Abstract
The congenital dyserythropoietic anemias (CDAs) are hereditary disorders characterized by distinct morphologic abnormalities of marrow erythroblasts. The unveiling of the genes mutated in the major CDA subgroups (I-CDAN1 and II-SEC23B) has now been completed with the recent identification of the CDA III gene (KIF23). KIF23 encodes mitotic kinesin-like protein 1, which plays a critical role in cytokinesis, whereas the cellular role of the proteins encoded by CDAN1 and SEC23B is still unknown. CDA variants with mutations in erythroid transcription factor genes (KLF1 and GATA-1) have been recently identified. Molecular diagnosis of CDA is now possible in most patients.
Introduction
The congenital dyserythropoietic anemias (CDAs) are a group of rare hereditary disorders characterized by congenital anemia, ineffective erythropoiesis with distinct morphologic features in bone marrow (BM) late erythroblasts, and the development of secondary hemochromatosis. Patients usually present with anemia, jaundice, splenomegaly, and suboptimal reticulocyte response for the degree of anemia. Aniso-poikilocytosis and basophilic stippling are commonly observed in the peripheral blood smear. The working classification initially proposed by Heimpel and Wendt is still used in clinical practice (CDA I, II, and III; Table 1).1,2 However, there are families that fulfill the general definition of the CDAs but do not conform to any of the 3 classical types (CDA variants, Table 1).
Characteristic features of different types of CDA
| CDA type . | I . | II . | III Familial . | III Sporadic . | Variants . |
|---|---|---|---|---|---|
| Inheritance | Autosomal recessive | Autosomal recessive | Dominant | Variable | Autosomal dominant or X linked or recessive |
| Cases reported | >300 | >450 | 2 families | <20 | ∼70 |
| BM morphology (light microscopy) | Abnormal chromatin structure, chromatin bridges | Binuclearity, multinuclearity of mature erythroblasts | Giant multinucleated erythroblasts | Giant multinucleated erythroblasts | CDA I–like, CDA II–like, others |
| BM EM findings | “Spongy” heterochromatin, invagination of cytoplasm into the nucleus | Peripheral cysternae beneath the plasma membrane | Clefts in heterochromatin, autophagic vacuoles, intranuclear cisternae | Various | Various |
| Mutated gene | CDAN1, C15ORF41 | SEC23B | KIF23 | Unknown | KLF1, GATA-1, unknown |
| Associated dysmorphology/organ involvement | Skeleton | Variable, rare | Monoclonal gammopathy, myeloma, angioid streaks | Variable | CNS, others |
| CDA type . | I . | II . | III Familial . | III Sporadic . | Variants . |
|---|---|---|---|---|---|
| Inheritance | Autosomal recessive | Autosomal recessive | Dominant | Variable | Autosomal dominant or X linked or recessive |
| Cases reported | >300 | >450 | 2 families | <20 | ∼70 |
| BM morphology (light microscopy) | Abnormal chromatin structure, chromatin bridges | Binuclearity, multinuclearity of mature erythroblasts | Giant multinucleated erythroblasts | Giant multinucleated erythroblasts | CDA I–like, CDA II–like, others |
| BM EM findings | “Spongy” heterochromatin, invagination of cytoplasm into the nucleus | Peripheral cysternae beneath the plasma membrane | Clefts in heterochromatin, autophagic vacuoles, intranuclear cisternae | Various | Various |
| Mutated gene | CDAN1, C15ORF41 | SEC23B | KIF23 | Unknown | KLF1, GATA-1, unknown |
| Associated dysmorphology/organ involvement | Skeleton | Variable, rare | Monoclonal gammopathy, myeloma, angioid streaks | Variable | CNS, others |
CNS, central nervous system; EM, electron microscopy.
The genes mutated in the majority of patients with CDA I and II have been previously described, whereas the CDA III gene was only recently identified.3-5 Additional genetic defects have been described in CDA variants. Because of these advancements, the diagnosis can now be confirmed by molecular testing in an increasing number of patients.
