Key Points
SH2B3 is a recessive tumor suppressor gene with germline and somatic mutations in ALL.
Abstract
The SH2B adaptor protein 3 (SH2B3) gene encodes a negative regulator of cytokine signaling with a critical role in the homeostasis of hematopoietic stem cells and lymphoid progenitors. Here, we report the identification of germline homozygous SH2B3 mutations in 2 siblings affected with developmental delay and autoimmunity, one in whom B-precursor acute lymphoblastic leukemia (ALL) developed. Mechanistically, loss of SH2B3 increases Janus kinase-signal transducer and activator of transcription signaling, promotes lymphoid cell proliferation, and accelerates leukemia development in a mouse model of NOTCH1-induced ALL. Moreover, extended mutation analysis showed homozygous somatic mutations in SH2B3 in 2 of 167 ALLs analyzed. Overall, these results demonstrate a Knudson tumor suppressor role for SH2B3 in the pathogenesis of ALL and highlight a possible link between genetic predisposition factors in the pathogenesis of autoimmunity and leukemogenesis.
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous disease in which multiple genetic alterations contribute to the malignant transformation of lymphoid progenitor cells. Although the pathogenesis of ALL is considered to be multifactorial, few environmental factors have been conclusively associated with increased risk.1 Recent genome-wide association studies of childhood ALL have suggested that common variation in CDKN2A/B, IKZF1, ARID5B, CEBPE, and BMI1-PIP4K2A may predispose to ALL.2-6 In addition, rare but recurrent cases of ALL have been reported in individuals affected with familial platelet disorder with a predisposition to myeloid malignancy (FPD/AML; OMIM #601399),7 Sotos syndrome (OMIM 117550),8 neurofibromatosis type 1 (OMIM 162200),9 and Börjeson-Forssman-Lehmann syndrome (OMIM 301900)10 resulting from germline mutations in the RUNX1, NSD1, NF1, and PHF6 tumor suppressor genes, respectively. On the basis of these observations, we hypothesized that leukemia development in the context of rare familial inherited disorders may indicate a tumor suppressor activity for the underlying genetic defect present in these kindreds.
In our study, we describe a consanguineous family of Eastern European Ashkenazi Jewish background with a germline mutation in the SH2B adaptor protein 3 (SH2B3) gene (also known as LNK) in which homozygous loss of SH2B3 associated with this allele showed growth retardation, mild developmental delay, chronic hepatitis, Hashimoto autoimmune thyroiditis, and B-precursor ALL. Here, we demonstrate that loss of SH2B3 is associated with increased signal transducer and activator of transcription 3 (STAT3) phosphorylation and augmented proliferation in patient-derived lymphoid cells and accelerated NOTCH1-induced leukemia in mice. These results, together with the identification of somatic loss of function mutations in SH2B3 in sporadic ALL cases, demonstrate a tumor suppressor role of SH2B3 in human leukemia and support a mechanistic role for deregulated cytokine signaling in the pathogenesis of this disease.
Materials and methods
Genotyping and homozygosity mapping
In accordance with the Declaration of Helsinki, all participants provided informed consent or assent for a protocol approved by the Columbia University Medical Center Institutional Review Board. Genomic DNA was extracted with Puregene kits (Qiagen, Valencia, CA) from whole blood samples taken from the 2 affected children and unaffected sibling and parents. DNA from all members of the family was genotyped with Affymetrix 250k single nucleotide polymorphism (SNP) microarray, GEO accession number GSE44025, (Affymetrix, Santa Clara, CA). Genotyping was performed according to the manufacturer’s protocol (Affymetrix Inc.). Raw genotyping data were processed with Genotype Console v3.2 (Affymetrix Inc.) and examined for Mendelian inconsistencies using the PEDCHECK program.11 Genome-wide homozygosity mapping was performed with the HomozygosityMapper program using the default settings for the Affymetrix 250k array12 to identify possible stretches of homozygous segments present within the 2 affected children, but absent in the unaffected child and parents.
Exome capture and whole-exome sequencing were performed at Knome (Cambridge, MA). The Agilent SureSelect Human All Exon Kit adapted for the Illumina sequencing platform was applied. Genomic DNA (5 µg) was randomly fragmented by sonication, treated with a Klenow fragment of DNA polymerase I to generate blunt ends, and then phosphorylated with polynucleotide kinase. Adaptor primers were annealed and ligated to the fragment ends. Ligated samples were hybridized with the baits for 48 hours, washed, and eluted using the Agilent protocol. After elution, the capture efficiency was evaluated via quantitative polymerase chain reaction. The resulting captured DNA samples were subjected to standard sample preparation procedures for Illumina Genome Analyzer sequencing according to the manufacturer’s instructions. The analysis pipeline included genome alignment followed by SNP/indel variant analysis. BWA (Burrows-Wheeler Aligner) was used to align 50-base reads to human genome GRCh37, allowing for a 3% error rate, 2 gaps, and a maximal edit distance of 5 bases. A custom module was used to select reads that most likely associate with the captured regions. SAMtools was used to call targeted bases, with valid-adjacent base calls that deviate from the reference treated as potential variations (SNP and indel [insertion or deletion]) and assigned a coverage-dependent Phred-scaled mutation probability.
