) The heat map representation of single-cell gene expression profiling was obtained by 2-dimensional hierarchical clustering analysis using Euclidean distance and average linkage using Qlucore Omics Explorer software. Each row corresponds to the expression level (Ct value) of a single gene and each column represents a single cell (black squares: naive conventional CD4+ T cells; green squares: RA+DR− Tregs; blue squares: RA−DR− Tregs; and red squares: RA−DR+ Tregs). (B) Dot plots were obtained by analyzing Ct values in GraphPad Prism software. Expression of transcripts for Treg markers FOXP3, CTLA4, FAS, IL2RA (CD25), IKZF2 (Helios), LRRC32 (GARP), TNFRSF18 (GITR), IL1R2, and chemokine receptors CCR8, CCR6, CCR4 were plotted for single RA−DR+ Tregs (red triangles), single RA−DR− Tregs (blue triangles), single RA+DR− Tregs (green squares), and CD4+CD25−CD45RA+ Tconvs (black dots). The expression of the housekeeping gene B2M was also analyzed in each T-cell subpopulation. One representative experiment of 3 from independent donors is shown.
Single-cell gene expression profiling reveals heterogeneity within Treg subpopulations. The 3 Treg subpopulations were sorted from adult peripheral blood of healthy donors. Each population was stimulated with CD3/CD28 coated beads for 24 hours then sorted as single cells into 96-well plates. Expression of the indicated mRNA transcript was assessed by single-cell quantitative RT-PCR using microfluidic arrays (Fluidigm). (A) The heat map representation of single-cell gene expression profiling was obtained by 2-dimensional hierarchical clustering analysis using Euclidean distance and average linkage using Qlucore Omics Explorer software. Each row corresponds to the expression level (Ct value) of a single gene and each column represents a single cell (black squares: naive conventional CD4+ T cells; green squares: RA+DR− Tregs; blue squares: RA−DR− Tregs; and red squares: RA−DR+ Tregs). (B) Dot plots were obtained by analyzing Ct values in GraphPad Prism software. Expression of transcripts for Treg markers FOXP3, CTLA4, FAS, IL2RA (CD25), IKZF2 (Helios), LRRC32 (GARP), TNFRSF18 (GITR), IL1R2, and chemokine receptors CCR8, CCR6, CCR4 were plotted for single RA−DR+ Tregs (red triangles), single RA−DR− Tregs (blue triangles), single RA+DR− Tregs (green squares), and CD4+CD25−CD45RA+ Tconvs (black dots). The expression of the housekeeping gene B2M was also analyzed in each T-cell subpopulation. One representative experiment of 3 from independent donors is shown.
Cytokine-secreting CD4+FOXP3+ T cells share features of Treg and effector T cells
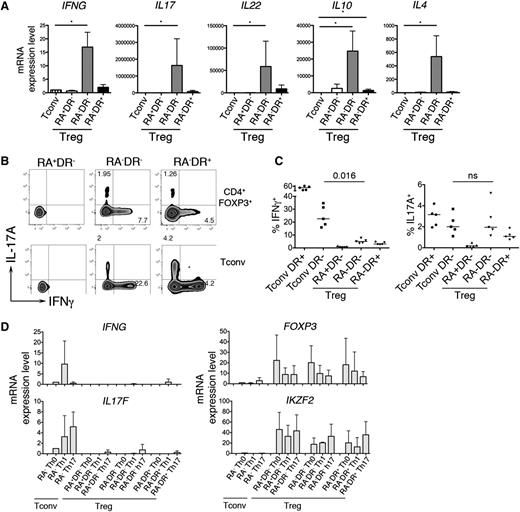
Our microarray analysis revealed an enrichment of transcripts encoding cytokines within the RA−DR− Treg population (supplemental Table 1). qPCR analysis confirmed that effector cytokines (IFNG, IL17, IL22, IL4) and the anti-inflammatory cytokine IL10 were expressed by RA−DR− Tregs, but not by naive Tconvs or RA+DR− and RA−DR+ Tregs (Figure 3A). Intracellular staining shows that a small fraction of RA−DR− Tregs expresses either IFN-γ or IL-17A (Figure 3B). The frequency of IFN-γ–secreting cells is significantly lower in Tregs compared with Tconvs (Figure 3C); however, the frequency of IL-17A–producing cells is similar in RA−DR− Treg and Tconv (Figure 3C). To test whether IFN-γ and IL-17 production by Treg is caused by lineage reprogramming of human Tregs into Th1 or Th17 effector cells,21-23 we cultured the 3 Treg populations and CD4+CD45RA− Tconvs for 2 weeks in the presence of Th1- or Th17-inducing cytokines. We did not observe a potent induction of cytokine transcripts in any Treg population in these conditions (Figure 3D).