The aim of this review is to update the molecular basis of this group of disorders and to outline the current diagnostic approach.
CDA I to III: clinical picture and BM findings
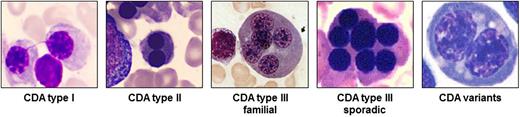
CDA I is an autosomal recessive disease associated with macrocytic anemia. Skeletal abnormalities of distal limbs have also been described.6 BM examination reveals erythroid hyperplasia with up to 10% binuclear erythroblasts. Pathognomonic chromatin bridges between nuclei are observed in up to 8% of erythroblasts (Figure 1).7 BM EM shows a spongy (“Swiss cheese”) appearance of heterochromatin.
Pathognomonic marrow erythroblasts. The BM erythroblasts morphology (light microscopy) of different types of CDA is shown. See Table 1 for the description.
Pathognomonic marrow erythroblasts. The BM erythroblasts morphology (light microscopy) of different types of CDA is shown. See Table 1 for the description.
CDA II is the most common form of the CDAs. BM light microscopy reveals more than 10% mature binuclear erythroblasts (Figure 1), and with EM, vesicles loaded with proteins of endoplasmic reticulum are found beneath the plasma membrane.2 The Ham test with red cell lysis in acidified allogenic sera has practically been abandoned as a diagnostic tool due to the need for multiple control samples. Analysis of red cell membrane proteins by sodium dodecyl sulfate polyacrylamide gel electrophoresis, identifying glycosylation abnormalities with fast-moving band 3 (the anion exchanger 1) and band 4.5 (glucose transporter 1), is highly sensitive and specific.
CDA III is the rarest among the 3 types. Although originally reported in an American family, most patients described to date belong to 1 large Swedish kindred. In both families, the inheritance is dominant. Few sporadic cases have also been described with possible autosomal recessive inheritance.2,8 Splenomegaly is usually absent. BM morphology is characterized by erythroid hyperplasia and large pathognomonic multinucleated erythroblasts (Figure 1). A large proportion of patients in the Swedish family exhibited monoclonal gammopathy, and some developed multiple myeloma.8
Therapeutic approach
Although most CDA patients are mildly or moderately affected and are not transfusion dependent, many do require blood transfusions during the first months of life, pregnancy, and/or sporadic infections. Iron overload with inappropriately low serum hepcidin was documented and correlates with age.9,10 A minority of patients are severely affected and are transfusion dependent. Patients with CDA I were found to respond to interferon α with increased hemoglobin levels and decreased iron overload.11 The role of splenectomy in the management of CDA is yet undetermined; however, patients with CDA type II seem to benefit from the procedure.12 CDA patients who are older than 10 years of age should be carefully monitored for the development of iron overload, which requires iron chelation therapy when identified. Few transfusion-dependent children with CDA underwent successful stem cell transplantation.13,14
Molecular insights
CDA I
The gene mutated in CDA I (CDAN1) was the first of the CDA genes to be described.3 It is localized to chromosome 15, spans 28 exons, and encodes a 134-kDa ubiquitous protein (codanin-1). The study of patients of different ethnic backgrounds identified more than 30 unique disease-causing mutations.3,6,15 Genotype-phenotype correlations could not be established. No patient was homozygous for null-type mutations, suggesting that a complete absence of codanin-1 is lethal. Indeed, CDA I knockout mice embryos die in utero at 6.5 days before erythropoiesis onset, probably due to the critical role of codanin-1 in developmental processes other than erythropoiesis.16 In ∼20% of families with the CDA I phenotype, no CDAN1 mutations in 1 or both alleles were found.17