SH2B3 mutation analysis in T-ALL and pre-B-ALL patient samples
T-ALL DNA samples were provided by the Eastern Cooperative Oncology Group (ECOG) and the Dana-Farber Cancer Institute in Boston, Massachusetts. Pre-B-ALL samples from the C10403 protocol were provided by the Hemato-Oncology Laboratory at the University of Padua in Padua, Italy. Informed consent to use leftover material for research purposes was obtained from all of the patients at the time of enrollment in the clinical trial according to the Declaration of Helsinki. All exon sequences from SH2B3 were amplified from genomic DNA by polymerase chain reaction and were analyzed by direct dideoxynucleotide sequencing.
Generation of lymphoblastoid cell lines
Peripheral blood samples were obtained from the Molecular Genetics Department at Columbia University Medical Center with informed consent and under the supervision of the local institutional review board committee. Mononuclear cells were collected by density centrifugation with Ficoll (GE Healthcare), and B lymphocytes were transformed with the Epstein-Barr virus (EBV) following standard procedures.13 Briefly, we resuspended peripheral blood mononuclear cells in EBV containing B95-8 conditioned media. We added cyclosporin A (2 µg/mL) to the media to inactivate natural killer cells and to prevent the activation of EBV-specific killer T lymphocytes. After 2 to 3 weeks, EBV-immortalized B cells were collected and subsequently grown in RPMI media supplemented with 10% fetal bovine serum with antibiotics.
Cell viability and proliferation studies
Cell growth was determined by measurement of the metabolic reduction of the tetrazolium salt MTT using the Cell Proliferation Kit I (Roche) following the manufacturer's instructions. Cell proliferation was analyzed with the CellTrace CFSE [carboxyfluorescein diacetate succinimidyl ester] Cell Proliferation Kit (Invitrogen) and by cell counts in cells grown at different serum concentrations. All experiments were done in triplicates.
Western blot analysis
Western blot analysis was performed following standard procedures using antibodies against SH2B3 (C20; Santa Cruz Biotechnology), phospho-Janus kinase 2 (JAK2), JAK2, phospho-STAT3, STAT3 (all from Cell Signaling), and against glyceraldehyde-3-phosphate dehydrogenase and actin beta as loading controls (Santa Cruz Biotechnology).
Plasmids
The pDONR-SH2B3 plasmid (Life Technologies) was used to subclone the hemagglutinin-tagged SH2B3 complementary DNA into the pLX301 Getaway destination vector (Addgene).
Lentiviral production and infection
pLX301 empty vector and pLX301-SH2B3 viral particles were produced on HEK293T cells by cotransfection with gag-pol and V-SVG–expressing vectors using the JetPEI transfection reagent (Polyplus). Viral supernatants were collected after 48 hours and were used for infection by spinoculation. After infection, cells were selected for 5 days in puromycin and ficolled the day before experiments.
Bone marrow transplants
Bone marrow from 8-week-old C57BL6 or Sh2b3−/− mice was harvested and lineage depleted using the Mouse Lineage Cell Depletion Kit (Milteny). Lin- progenitor cells were subjected to 2 rounds of infection with MigR1-ΔE-Notch1 retroviral particles. Fifty thousand Lin-Sca1+–infected cells were transplanted into irradiated C57BL6 recipients via intravenous injection. Transplanted animals were monitored for leukemia development by flow cytometry analysis of green fluorescent protein (GFP) and CD4/CD8 expression in peripheral blood.
Microarray analysis
RNA isolation from mice tumors was performed in triplicate using the RNeasy Micro kit (QIAGEN) according to the manufacturer’s protocol. Next, 10 ng of RNA from each sample was amplified using the Ovation PicoSL WTA System (NuGen), labeled by the Encore BiotinIL Module (NuGen), and hybridized to the Mouse430_2 (Affymetrix) microarray chips according to the manufacturer’s protocol. Expression values were normalized with GeneChip Robust Multiarray Averaging using the open-source Bioconductor project (www.bioconductor.org) within the statistical computing environment R (http://cran.r-project.org). Differentially expressed genes were selected by the Student t test (P < .001) and fold change greater than 1.5. Gene Set Enrichment Analyses (GSEAs) were carried out in MATLAB, including the C2 molecular signature database gene sets14 and hematopoietic stem cell signature genes from the D-MAP data set (top 200 upregulated or downregulated genes in hematopoietic stem cell populations vs all other hematopoietic populations.15
Statistical analysis
Statistical assessment of significance in parametric tests was performed using the Student t test. Statistical significance of survival was calculated using the log-rank test.