IL-17– and IFN-γ–producing Tregs. (A) The 3 Treg subpopulations and naive Tconvs were sorted and stimulated with anti-CD3/CD28 coated beads for 24 hours. mRNA expression of the indicated cytokines was determined by qRT-PCR using TaqMan low-density arrays. Mean values and SEM of 6 different donors are shown. (B-C) For intracellular cytokine staining, purified CD4 T cells were stimulated 5 hours with PMA/ionomycin (50 ng/mL, 1 μg/mL). (B) After 1 hour of stimulation, Brefeldin A was added to the culture media. After staining for extracellular antigens, cells were fixed, permeabilized, and intracellular staining of FOXP3, Helios, Ki-67, IL-17A, and IFN-γ was analyzed by flow cytometry. (C) Plots show the percentage of IFN-γ+ and IL17A+ T cells for conventional CD45RA−HLADR- and CD45RA−HLADR+ T cells and the 3 Treg subpopulations. P values were obtained with the a-tailed nonparametric Mann-Whitney test. (D) For polarization assays, the 3 Treg subpopulations and conventional memory T cells were sorted and cultured for 2 weeks in Th0, Th1, or Th17 polarizing conditions. T cells were cultured in complete RPMI with CD3/CD28 beads (Invitrogen) and 100 U/mL IL-2. No additional cytokines or anti-cytokine antibodies were added to the Th0 condition. Th1 condition: IL-12 (10 ng/mL), anti–IL-4 (1 µg/mL). Th17 condition: IL-23 (50 ng/mL), IL-1β (10 ng/mL), IL-21 (10 ng/mL), TGF-β (10 ng/mL), anti–IL-4 (1 µg/mL), and anti–IFN-γ (1 µg/mL). Cytokines were added again to the media 48 hours after the first stimulation. Cells were restimulated with cytokines (day 7) and anti-CD3/CD28 beads (days 7 and 13). At day 14, the mRNA levels of IFNG, IL17F, FOXP3, and IKZF2 (Helios) were determined using 48.48 Dynamic Arrays.
IL-17– and IFN-γ–producing Tregs. (A) The 3 Treg subpopulations and naive Tconvs were sorted and stimulated with anti-CD3/CD28 coated beads for 24 hours. mRNA expression of the indicated cytokines was determined by qRT-PCR using TaqMan low-density arrays. Mean values and SEM of 6 different donors are shown. (B-C) For intracellular cytokine staining, purified CD4 T cells were stimulated 5 hours with PMA/ionomycin (50 ng/mL, 1 μg/mL). (B) After 1 hour of stimulation, Brefeldin A was added to the culture media. After staining for extracellular antigens, cells were fixed, permeabilized, and intracellular staining of FOXP3, Helios, Ki-67, IL-17A, and IFN-γ was analyzed by flow cytometry. (C) Plots show the percentage of IFN-γ+ and IL17A+ T cells for conventional CD45RA−HLADR- and CD45RA−HLADR+ T cells and the 3 Treg subpopulations. P values were obtained with the a-tailed nonparametric Mann-Whitney test. (D) For polarization assays, the 3 Treg subpopulations and conventional memory T cells were sorted and cultured for 2 weeks in Th0, Th1, or Th17 polarizing conditions. T cells were cultured in complete RPMI with CD3/CD28 beads (Invitrogen) and 100 U/mL IL-2. No additional cytokines or anti-cytokine antibodies were added to the Th0 condition. Th1 condition: IL-12 (10 ng/mL), anti–IL-4 (1 µg/mL). Th17 condition: IL-23 (50 ng/mL), IL-1β (10 ng/mL), IL-21 (10 ng/mL), TGF-β (10 ng/mL), anti–IL-4 (1 µg/mL), and anti–IFN-γ (1 µg/mL). Cytokines were added again to the media 48 hours after the first stimulation. Cells were restimulated with cytokines (day 7) and anti-CD3/CD28 beads (days 7 and 13). At day 14, the mRNA levels of IFNG, IL17F, FOXP3, and IKZF2 (Helios) were determined using 48.48 Dynamic Arrays.