EM of control and CDA I BM stained with gold-labeled anti–codanin-1 antibody demonstrated that codanin-1 mainly localizes to heterochromatin.18 A comparison of controls and CDA I erythroblasts revealed that CDA I intermediate erythroblasts had abnormal accumulation of heterochromatin protein (HP-1α) in their Golgi apparatus. Furthermore, partial colocalization of codanin-1 and SEC23B, the protein mutated in CDA II, was observed in CDA I intermediate erythroblasts, suggesting intracellular transport pathway defects in both CDA I and II.16 Noy-Lotan et al found in nonerythroid cells that E2F1 (a main regulator of G1/S transition) binds to the CDAN1 promoter and initiates codanin-1 transcription, resulting in a rise in its levels during S phase.18 It was further demonstrated that codanin-1 binds the anti-silencing factor-1 (Asf1), an H3-H4 histone chaperone involved in nucleosome assembly and disassembly.19 More recently, Ask et al showed that codanin-1 is part of a cytosolic Asf1–H3-H4–importin-4 complex20 and acts as a negative regulator of Asf1. Only after codanin-1 dissociated from the Asf1–H3.1-H4 complex in the nucleus was Asf1 able to bind other histone chaperons (chromatin assembly factor-1 and histone cell cycle regulator) and deliver histones for chromatin assembly.
Recently, whole genome sequencing of a CDA I patient belonging to a Pakistani family identified a new CDA I gene, C15ORF41.21 The cellular role of the restriction endonuclease encoded by this gene is yet unknown. Interestingly, it has been previously shown that C15ORF41 interacts with Asf1b, possibly supporting the hypothesis that the primary defect in CDA I is in DNA replication and chromatin assembly.
In erythroid lineage, terminal differentiation is coupled to proliferation with few cell cycle divisions through which cell size is also reduced.22,23 Cell cycle defects in mice result in S phase arrest, ineffective erythropoiesis, and macrocytosis.24 Based on the possible involvement of CDA I genes in DNA replication and chromatin assembly, it can be speculated that CDA I pathogenesis may involve disruption of the intrinsic connection between cell cycle dynamics and terminal erythroid differentiation.
CDA II
SEC23B, the gene mutated in CDA II, was originally mapped to chromosome 20 and identified by means of functional and homozygosity mapping.4,25 This gene encodes the cytoplasmic COPII (coat protein) component SEC23B, which is involved in the secretory pathway of eukaryotic cells. This multisubunit complex mediates accumulation of secretory cargo, deformation of the membrane, and anterograde transport of correctly folded cargo for budding from the endoplasmic reticulum toward the Golgi apparatus.26
To date, 157 cases from 137 different CDA II families with SEC23B mutations were molecularly characterized.27 The majority (52%) of CDAII cases are caused by missense mutations, whereas 20% are due to nonsense, 13% to intronic, and 13% to small indel mutations. The disease inheritance is typically autosomal recessive; yet, in 13% of cases, only 1 SEC23B mutation was found. This suggests the presence of a second still unidentified mutational event, likely in a noncoding regulatory region of the gene that has not yet been examined27 (A. Iolascon, Federico II University, oral communication, December 8, 2012). These results demonstrate that mutations occur along the whole gene. In contrast, in Southern Italy, 2 mutations (R14W and E109K) account for ∼50% of all cases.28 In rare cases, classified as CDA II on the basis of BM and biochemical analyses, no SEC23B mutation was found. This suggests the existence of another causative gene as demonstrated by linkage exclusion.29 Compound heterozygosity for missense and nonsense mutations tends to produce more severe clinical presentations.30 Homozygosity or compound heterozygosity for null mutations was never found and is presumably lethal. Recently, the presence of 2 hypomorphic alleles accounting for mild CDA II clinical forms was described. It has been suggested that compensatory expression of SEC23A, a related gene, could ameliorate the effect of reduced SEC23B expression.31
Sec23b-deficient mice (Sec23b gt/gt) have no apparent anemia phenotype, but they die shortly after birth with degeneration of secretory tissues, including pancreas and salivary glands.32 The disparate mouse and human phenotypes may result from either residual SEC23B function associated with the hypomorphic mutations found in humans or a species-specific shift in function between the closely related paralogues SEC23A and SEC23B.