Results
Clinical description
Here, we studied a 6-year-old boy with a clinical history of growth retardation, mild developmental delay, chronic hepatitis of probable autoimmune origin, and Hashimoto thyroiditis. He had an uncomplicated prenatal history except for the ultrasonographic finding of a single umbilical artery without other associated anomalies. He was small for gestational age with a birth weight of 2324 g. He remained in the hospital for 2 weeks after birth because of jaundice and anemia, thrombocytopenia, and high white cell counts. Results of a bone marrow biopsy at age 4 weeks were normocellular with 10% to 15% blasts and no megakaryocytes. At that time, hepatosplenomegaly was noted. White cell blood counts normalized without intervention by age 14 months; however, the hepatosplenomegaly persisted. A needle liver biopsy result demonstrated fragmented nodules of cirrhotic liver. Histologic findings included chronic inflammatory cells within the portal tracts and fibrous septa combined with foci of periportal interface hepatitis and parenchymal necroinflammation. No globules of α-1-antitrypsin were evident on diastase pretreated periodic acid-Schiff stain, and histologic analysis revealed no ground-glass inclusions, cholestasis, or excess iron accumulation. Results on antinuclear antibody, F-actin antibody, and liver-kidney-microsome antibody were all negative. At age 4 years, the patient was diagnosed with Hashimoto thyroiditis. At age 6 years, he is currently 100.5 cm tall (<3%), weighs 16.3 kg (10%), and has a head circumference of 48.5 cm (<3%). He had mild developmental delays, being able to sit without support at age 8 months and walk at age 18 months. He also presented with speech delay and required speech therapy, but he attends school in a regular first-grade class.
His family history is significant for a sister who shares many similar features and 1 healthy brother. His parents are third cousins of Eastern European Ashkenazi Jewish ancestry (Figure 1). His younger sister is 2.5 years old with a similar history of growth retardation, mild developmental delay, and chronic hepatitis in whom normal karyotype B-precursor ALL developed at age 18 months. She was born after an uncomplicated prenatal course with no ultrasonographic anomalies. She was small for gestational age with a birth weight of 1842 g, 45 cm of length, and 31.5 cm of head circumference. As an infant she experienced hepatosplenomegaly and showed mild delay in her development, with height within 3% and 25% since birth, weight at 3% to 10%, and a head circumference at 3% to 10%. At age 18 months, she was diagnosed with B-precursor ALL. Blood smear revealed 10% blasts with elevated white blood cell counts (5×109 L−1), anemia (hemoglobin 55 g L−1), and thrombocytopenia (platelet count 55×109 L−1). Bone marrow aspirate and biopsy results showed 70% lymphoblasts. Immunophenotypic analysis revealed approximately 81% of the blasts expressing TdT, dim CD45, HLA DR, CD38, CD34, CD19, CD10, dim CD22, and CD7. Bone marrow karyotype was normal (46, XX). Fluorescence in situ hybridization showed a CDKN2A deletion in 42.6% of cells (33.1% of cells with heterozygous deletion and 9.5% with homozygous deletion).
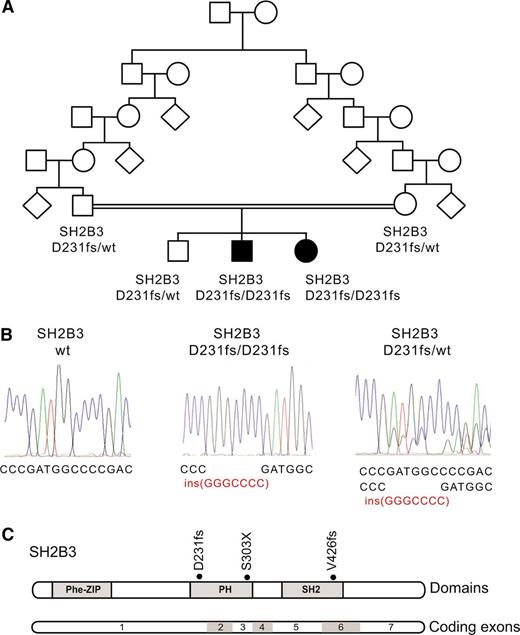
Identification of a SH2B3 mutation in a familial autoimmunity and acute leukemia kindred. (A) Family pedigree. Affected individuals are shaded in black. (B) DNA sequencing chromatograms showing the SH2B3 D231fs mutation in the proband and a heterozygous carrier compared with a control SH2B3 wild-type DNA sample. (C) Schematic representation of the structure of the SH2B3 protein. The phenylalanine zipper (Phe-ZIP), the Plekstrin homology domain (PH), and the Scr Homology 2 domains (SH2) are indicated. The SH2B3 D231fs mutation identified in this family and somatic SH2B3 mutations found in ALL patient samples are shown.
Identification of a SH2B3 mutation in a familial autoimmunity and acute leukemia kindred. (A) Family pedigree. Affected individuals are shaded in black. (B) DNA sequencing chromatograms showing the SH2B3 D231fs mutation in the proband and a heterozygous carrier compared with a control SH2B3 wild-type DNA sample. (C) Schematic representation of the structure of the SH2B3 protein. The phenylalanine zipper (Phe-ZIP), the Plekstrin homology domain (PH), and the Scr Homology 2 domains (SH2) are indicated. The SH2B3 D231fs mutation identified in this family and somatic SH2B3 mutations found in ALL patient samples are shown.