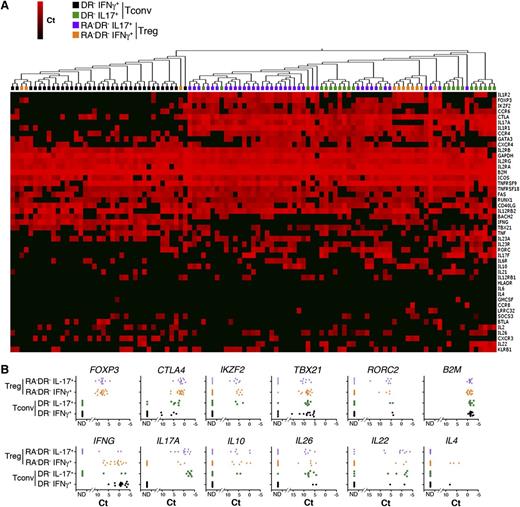
To determine whether cytokine-producing CD4+FOXP3+ T cells maintain the molecular profiles of Tregs, we analyzed whether a single Treg cell can coexpress Treg marker genes and effector cytokines. We found that IFN-γ– and IL-17A–expressing Tregs maintain the expression of Treg markers such as FOXP3 and CTLA4. A substantial fraction of IFN-γ– and IL-17A–secreting Tregs express IKZF2 (Helios) (Figure 4; supplemental Figure 2A-B). Consistent with a recent study,39 CD161 transcripts (KLRB1) were mainly expressed by IL-17–producing Tregs. In addition, cytokine-producing Tregs express the Th1-specific transcription factor TBX21 (T-bet) or the Th17-specific factor RORC, together with effector cytokines, without upregulating IL2 expression (supplemental Figure 2C).
Single-cell analysis of cytokine-producing Tregs. (A) IL-17A– and IFN-γ–producing Tregs and Tconvs were isolated using a cytokine capture assay on sorted populations of CD4+CD25highHLADR−CD45RA− (RA−DR− Tregs) Tregs and CD4+CD25−CD45RA−HLADR− Tconvs (RA−DR− Tconv). Single IFN-γ– and IL-17A–producing cells were sorted into 96 cells plates and single-cell gene expression profiling was performed using 48.48 Dynamic Arrays (18 cycles preamplification). The heat map represents 2-dimensional hierarchical clustering using Euclidean distance and average linkage. (B) mRNA expression of Treg markers (FOXP3, CTLA4, Helios), Th1 and Th17 transcription factors (TBX21, RORC2), and cytokines (IFN-γ, IL17A, IL10, IL26, IL22, IL4) in single RA−DR− Tregs and RA−DR− Tconvs. A representative experiment performed with 4 independent donors is shown.
Single-cell analysis of cytokine-producing Tregs. (A) IL-17A– and IFN-γ–producing Tregs and Tconvs were isolated using a cytokine capture assay on sorted populations of CD4+CD25highHLADR−CD45RA− (RA−DR− Tregs) Tregs and CD4+CD25−CD45RA−HLADR− Tconvs (RA−DR− Tconv). Single IFN-γ– and IL-17A–producing cells were sorted into 96 cells plates and single-cell gene expression profiling was performed using 48.48 Dynamic Arrays (18 cycles preamplification). The heat map represents 2-dimensional hierarchical clustering using Euclidean distance and average linkage. (B) mRNA expression of Treg markers (FOXP3, CTLA4, Helios), Th1 and Th17 transcription factors (TBX21, RORC2), and cytokines (IFN-γ, IL17A, IL10, IL26, IL22, IL4) in single RA−DR− Tregs and RA−DR− Tconvs. A representative experiment performed with 4 independent donors is shown.