CDA III
The gene causing CDA III in a Swedish family has been previously mapped to a region on chromosome 15q23.33 This region was targeted by array-based sequence capture, and a novel mutation, c.2747C>G(p.P916R) in the KIF23 gene, was shown to be associated with the autosomal dominant form of CDA III.5 The same mutation was also found in CDA III patients from an American family without any known relation to the Swedish kindred.5 KIF23 encodes a kinesin-superfamily molecule, mitotic kinesin-like protein 1 (MKLP1), a mitotic protein essential for cytokinesis.34,35 MKLP1 interacts with Arf6 (adenosine 5′-diphosphate–ribosylation factor 6), ultimately forming an extended b sheet, which interacts with the membrane surface at the cleavage furrow. The Arf6-MKLP1 complex has a crucial role in cytokinesis by connecting the microtubule bundle and membranes at the cleavage plane.34 Knockdown of Arf6 results in binucleated and multinucleated cells, which is a clinical hallmark of CDA III erythroblasts.33 In knockdown and rescue experiments with HeLa cells, cytokinesis failure and binucleated cells were seen more frequently with the P916R mutant than the wild-type GFP-MKLP1, indicating that the P916R mutation impairs the function of MKLP1 in cytokinesis.5 Two isoforms of MKLP1 were also identified, with varying expression in different tissues, possibly explaining the mutation’s heterogenic effects in between tissues.5
CDA variants
Several forms of CDAs do not fulfill classical BM morphologic or biochemical criteria.2,7 Based on light microscopy and EM of BM, Wickramasinghe classified them into 4 additional CDA groups (IV-VII), each one including relatively few patients.2 Nevertheless, there were still descriptions of patients not belonging to any of the groups,2,36,37 suggesting further that CDA subtypes are extremely heterogeneous and probably represent multiple unrelated genetic disorders. Indeed, in recent years additional genetic defects associated with CDA phenotypes have been identified. They include mutations in erythroid transcription factor genes (GATA-1 and KLF1) and mutations in other genes where CDA is part of a broader clinical syndrome.
Mutations in erythroid transcription factors GATA-1 and KLF1.
GATA-1 plays an integral role in the development of erythroid and megakaryocytic lineages.38 With the help of its cofactor FOG-1 (friend of GATA-1), it coordinates hematopoietic cell differentiation by activating linage-specific mature forms and repressing genes associated with undifferentiated states. There are currently a few documented familial inherited forms of X-linked thrombocytopenia characterized by macrothrombocytopenia, bleeding tendency with varying degrees of dyserythropoiesis ranging from hydrops fetalis and transfusion dependency to dyserythropoiesis without anemia.39 All identified mutations affect the GATA-1:FOG-1 interaction. One additional mutation affecting DNA binding is associated with X-linked thrombocytopenia and thalassemia with unbalanced α/β hemoglobin chain production resembling β-thalassemia.39
KLF1 is an erythroid transcription factor involved in promoting erythropoiesis and attenuating megakaryocytic differentiation.40 Additionally, it takes part in the developmental switch between fetal and adult hemoglobin. KLF1 is also required for cell cycle divisions preceding terminal erythroid differentiation and cell cycle exit during erythroid maturation. One KLF1 mutation (c.937G>A), resulting in substitution of the evolutionary conserved glutamate 325 by lysine (E325K), has been identified in more than 5 CDA patients.41-45 In some of these patients other red blood cell abnormalities have been documented including persistent expression of embryonic ς and ε globin chains, high fetal hemoglobin (40%), novel intra erythroblastic and intra erythrocytic inclusions, deficiency of erythroid CD44 and aquaporin 1.41,42 Interestingly, one of those patients41,42 was first identified by Wickramasinghe in 1991 and was classified as having a distinct form of CDA (non-IV–VII group).46 She was born with hydrops fetalis, required red cell transfusions over the first year of life, and thereafter developed a non-transfusion dependent anemia.