Identification of a mutation in SH2B3
The presence of parental consanguinity and the occurrence of male and female affected children from unaffected parents in this family suggested an autosomal recessive mode of inheritance (Figure 1A). In addition, we proposed that a common underlying genetic defect resulting in dysregulation of the development and function of the immune system could contribute to the concurrence of a lymphoid neoplasia and an autoimmune disorder in this family. Thus, to identify the possible causative genetic alteration underlying these defects, we performed genome-wide homozygosity mapping to identify extended homozygous regions shared among the affected children and not shared with their unaffected sibling. SNP microarray analysis and homozygosity mapping identified 2 regions of shared homozygosity on chromosome 12 (106 228,244-119 573,834) and chromosome 3 (76 932,137-80 493,393) in the genome of the 2 affected children and absent in their unaffected sibling. These regions of homozygosity include 186 known or predicted genes (180 on chromosome 12 and 6 on chromosome 3) based on NCBI build 39 and ENSEMBL annotations.
Next, we performed whole-exome sequencing of genomic DNA of each of the 2 affected siblings, which identified 12 predicted coding, nonsynonymous variants in 11 genes mapping to these regions of linkage and homozygosity (12 on chromosome 12 and none on chromosome 3), including a homozygous frameshift c.671insGGCCCCG p. Asp231Gly fs*38 mutation in the SH2B3 gene (Figure 1B-C). Dideoxynucleotide sequencing of SH2B3 confirmed that both affected siblings were homozygous for this mutation, whereas the unaffected brother and both parents were carriers. Analysis of 2000 normal Ashkenazi Jewish adults identified 1 SH2B3 c.671insGGCCCCG carrier, and no individuals who were homozygous for the insertion.
Impaired SH2B3 expression, dysregulated JAK-STAT signaling, and increased cell proliferation in SH2B3 mutant lymphoid cells
SH2B3 encodes an adaptor protein involved in the negative regulation of cytokine-signaling pathways in the hematopoietic and lymphoid systems.16 Notably, Sh2b3-deficient mice show accumulation of B-cell progenitors and immature B cells in the bone marrow and spleen and expansion of the hematopoietic stem cell pool with increased self-renewal: phenotypes potentially related to the development of autoimmunity and leukemic transformation.17-20
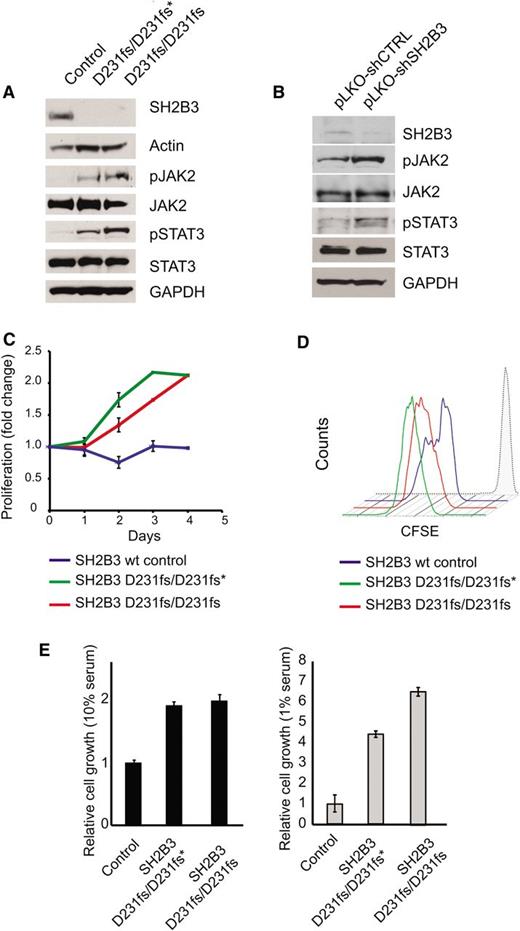
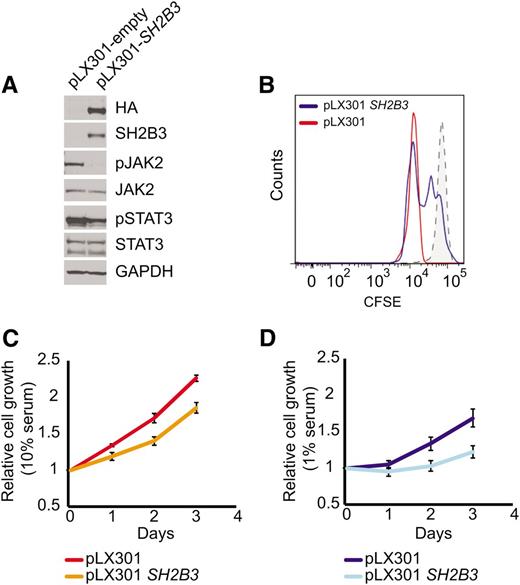
Immunosuppressive therapy precluded a direct analysis of lymphoid populations in the 2 affected siblings harboring the SH2B3 c.671insGGCCCCG allele. Thus, we generated immortalized lymphoblastoid cell lines to test the functional consequences of this mutation. Western blot analysis with a SH2B3 C-terminal antibody showed complete loss of SH2B3 protein expression in SH2B3 c.671insGGCCCCG mutant cells compared with cells derived from a SH2B3 wild-type control (Figure 2A). Consistently, analysis of JAK-STAT signaling in SH2B3-deficient cells showed increased JAK2 and STAT3 phosphorylation compared with SH2B3 wild-type controls (Figure 2A). Similarly, short hairpin RNA (shRNA) knockdown of SH2B3 in JURKAT ALL cells resulted in increased JAK2 and STAT3 phosphorylation compared with control shRNA–expressing cells (Figure 2B). Phenotypically, patient-derived SH2B3 mutant lymphoblastoid cells showed increased growth and proliferation compared with controls, particularly when grown in low serum conditions (Figure 2C-E). Importantly, lentiviral expression of wild-type SH2B3 in patient-derived SH2B3-deficient lymphoblastoid cells resulted in decreased JAK2 and STAT3 phosphorylation (Figure 3A) and reduced cell proliferation (Figure 3B-D).