However, the messenger RNA (mRNA) levels of TBX21/IFNG and RORC/IL17A are lower in Tregs compared with Th1 and Th17 cells (Figure 4). Hierarchical clustering based on genes from the Treg signature and genes associated with Th1 or Th17 cells indicates that Th17 cells are more closely related to cytokine-producing Tregs than to Th1 cells. Cytokine-secreting Tregs, and Th17 Tconvs are characterized by higher levels of expression of the IL-10 family members IL10, IL22, and IL26, the chemokine receptors CCR4 and CCR6, and IL1R1 and CTLA4, compared with Th1 Tconvs (Figure 4). Cytokine-secreting Tregs express lower levels of the Treg markers FOXP3, IKZF2, and FAS compared with Tregs that do not produce IFN-γ or IL-17A (supplemental Figure 2A-B). These data demonstrate the coexistence within the same cell of transcriptional programs controlling Tregs and Th1 or Th17 cell functions, and provide further evidence for the functional diversity of the human CD4+FOXP3+ T-cell compartment.
Altered homeostasis of CD4+FOXP3+ T-cell populations in patients developing aGVHD after alloHSCT
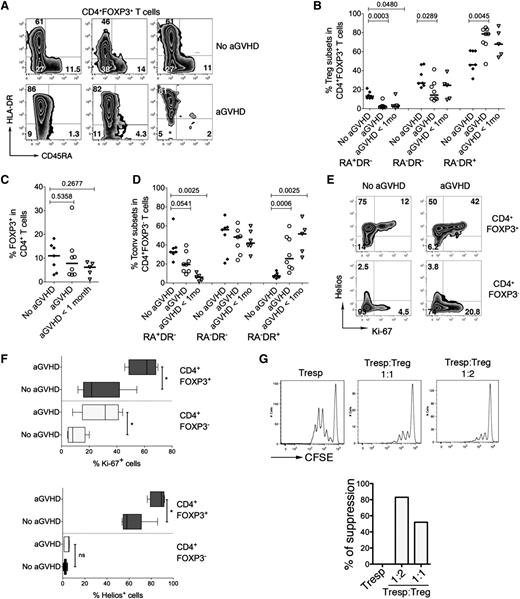
Several studies suggested that conditions similar to the profound lymphopenic and highly inflammatory environment generated by the conditioning regimen prior to alloHSCT may facilitate lineage reprogramming of Tregs into proinflammatory effector T-cell subsets.21-23,40 To provide an in vivo example for human Treg subpopulations in an inflammatory and lymphopenic environment, we designed an exploratory study to analyze CD4+FOXP3+ T-cell populations frequencies in 20 consecutive alloHSCT patients at the time of engraftment (supplemental Table 2). Chimerism analyses using qPCR of polymorphic short tandem repeats show early donor reconstitution (90%-100%), suggesting that the majority of CD4+FOXP3+ T cells are donor-derived (data not shown). We found that our cohort of patients developing aGVHD almost completely lacked RA+DR−CD4+FOXP3+ T cells but had a strong enrichment of RA−DR+CD4+FOXP3+ T cells compared with the patients that had not developed aGVHD (Figure 5A). RA−DR+CD4+FOXP3+ T cells were significantly more frequent in aGVHD patients than in those without aGVHD (P = .0045). Inversely, RA−DR− and RA+DR− CD4+FOXP3+ T cells are more frequent in patients without aGVHD than in patients with aGVHD (P = .029 and P = .0003, respectively) (Figure 5B). We also observed a trend toward an activated phenotype in FOXP3−CD4+ T cells in aGVHD patients (Figure 5D). Five patients who did not have aGVHD at the time of hematopoietic recovery but developed aGVHD within 1 month after sampling (“aGVHD <1 mo”, Figure 5B), had similar Treg profiles to patients with aGVHD at the time of sampling. The overall frequency of FOXP3+ T cells within the CD4+ T-cell compartment was not significantly different in the 3 groups (Figure 5C). Supplemental Figure 3 shows the percentages of Treg subpopulations for each patient in the 3 groups. We also found that the fraction of proliferating CD4+FOXP3+ T cells is substantially higher in aGVHD patients than in patients not developing aGVHD (Figure 5E-F). Higher frequencies of proliferating cells were also found in the FOXP3−CD4+ T-cell compartment in aGVHD patients. CD4+FOXP3+ T cells expressing high levels of Ki-67 also expressed high levels of HLA-DR. However, the HLA-DR+ Treg population also included cells that do not express Ki-67, indicating that HLA-DR is not simply a marker for activation in these cells (supplemental Figure 4). We also noted that a substantial fraction of CD4+FOXP3+ T cells in alloHSCT patients expressed Helios, a transcription factor initially shown to be selectively expressed by thymus-derived Tregs (Figure 5E-F).15,41