CDA as part of a broader syndrome.
Three different syndromes including CDA have been molecularly studied so far:
Majeed syndrome is a rare autosomal recessive disorder characterized by early onset of chronic recurrent multifocal osteomyelitis, inflammatory dermatosis, and microcytic dyserythropoietic anemia with increased marrow erythropoiesis and up to 25% binucleated and trinucleated erythroblasts.47 The disease is caused by LPIN2 mutations. Lipin2 is involved in lipid metabolism by affecting phosphatide phosphatase activity.48,49 The link between lipin2 and the hematologic features or the autoinflammatory bone disease is unclear.
CDA with exocrine pancreatic insufficiency and calvarial hyperostosis caused by a mutation in COX4I2 was also described.50 Blood smear revealed aniso-pokilocyosis, basophilic stippling, and few normoblasts. In BM erythroid hyperplasia, megaloblastic changes and bi- and multinucleated erythroblasts were present. The COX4I2 isoform of the COX4 protein is an essential structural subunit of cytochrome c oxidase complex.
Mevalonate kinase deficiency is a rare inborn error of metabolism caused by mutations in the mevalonate kinase gene. A recent report described a patient with an intermediate form of the disease and 6% to 8% of marrow erythroblasts demonstrating dyserythropoietic features.51
Current diagnostic approach
The identification of the mutated genes involved in the majority of CDA patients improved the diagnostic possibilities. When the clinical picture is suggestive and the findings in peripheral blood and BM light microscopy are compatible with one of the classical I to III types (Table 1; Figure 1), the next diagnostic step should be to sequence the appropriate gene. For these patients, additional specialized tests including BM EM and/or sodium dodecyl sulfate polyacrylamide gel electrophoresis of red cell membranes are not required. Whenever clinical and hematologic findings are generally compatible with CDA despite the absence of specific microscopic features of CDA I to III, a variant CDA should be considered. In these selected cases, mutations in KLF1 and GATA-1 should be explored by DNA sequencing. Future analysis will probably include high-throughput genomic analyzing technology, which will eventually lead to identification of new causative genetic factors.
Conclusions
The hallmark of the CDAs is failure of terminal erythropoiesis. The identification of several CDA genes has improved the diagnostic aspect of this disease; however, no comprehensive explanation for the mechanism of erythropoietic disruption has been disclosed. CDA type III gene (KIF23) encodes a protein playing a critical role in cytokinesis, suggesting a possible mechanism for the development of multinucleated erythroblasts. Codanin-1 and possibly C15ORF41 may facilitate histone assembly into chromatin during cell cycle. The fact that the proteins encoded by the CDA I, II, and III genes are ubiquitously expressed while the disease manifestations are mainly erythroid restricted remains a quandary. Further studies on CDA-related proteins and identification of new genes causing CDA variants will continue to offer insight into different pathways underlying erythropoiesis.
Acknowledgments
The authors thank Lucio Luzzatto, Roberta Russo, and Mitchell J. Weiss from the Children’s Hospital of Philadelphia for helpful discussions. The authors also thank Dr Joanne Yacobovich from the Schneider Children’s Medical Center of Israel for her help in preparation of this manuscript.
This work was supported by the Italian Ministero dell’Università e della Ricerca; by Telethon, Italy (GGP09044) (A.I.); by contract grant MUR-PS 35-126/Ind; and by Regione Campania (grant DGRC 1901/200) (A.I.).
Authorship
Contribution: A.I., H.H., A.W., and H.T. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Achille Iolascon, CEINGE, Biotecnologie Avanzate, Via Gaetano Salvatore, 486, 80145 Naples, Italy; e-mail: achille.iolascon@unina.it.