Functional characterization of the SH2B3 D231fs allele. (A) Western blot analysis of SH2B3 expression and JAK-STAT signaling in lymphoblastoid cells from the proband (noted as SH2B3 D231fs/D231fs*), the affected sibling (SH2B3 D231fs/D231fs), and an SH2B3 wild-type healthy control. (B) Western blot analysis of SH2B3 expression and JAK-STAT signaling in JURKAT T-ALL cells expressing SH2B3 targeting shRNAs (pLKO-shSH2B3) or an inactive control shRNA (pLKO-shCTRL). (C) Differential growth curves of SH2B3 wild-type and patient-derived SH2B3 mutant lymphoblastoid cells grown in 1% fetal bovine serum (FBS) containing media. (D) CFSE dilution analysis of cell cycle kinetics in SH2B3 patient-derived SH2B3 mutant lymphoblastoid cells in 1% FBS containing media. (E) Differential cell growth of SH2B3 wild-type and patient-derived SH2B3 mutant lymphoblastoid cells in standard (10% FBS) and low serum (1% FBS) conditions. Experiments shown were performed at least twice, and quantitative assays were performed in triplicate.
Functional characterization of the SH2B3 D231fs allele. (A) Western blot analysis of SH2B3 expression and JAK-STAT signaling in lymphoblastoid cells from the proband (noted as SH2B3 D231fs/D231fs*), the affected sibling (SH2B3 D231fs/D231fs), and an SH2B3 wild-type healthy control. (B) Western blot analysis of SH2B3 expression and JAK-STAT signaling in JURKAT T-ALL cells expressing SH2B3 targeting shRNAs (pLKO-shSH2B3) or an inactive control shRNA (pLKO-shCTRL). (C) Differential growth curves of SH2B3 wild-type and patient-derived SH2B3 mutant lymphoblastoid cells grown in 1% fetal bovine serum (FBS) containing media. (D) CFSE dilution analysis of cell cycle kinetics in SH2B3 patient-derived SH2B3 mutant lymphoblastoid cells in 1% FBS containing media. (E) Differential cell growth of SH2B3 wild-type and patient-derived SH2B3 mutant lymphoblastoid cells in standard (10% FBS) and low serum (1% FBS) conditions. Experiments shown were performed at least twice, and quantitative assays were performed in triplicate.
SH2B3 expression rescues deregulated cell signaling growth and proliferation in patient-derived SH2B3 mutant lymphoblastoid cells. (A) Western blot of patient-derived SH2B3-deficient lymphoblastoid cells reconstituted with empty vector (pLX301) or SH2B3 expressing lentiviruses (pLX301 SH2B3). (B) CFSE dilution assay analysis of cell cycle kinetics in SH2B3 mutant cells infected with empty vector or SH2B3 expressing lentiviruses (C,D). Differential cell growth of SH2B3 mutant cells infected with empty vector or SH2B3 expressing lentiviruses in standard (10% FBS) (C) and low-serum (1% FBS) (D) culture conditions. Experiments shown were performed twice, and quantitative assays were performed in triplicate.
SH2B3 expression rescues deregulated cell signaling growth and proliferation in patient-derived SH2B3 mutant lymphoblastoid cells. (A) Western blot of patient-derived SH2B3-deficient lymphoblastoid cells reconstituted with empty vector (pLX301) or SH2B3 expressing lentiviruses (pLX301 SH2B3). (B) CFSE dilution assay analysis of cell cycle kinetics in SH2B3 mutant cells infected with empty vector or SH2B3 expressing lentiviruses (C,D). Differential cell growth of SH2B3 mutant cells infected with empty vector or SH2B3 expressing lentiviruses in standard (10% FBS) (C) and low-serum (1% FBS) (D) culture conditions. Experiments shown were performed twice, and quantitative assays were performed in triplicate.