Altered homeostasis of CD4+FOXP3+ T cells in patients developing acute graft-versus-host disease after alloHSCT. Analysis of Treg subpopulations was performed in 20 consecutive alloHSCT recipients. Patients were analyzed at the time of engraftment (2-3 weeks after alloHSCT, see supplemental Table 2 for patient characteristics). In the control group (No aGVHD), a blood sample was collected at time of hematopoietic recovery (group 2, n = 7). In patients with aGVHD (n = 8), a blood sample was collected at diagnosis of aGVHD, before steroid treatment initiation. Patients in the group “aGVHD <1 mo” (n = 5) did not display symptoms of aGVHD at the time of analysis at the time of engraftment but developed aGVHD within 1 month after sampling (ie, 2 months after alloHSCT). (A) Expression of HLADR and CD45RA in the peripheral blood CD4+FOXP3+ T-cell compartment was analyzed by flow cytometry. (B-C) Plots represent the frequencies of the three Treg subpopulations within CD4+FOXP3+ T cells in 7 patients without aGVHD, 8 patients developing aGVHD, and 5 patients developing aGVHD within 1 month after sampling (B) and percentages of total CD4+FOXP3+ T cells within CD4+ T cells (C). (D) Expression of HLA-DR and CD45RA in the CD4+FOXP3− T-cell compartment. P values were obtained using the 2-tailed nonparametric Mann-Whitney test. (E) Tregs and Tconvs were analyzed by flow cytometry for the expression of Helios and Ki-67. (F) Percentage of Ki-67+ (top panel) and Helios+ cells (bottom panel) in CD4+FOXP3+ Tregs and conventional CD4+FOXP3− T cells in patients with aGVHD (n = 5) and patients not developing aGVHD (n = 6, No aGVHD). Five patients with aGVHD and 6 patients without aGVHD were analyzed. (G) Suppression assays were performed with Tregs and Tconvs from a patient with aGVHD. Ratios of 1:1 and 1:2 (Tresp:Treg) were used.
Altered homeostasis of CD4+FOXP3+ T cells in patients developing acute graft-versus-host disease after alloHSCT. Analysis of Treg subpopulations was performed in 20 consecutive alloHSCT recipients. Patients were analyzed at the time of engraftment (2-3 weeks after alloHSCT, see supplemental Table 2 for patient characteristics). In the control group (No aGVHD), a blood sample was collected at time of hematopoietic recovery (group 2, n = 7). In patients with aGVHD (n = 8), a blood sample was collected at diagnosis of aGVHD, before steroid treatment initiation. Patients in the group “aGVHD <1 mo” (n = 5) did not display symptoms of aGVHD at the time of analysis at the time of engraftment but developed aGVHD within 1 month after sampling (ie, 2 months after alloHSCT). (A) Expression of HLADR and CD45RA in the peripheral blood CD4+FOXP3+ T-cell compartment was analyzed by flow cytometry. (B-C) Plots represent the frequencies of the three Treg subpopulations within CD4+FOXP3+ T cells in 7 patients without aGVHD, 8 patients developing aGVHD, and 5 patients developing aGVHD within 1 month after sampling (B) and percentages of total CD4+FOXP3+ T cells within CD4+ T cells (C). (D) Expression of HLA-DR and CD45RA in the CD4+FOXP3− T-cell compartment. P values were obtained using the 2-tailed nonparametric Mann-Whitney test. (E) Tregs and Tconvs were analyzed by flow cytometry for the expression of Helios and Ki-67. (F) Percentage of Ki-67+ (top panel) and Helios+ cells (bottom panel) in CD4+FOXP3+ Tregs and conventional CD4+FOXP3− T cells in patients with aGVHD (n = 5) and patients not developing aGVHD (n = 6, No aGVHD). Five patients with aGVHD and 6 patients without aGVHD were analyzed. (G) Suppression assays were performed with Tregs and Tconvs from a patient with aGVHD. Ratios of 1:1 and 1:2 (Tresp:Treg) were used.