Recurrent somatic mutations in SH2B3 in ALL
To better establish the potential tumor suppressor role of SH2B3 in the pathogenesis of ALL, we performed dideoxynucleotide sequencing of a cohort of 167 ALL samples, including 119 B-precursor ALLs and 96 T-cell ALLs. This analysis identified a homozygous c.1279insCTGTTGCCGTGTGC p.Gln427Pro fs*40 in a patient with pre-B ALL and a homozygous nonsense SH2B3 mutation c.908C>A p.Ser303* in a pediatric T-ALL sample (Figure 1B). Notably, analysis of genomic DNA obtained from peripheral blood leukocytes from this patient with T-ALL at the time of clinical remission recovered the wild-type SH2B3 sequence demonstrating the somatic origin of the c.908C>A p.Ser303* SH2B3 mutation. In addition, mutation analysis of the NOTCH1 oncogene showed the co-occurrence of a prototypical NOTCH1-activating mutation (NOTCH1 L1601P) in the leukemic DNA from this patient, suggesting a cooperative role of NOTCH1 signaling and SH3B2 loss in the pathogenesis of T-ALL.
Loss of Sh2b3 accelerates the development of NOTCH-induced leukemias in mice
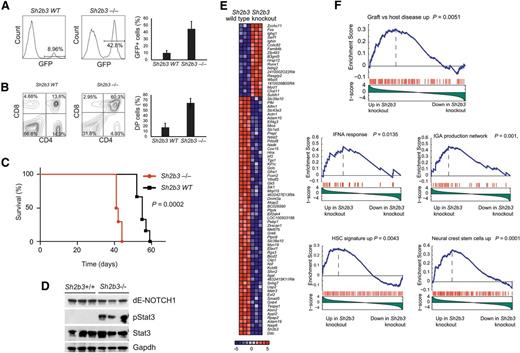
Following on this result, and to formally address the interaction between oncogenic NOTCH1 and Sh2b3 loss in T-cell transformation, we analyzed the effects of genetic inactivation of Sh2b3 in a retroviral-transduction bone marrow transplantation model of NOTCH1-induced T-ALL.21 In this experiment, we infected Lin- hematopoietic progenitor cells from Sh2b3 knockout and wild-type littermate controls with bicistronic GFP retroviruses driving expression of ΔE-NOTCH1, a truncated and constitutively active form of a human NOTCH1 oncogene,22 and transplanted them into irradiated isogenic recipients. Forced expression of activated NOTCH1 in this model typically results in a wave of extrathymic T-cell lymphopoiesis 3 weeks after transplantation with the transient release of immature CD4/CD8 double-positive cells into peripheral blood, which is followed by full leukemia transformation 5 to 10 weeks later.21 Notably, mice transplanted with Sh2b3−/− ΔE-NOTCH1–expressing progenitors in this experiment showed a marked increase in the number of GFP-positive and CD4/CD8-circulating cells in peripheral blood 3 weeks after transplantation compared with animals transplanted with Sh2b3 wild-type ΔE-NOTCH1–infected progenitor cells (Figure 4A-B). This mechanism was followed by accelerated overt leukemia development and leukemia-associated mortality compared with controls (Figure 4C). Notably, western blot analysis of Stat3 phosphorylation in these tumors revealed a marked increase in phospho-Stat3 levels in Sh2b3 null T-ALL lymphoblasts compared with wild-type controls (Figure 4D). Following these results, and to gain further insight on the role and mechanisms of Sh2b3 loss in T-cell transformation, we performed microarray gene expression analysis of Sh2b3 wild-type and Sh2b3−/−NOTCH1–induced leukemias. In this analysis, Sh2b3 NOTCH1–induced tumors showed a characteristic gene expression signature consisting of 19 upregulated and 62 downregulated genes (>1.5-fold change, P < .005) (Figure 4E). GSEA of this signature against the C2 molecular signature database, a collection of 4850 curated genesets assembled from online pathway databases, publications, and knowledge of domain experts,14 revealed a significant enrichment of genes upregulated in graft-versus-host disease and of genes implicated in interferon-α response and IgA production in the intestine in Sh2b3 knockout leukemia upregulated transcripts (Figure 4F). These results further support a mechanistic link between SH2B3 loss, leukemia transformation, and the development of autoimmunity. Moreover, and consistent with the proposed role of SH2B3 in the regulation of stem cell homeostasis,17-20 Sh2b3 knockout leukemia signatures were also enriched in genes upregulated in neural crest stem cells and in a hematopoietic stem cell signature generated from the D-MAP data set, a collection of gene expression profiles from multiple human hematopoietic populations15 (Figure 4F).
Genetic loss of Sh2b3 accelerates NOTCH1-induced T-ALL. (A) Representative plots (of one of 6 animals per group) and quantification of GFP-expressing preleukemic cells in peripheral blood of mice transplanted with either Sh2b3−/− or wild-type hematopoietic progenitors infected with retroviruses expressing ΔE-NOTCH1. (B) Representative plots (of one of 6 animals per group) and quantification of double-positive (CD4+CD8+; DP) cells in mice 3 weeks after transplantation. (C) Kaplan-Meier survival curves of mice transplanted with Sh2b3−/− (n = 6) or wild-type (n = 6) hematopoietic progenitors expressing ΔE-NOTCH1. (D) Western blot analysis of Stat3 phosphorylation in NOTCH1-induced tumors generated from hematopoietic precursors from Sh2b3 wild-type and Sh2b3−/− mice. (E) Heat map diagram of top ranking differentially expressed genes by the Student t test in Sh2b3−/− (n = 3) vs Sh2b3 wild-type (n = 3) NOTCH1-induced leukemias. Genes in the heat map are shown in rows, and each individual sample is shown in 1 column. The scale bar shows color-coded differential expression, with red indicating higher levels of expression and blue indicating lower levels of expression. (F) GSEA enrichment plots of gene sets related to autoimmunity, immune function, and stem cells associated with the Sh2b3−/− NOTCH1–induced leukemia signature.
Genetic loss of Sh2b3 accelerates NOTCH1-induced T-ALL. (A) Representative plots (of one of 6 animals per group) and quantification of GFP-expressing preleukemic cells in peripheral blood of mice transplanted with either Sh2b3−/− or wild-type hematopoietic progenitors infected with retroviruses expressing ΔE-NOTCH1. (B) Representative plots (of one of 6 animals per group) and quantification of double-positive (CD4+CD8+; DP) cells in mice 3 weeks after transplantation. (C) Kaplan-Meier survival curves of mice transplanted with Sh2b3−/− (n = 6) or wild-type (n = 6) hematopoietic progenitors expressing ΔE-NOTCH1. (D) Western blot analysis of Stat3 phosphorylation in NOTCH1-induced tumors generated from hematopoietic precursors from Sh2b3 wild-type and Sh2b3−/− mice. (E) Heat map diagram of top ranking differentially expressed genes by the Student t test in Sh2b3−/− (n = 3) vs Sh2b3 wild-type (n = 3) NOTCH1-induced leukemias. Genes in the heat map are shown in rows, and each individual sample is shown in 1 column. The scale bar shows color-coded differential expression, with red indicating higher levels of expression and blue indicating lower levels of expression. (F) GSEA enrichment plots of gene sets related to autoimmunity, immune function, and stem cells associated with the Sh2b3−/− NOTCH1–induced leukemia signature.
Discussion
The SH2B3 gene encodes an adaptor protein involved in the negative regulation of multiple signaling molecules and tyrosine kinase receptors with important roles in hematopoietic development, including JAK2, c-KIT, MPL, EPOR, PDFGRA, PDGFRB, FLT3, and c-FMS.20,23-33 As is the case for other negative regulators of cytokine signaling such as suppressor of cytokine signaling proteins, SH2B3 is transcriptionally upregulated upon JAK-STAT activation and has been proposed to function as a central node in the negative control of different signaling pathways.28,29 Expression analyses and functional characterization of Sh2b3 in the hematopoietic system have revealed a critical role in the in the control of self-renewal and quiescence in hematopoietic stem cells.17,19,30,31,34,35 Thus, Sh2b3 null mice show an expansion of the hematopoietic stem cell compartment, together with increased erythroid and megakaryocytic precursors in the bone marrow and elevated white cell and platelet counts in peripheral blood.17,19,30,31,34,35 These phenotypes are the result of increased thrombopoietin and thrombopoietin plus erythropoietin signaling in hematopoietic stem cells and progenitors, respectively.32,33 In addition, Sh2b3 knockout mice have marked splenomegaly resulting from the expansion and accumulation of B cells in response to stem cell factor stimulation, revealing a particularly prominent role of Sh2b3 in the control of B-cell homeostasis.17,18 In contrast, and despite a proposed role as a negative regulator of T-cell receptor signaling, T-cell development seems to be unperturbed in Sh2b3-deficient mice.23,36
Increased and dysregulated cytokine signaling is a major mechanism in the pathogenesis of multiple hematopoietic malignancies. Thus, activating mutations in cytokine receptors including c-KIT and FLT3 are highly prevalent in acute myeloid leukemias, and gain-of-function mutations in IL7R and rearrangement and consequent overexpression of CRLF2 are frequently found in ALLs.37-41 In addition, the JAK2 V617F–activating allele is a hallmark of polycythemia vera and essential thrombocytosis.42 Notably, SH2B3 has been shown to interact with several of these mutant signaling factors including the KIT D816V, FLT3 ITD, and JAK2 V617F proteins.28,29,43 Moreover, loss of Sh2b3 can increase the oncogenic effects of JAK2 V617F expression in a mouse model of myeloproliferative disease.44 Consistently, and in support of a tumor suppressor role of SH2B3 in myeloid tumors, heterozygous mutations clustered in the PH domain of SH2B3 have been described in myeloproliferative neoplasms including 3% of essential thrombocytosis, 4% of primary myelofibrosis, and 7% of chronic myelomonocytic leukemias.45 In addition, germline heterozygous SH2B3 mutations involving the PH domain have also been found in 2 patients with essential erythrocytosis.46