To assess the molecular characteristics of these cells, we performed single-cell gene expression profiling of DR+ and DR− CD4+CD25highCD127− T cells and Tconvs from several aGVHD patients. Hierarchical clustering clearly segregates CD4+CD25highCD127− T cells from Tconvs, but expression of Treg markers was similar in DR+ and DR− CD4+CD25highCD127− T cells (Figure 6A-B). Similarly to healthy donors, gene expression in Tregs from aGVHD patients is highly heterogeneous (Figure 6A-B). CD4+CD25highCD127− T cells express high levels of FOXP3, while FOXP3 expression cannot be detected in Tconvs from the same donor (Figure 6B). Consistent with flow cytometry data (Figure 5E-F), the majority of CD4+CD25highCD127− T cells in aGVHD patients express IKZF2. Furthermore, CD4+CD25highCD127− T cells express IKZF4 (Eos) and high levels of CTLA4 and LGALS1 (Galectin-1) (Figure 6B), demonstrating that Tregs maintain the expression of Treg signature genes in the highly lymphopenic and inflammatory environment of aGVHD. Expression of cytokine genes could not be detected in Tregs isolated from aGVHD patients, possibly as a result of the immunosuppressant treatment of GVHD prophylaxis (data not shown).
Single-cell analysis of Treg patients with acute graft-versus-host disease. (A) Single cells from the indicated CD4+ T-cell populations from aGVHD patients were sorted into 96 cells plates without prior stimulation and analyzed using 48.48 Dynamic Arrays. The heat map of the single-cell gene expression profiling was obtained by 2-dimensional hierarchical clustering using Euclidean distance and average linkage. (B) Single-cell mRNA expression of Treg markers (FOXP3, IL2RA, CTLA4, Helios, LSGALS, and IKZF4), chemokine receptor CCR8 and the housekeeping gene B2M were plotted for RA−DR− and RA−DR+ Tregs and Tconvs. One of 3 analyses of 3 different patients is shown.
Single-cell analysis of Treg patients with acute graft-versus-host disease. (A) Single cells from the indicated CD4+ T-cell populations from aGVHD patients were sorted into 96 cells plates without prior stimulation and analyzed using 48.48 Dynamic Arrays. The heat map of the single-cell gene expression profiling was obtained by 2-dimensional hierarchical clustering using Euclidean distance and average linkage. (B) Single-cell mRNA expression of Treg markers (FOXP3, IL2RA, CTLA4, Helios, LSGALS, and IKZF4), chemokine receptor CCR8 and the housekeeping gene B2M were plotted for RA−DR− and RA−DR+ Tregs and Tconvs. One of 3 analyses of 3 different patients is shown.
The low number of T cells in the periphery of aGVHD patients early after alloHSCT precluded a direct analysis of Treg suppressive activity for all patients. However, we could analyze the suppressive activity of sorted CD4+CD25hiCD127− T cells from 1 patient developing aGVHD 15 days after alloHSCT, demonstrating that Tregs from this patient are functionally active (Figure 5G).
These data indicate that a suppressive Treg population is maintained in aGVHD patients, and that aGVHD patients may have a lower frequency of Tregs with a “naive” phenotype (CD45RA+ HLA-DR−).
Discussion
Treg heterogeneity has been assessed in different studies by gene expression analysis of sorted bulk populations using microarrays.6-9,13,14,42 A limitation of these studies is that they may not reveal signals from low frequency, but functionally relevant, cell subsets. Decrypting the transcriptional networks and understanding the functional diversity of Tregs requires multiparameter gene expression profiling of single cells.
Our single-cell analysis of the human Treg compartment revealed an unexpected level of complexity and functional heterogeneity of human Tregs. We found that genes belonging to our Treg signatures showed, at the single-cell level, a highly variegated gene expression pattern that could not be appreciated by the analysis of bulk populations or by flow cytometry. Similar to CD4+ FOXP3− effector T cells, the mosaic-like expression of transcription factors, signaling molecules and homing receptors is likely to endow individual Tregs with specific functions and migratory properties. Despite the remarkable qualitative and quantitative differences in gene expression from cell to cell, we also noted 2 invariable features shared by all human Tregs. Single-cell analysis revealed that all Tregs express FOXP3 and lack IL-2 expression emphasizing their key role in regulating Treg function. Consistent with flow cytometry analysis, we noted that the expression level of FOXP3 and CTLA4 and other genes associated with the function of Tregs varies by 2 to 3 orders of magnitude from cell to cell, indicating that these molecules may act as important rheostats to tune the immunosuppressive function of individual cells. An important question that remains to be solved concerns the mechanisms accounting for the remarkably strong variation in the expression of FOXP3 and other Treg signature genes from cell to cell. In particular, it will be important to understand the relative contributions of stochasticity in gene expression on the one hand, and environmental factors such as T-cell receptor signal strength, costimulatory signals, and cytokines on the other.
We have characterized at the single-cell level a very small population of IFN-γ– and IL-17A–producing Tregs by combining the isolation of viable cytokine-secreting cells with single-cell gene profiling. This analysis revealed that IFN-γ– and IL-17A–producing Tregs are characterized by an overlap within the same cell of gene expression signatures of Th1 or Th17 cells and of Tregs. The lack of IL-2 expression as well as the expression of high levels of CTLA-4 and other molecules typically associated with Treg functions suggests immunoregulatory properties for these cells, consistent with previous studies.43,44 We noted that IL-17A and IFN-γ transcripts are expressed at lower levels in cytokine-secreting Tregs than in conventional CD4+ effector T cells, which may indicate a lower proinflammatory potential. Interestingly, IL-17A– and IFN-γ–secreting Tregs expressed lower levels of FOXP3 than Tregs that do not produce these cytokines. These data suggest that FOXP3 controls not only the immunosuppressive activity of Tregs, but may also suppress their “inflammatory” functions.
Our observation that HLA-DR+CD4+FOXP3+ T cells are preserved in patients developing aGVHD is seemingly at odds with the notion that this Treg subpopulation is endowed with the highest immunosuppressive activity in vitro. However, previous results obtained in a mouse model of GVHD have demonstrated that Tregs with a naive phenotype (CD62L+CD4+CD25+) but not CD62L− Tregs can prevent aGVHD.45,46 This suggests that Treg homing to secondary lymphoid organs is a key factor to control the excessive immune activation seen in aGVHD patients. Consistently, aberrant expression of homing receptors has been reported in a recent study comparing Tregs in patients developing or not GVHD after alloHSCT.47 Alternatively, CD4+FOXP3+ T cells may be functionally impaired in aGVHD patients. However, our preliminary results suggest that even in the setting of active GVHD Tregs may retain suppressive activity, arguing against the latter hypothesis. In this exploratory study of aGVHD patients at disease onset, we did not observe significant differences in the overall frequency of CD4+FOXP3+ T cells within the CD4+ T-cell compartment in aGVHD patients, compared with tolerant patients early after HSCT. Furthermore, our analysis of gene expression of Tregs in aGVHD patients revealed no aberrant expression of molecules associated with the functional activity of Tregs, compared with healthy donor Tregs. However, we observed a decrease in aGVHD patients, of the fraction of CD4+FOXP3+ T cells with a CD45RA+ phenotype. These data raise the possibility that the frequency of specific subpopulations of CD4+FOXP3+ T cells, rather than the frequency of the total pool of CD4+FOXP3+ T cells may be altered in patients developing aGVHD. Because our data were obtained with only 20 alloHSCT patients an appropriately designed clinical study will be necessary to investigate the hypothesis that the specific loss of Tregs with a “naive” phenotype may be associated with the breakdown of tolerance in patients with aGVHD. If validated, these findings would indicate that analysis of Treg subpopulations at the time of engraftment may help identify patients at risk of developing aGVHD.
We found that the fraction of proliferating CD4+FOXP3+ T cells is significantly higher in aGVHD patients than in patients not developing aGVHD. A previous clonotype analysis of Tregs demonstrated that “activated” (CD45RA−) Tregs derive from Tregs with a “naive” (CD45RA+) phenotype.14 Together, these findings suggest that increased proliferation of Tregs displaying a highly activated phenotype may cause the exhaustion of the pool of naive Tregs in aGVHD patients.