Here, we describe a consanguineous family in which homozygous loss of SH2B3 was associated with developmental delay, growth retardation, and chronic hepatitis in 2 siblings. In addition, Hashimoto thyroiditis, an autoimmune disorder typically affecting middle-aged adult women, developed in one of the affected children at age 4 years and the other sibling was diagnosed with B-precursor ALL at age 18 months. The cause of developmental delay, growth retardation, and hepatitis in this kindred remains to be firmly established. However, a possible autoimmune mechanism has been proposed for the progressive liver damage in the 2 affected children based on the negative results of an extensive battery of tests ruling out other causes, the co-occurrence of Hashimoto thyroiditis in one of the patients, and the clinical amelioration of the hepatitis in one of the children during receipt of chemotherapy for the treatment of ALL. SNP array analysis and exome sequencing in this family identified a loss of function allele in SH2B3 as the only coding deleterious variant segregating with these phenotypes. However, it is possible that an as-yet unidentified noncoding pathogenic mutation may account for the developmental delay and growth retardation in these children. A role for mutational loss of SH2B3 in the deregulation of the immune system is supported by our results, demonstrating an increased proliferation advantage of lymphoblastoid cells from these patients, and correction of this cellular phenotype by re-expression of wild-type SH2B3. Notably, several recent genome-wide association studies have associated a coding variant in the PH domain of SH2B3 (SNP rs3184504, SH2B3 R262W) with increased risks for celiac disease, type 1 diabetes, asthma, and multiple sclerosis, supporting a role of SH2B3 in the pathogenesis of inflammatory and autoimmune disorders.47-50
In this context, the development of B-precursor ALL in one of the 2 children harboring homozygous loss of SH2B3 supported a potential pathogenic role for deregulated cytokine signaling resulting from SH2B3 loss in this malignancy. Somatic mutations in SH2B3 have been described albeit at low frequency in the context of genomic studies of high-risk B-precursor ALLs51 and early T-cell precursor ALL.52 In addition, a recent report has shown a primary leukemia xenograft harboring an SH2B3 deletion and an activating IL7RA mutation with associated high levels of STAT5 phosphorylation and response to JAK inhibition with ruxolitinib.53 However, a comprehensive mutation analysis of SH2B3 in this disease has not been reported, and a direct pathogenic role of SH2B3 loss in ALL has not been formally established before. Here, we describe 2 additional ALL cases, one B-precursor ALL and one T-ALL, harboring homozygous truncating mutations in SH2B3. In one case with available material, analysis of remission DNA demonstrated the somatic origin of the SH2B3 mutation. These findings, together with the accelerated development of T-ALL on expression of oncogenic NOTCH1 in Sh2b3 null hematopoietic progenitors, support a recessive tumor suppressor role for SH2B3 in ALL. In this context, heterozygous carriers of the familial SH2B3 D231fs allele identified here were asymptomatic and have no discernible alterations in the hematopoietic system. This finding is in contrast with the development of essential erythrocytosis in germline carriers of SH2B3 PH mutations. This observation, together with the heterozygous nature of somatic SH2B3 PH mutations found in myeloid tumors, suggest a dominant effect of SH2B3 PH alleles in the disruption of hematopoietic homeostasis.
Overall, the results shown in our study demonstrate a tumor suppressor role for SH2B3 in the pathogenesis of ALL and associate germline homozygous loss of this negative regulator of cytokine signaling with the development of autoimmunity.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Peter H. Wiernik as ECOG leukemia committee chair at the time the E2993 study was initiated.
This work was supported by a research grant from Bonei Olam; an Innovative Research Award by the Stand Up to Cancer Foundation (A.A.F.); and the National Institutes of Health (ECOG Leukemia Tissue Bank grants CA21115, CA14958, and CA17145).
A.P.-G. is a postdoctoral researcher funded by the Rally Foundation.
Authorship
Contribution: A.P.-G. performed mutation analysis and in vitro and in vivo functional analysis of SH2B3 and wrote the manuscript; A.A.-I. performed bioinformatic analyses; M.H. performed sequence analysis; I.R. generated and analyzed lymphoblastoid cell lines; K.K., E.P., J.R., J.M.R., M.S.T., M.P., and G.B. provided clinical samples and clinical information; C.A.L. and C.J. analyzed exome sequencing data; W.T. provided SH2B3 knockout mice; W.K.C. designed the study, directed genetic studies, identified the SH2B3 c.671insGGCCCCG p. Asp231Gly fs*38 mutation, and wrote the manuscript; and A.A.F. designed and directed functional and genetic studies, supervised research, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adolfo A. Ferrando, Institute for Cancer Genetics, Columbia University, Irving Cancer Research Center 4-402A, 1130 St Nicholas Ave, New York, NY, 10032; e-mail: af2196@columbia.edu.
References
Author notes
W.K.C. and A.A.F. contributed equally to this work.