Previous reports have demonstrated that patients with chronic GVHD (cGVHD) have fewer Tregs in peripheral blood than patients who do not develop cGVHD after alloHSCT.29,30 Matsuoka et al recently analyzed the reconstitution of Tregs and conventional CD4+ T cells in patients in the first year after alloHSCT and found that abnormal homeostasis of Tregs was directly correlated with increased incidence and severity of cGVHD,48 consistent with our results in the setting of aGVHD. Matsuoka et al showed that reconstitution of Tconvs was mainly driven by thymus-derived CD4+ T cells, whereas thymic generation of Tregs was markedly impaired, and Tregs expanded mainly by lymphopenia-driven proliferation of Tregs present in the graft. In this study, a large proportion of Tregs expressed FAS (CD95), and Tregs from alloHSCT patients were highly susceptible to apoptosis. We cannot rule out that in aGVHD naive Tregs undergo increased cell death. However, our data on healthy donors and patients not developing GVHD show that the CD45RA+HLA-DR− Treg population is the least susceptible to activation-induced cell death (supplemental Figure 5).
Previous reports have proposed that the transcription factor Helios is selectively expressed by thymus-derived Tregs.15,41 More recent data, however, showed that Helios can be expressed by peripherally induced mouse CD4+Foxp3+ T cells49 and may represent a marker of T-cell activation and proliferation rather than a specific marker for thymic Tregs.50 However, in our study, we did not find a correlation between T-cell activation and Helios expression. In particular, 95% of resting, Ki-67–negative CD4+FOXP3+ T cells in thymus and cord blood with a nonactivated phenotype (CD45RA+ HLADR−) expressed Helios. We also noted that a large fraction of CD4+FOXP3+ T cells in alloHSCT patients at the time of engraftment were Helios-positive. Because thymus function is impaired at this early time point after transplantation, we favor the hypothesis that Helios-positive Tregs in these patients were generated in the donor’s thymus. However, it remains to be investigated whether peripheral conversion of conventional CD4+ T cells early after alloHSCT may contribute significantly to Treg homeostasis.
In conclusion, our cross-sectional study of CD4+FOXP3+ T cells in alloHSCT patients at the time of engraftment suggests that CD4+CD25highCD127− T cells maintain suppressive activity and high expression levels of Treg markers such as FOXP3, CTLA4, and IKZF2, despite a severely inflammatory and lymphopenic environment. This argues in favor of the Treg being a stable lineage, consistent with recent results obtained in mice.25 We therefore propose that heterogeneity at the single-cell level, rather than reprogramming of CD4+FOXP3+ T cells, explains the remarkable complexity and functional diversity of human Tregs.
Finally, our study demonstrates that multiparameter single-cell analysis techniques not only uncover previously unappreciated levels of heterogeneity within cell “subsets”, but can also provide valuable information about transcriptional networks in single cells. The use of single-cell analyses offers the unique opportunity to investigate the early events that shape immune reconstitution after alloHSCT, at a time point when it may still be possible to tilt the balance between excessive inflammation and tolerance induction.35
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
S.M. was supported by a fellowship from the Ligue Nationale Contre le Cancer. L.W. was supported by a Marie Curie Intra European Fellowship within the 7th European Community Framework Programme and fellowships from the Fondation Association pour la Recherche sur le Cancer and Cantarini-Pasteur. C.B. was a recipient of a fellowship from the Agence Nationale de Recherches sur le Sida. This work was supported by the Agence Nationale de Recherche (ANR-GENOPAT, ANR-PHYSIO), the Agence Nationale de Recherches sur le Sida, the Comité 75 from the Ligue Nationale Contre le Cancer, and institutional funds from Institut Pasteur.
Authorship
Contribution: S.D. designed, performed, and analyzed the experiments, and wrote the manuscript; S.M., A.X., Y.P., and L.W. performed and analyzed experiments; A.X. and J.L. provided critical reagents and edited the manuscript; S.M., C.B., and A.B. performed and analyzed microarray data; and E.B., G.S., and L.R. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lars Rogge, Immunoregulation Unit, Institut Pasteur, 25 rue du Docteur Roux, 75724 Paris, France; e-mail: lars.rogge@pasteur.fr; and Gérard Socié, Service d'Hématologie-Greffe, Hôpital Saint-Louis, Assistance Publique-Hopitaux de Paris, Université Paris VII Denis-Diderot, 1 Ave Claude Vellefaux, 75010 Paris, France; e-mail: gerard.socie@sls.aphp.fr.
References
Author notes
S.M., A.X., and Y.P. contributed equally to this study.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal