Key Points
Low free protein S and low total protein S levels could not identify subjects at risk for venous thrombosis in a population-based study.
Protein S testing and subsequent testing on PROS1 mutations should not be considered in unselected patients with venous thrombosis.
Abstract
In thrombophilic families, protein S deficiency is clearly associated with venous thrombosis. We aimed to determine whether the same holds true in a population-based case-control study (n = 5317). Subjects were regarded protein S deficient when protein S levels were <2.5th percentile of the controls. Free and total protein S deficiency was not associated with venous thrombosis: free protein S < 53 U/dL, odds ratio [OR] 0.82 (95% confidence interval [CI], 0.56-1.21) and total protein S < 68 U/dL, OR 0.90 (95% CI, 0.62-1.31). When lower cutoff values were applied, it appeared that subjects at risk of venous thrombosis could be identified at levels <0.10th percentile of free protein S (<33 U/dL, OR 5.4; 95% CI, 0.61-48.8). In contrast, even extremely low total protein S levels were not associated with venous thrombosis. PROS1 was sequenced in 48 subjects with free protein S level <1st percentile (<46 U/dL), and copy number variations were investigated in 2718 subjects, including all subjects with protein S (free or total) <2.5th percentile. Mutations in PROS1 were detected in 5 patients and 5 controls reinforcing the observation that inherited protein S deficiency is rare in the general population. Protein S testing and PROS1 testing should not be considered in unselected patients with venous thrombosis.
Introduction
Protein S is a vitamin K-dependent glycoprotein that is synthesized in hepatocytes and endothelial cells.1 Protein S circulates at a concentration of ∼350 nM, of which 40% is free and 60% is bound to C4b-binding protein.2 Protein S assists in the downregulation of thrombin formation by stimulating the activity of both activated protein C and tissue factor pathway inhibitor.3,4
Hereditary protein S deficiency is a haploinsufficiency disorder associated with an increased risk of venous thrombosis.5 Protein S deficiency can be classified as type I (decreased levels of both total and free protein S antigen), type II (decreased activated protein C–cofactor activity but total and free protein S antigen levels within their normal ranges), and type III (decreased levels of free protein S antigen levels only).6 The genetic basis of protein S deficiency is heterogeneous. More than 200 mutations in PROS1 have been described, the vast majority missense or nonsense mutations.7-9 Sequencing exons and splicing junctions of PROS1 has been successful in the identification of a mutation in only approximately 50% of families with protein S deficiency.8 We, along with others, have shown that gross deletions and insertions of PROS1 are present in approximately 30% of the point mutation-negative families.5,10,11
Most of the insights regarding the association between protein S deficiency and venous thrombosis have been derived from studies of thrombophilic families. This has led to the notion that the risk of venous thrombosis in protein S deficient subjects is 5 to 10 times higher than in nonprotein S deficient relatives.5,12-14 The clinical relevance for the individual patient is substantial, as subjects with familial protein S deficiency have a first venous thrombosis incidence of 0.7% per year15 and an annual recurrence risk of 6% to 10%.14,16,17 It is not clear whether the conclusions from these family studies can be extrapolated to the general population. The coinheritance of other defects may contribute to or explain the risks in families, as was shown for factor V Leiden, and protein C deficiency.18,19 Relatively small population-based studies (with a maximum number of participants of 327)19 point toward a lower risk of thrombosis associated with protein S deficiency. Some studies reported up to 2.5 times increased risks, whereas others found no increase at all.19-21
We set out to determine whether low levels of free or total protein S were associated with an increased risk of venous thrombosis in a large population-based case-control study. The molecular basis for protein S deficiency was also investigated by analysis of copy number variation of PROS1 and resequencing of subjects with the lowest levels of protein S to support the risk estimates based on plasma levels.
Methods
Subjects
The Multiple Environmental and Genetic Assessment of risk factor for venous thrombosis (MEGA) case-control study has been previously described.22 Consecutive patients aged 18 to 70 years, with a first episode of deep venous thrombosis or pulmonary embolism between March 1, 1999 and August 31, 2004, were included, with a total of 4956. Age- and gender-matched controls were partners of patients (n = 3297) or recruited by random digit dialing (RDD) between January 1, 2002 and December 1, 2004 (n = 3000). The participants filled out a questionnaire on potential risk factors for venous thrombosis. Blood sampling was performed for participants through May 2002.
All subjects provided informed consent in accordance with the Declaration of Helsinki. The study was approved by the Medical Ethics Committee of the Leiden University Medical Center.
Blood samples
Patients who were diagnosed by May 31, 2002 were invited, together with their partners, for a blood sample at least 3 months after discontinuation of the oral anticoagulant therapy. In patients who continued to take oral anticoagulant therapy for more than 1 year after the event, blood was drawn during therapy. Blood was collected into tubes containing trisodium citrate 0.106 mol/L as previously described.22 An additional control group was recruited from the population by RDD. DNA was obtained by standard methods and was available for 4485 patients and 4889 control subjects.
Total and free protein S were measured on a STA-R automated coagulation analyzer (Diagnostica Stago, Asnières, France). Total protein S was measured by an enzyme-linked immunosorbent assay, and free protein S was measured by an immunoturbidimetric method (both from Diagnostica Stago).
Cutoff points
The range from 2.5th to 97.5th percentile of both total and free protein S in control subjects were considered as reference values. We also categorized cutoff points into different percentiles (ie, 97.5th to 75th percentile as the reference group compared with the 50th to 75th percentile, 50th to 25th percentile, 10th to 25th percentile, and <10th percentile of the control population), to see if this would change the results. Furthermore, a sensitivity analysis was performed in which very low protein S levels (ie, <0.5th percentile) were compared with the 2.5th to 97.5th percentile of (free) protein S. We did not include the 97.5 to 100th percentile in the reference population, as other studies suggest that high levels protein S may be indicative of underlying comorbidity.23
We also constructed cumulative distribution functions to visualize protein S levels in patients and controls. We excluded controls and patients at blood draw who used vitamin K antagonists, who were pregnant, or who used oral contraceptives, and we made a distinction between men, women, and patients who had provoked or unprovoked venous thrombosis.
Genetic analysis of PROS1
Copy number variations were analyzed by multiplex ligation-dependent probe amplification (MLPA)24 using the SALSA MLPA KIT P112 PROS1 (MRC-Holland, Amsterdam, The Netherlands) as previously described.11
Exons and their flanking regions, and the 5′- and 3′-UT regions were resequenced using primers designed to avoid the amplification of PROSP. After amplification, polymerase chain reaction products were sequenced using an ABI Prism 3730 DNA Analyzer (Applied Biosystems, Carlsbad, CA). Primers and polymerase chain reaction conditions are available on request.
Statistical analysis
With linear regression methods, we estimated the effect of vitamin K antagonist use, estrogen use, pregnancy, and puerperium on protein S levels. Based on previously reported associations,25 we also estimated the effect of obesity on protein S levels. Mean differences were adjusted for age and gender, as both age and gender influence protein S levels,26 and for each other in a multivariate analysis. Because there is no reason to assume that low levels of protein S are more common among partners of patients than in the general population, both RDD subjects and partners of patients were considered as control subjects and all analyses were unmatched.
Because protein S levels were collected after venous thrombosis, it is conceivable that the event itself influenced protein S levels. To analyze this possibility we performed a sensitivity analysis: scatter diagrams were constructed with regression lines, wherein protein S level was plotted against time between venous thrombosis and blood sampling (in patients not on vitamin K antagonists).
Odds ratios (ORs) with 95% confidence intervals (CIs) were calculated, and were adjusted for age and gender by logistic regression methods. In the MEGA study, because the cases were patients with a first venous thrombosis who were sampled from a stable, dynamic population, because partner controls were matched on time to cases, and because random controls were selected from the same population as the cases, these ORs can be interpreted as relative risks.27 Subjects who used vitamin K antagonists at the time of the blood draw were excluded when obtaining relative risk estimates. A preplanned sensitivity analysis was performed in which we excluded estrogen users and pregnant women at the time of the blood draw (as estrogen use and pregnancy decrease protein S levels).28,29 To further diminish the influence of estrogen-related hormones to protein S levels, we restricted an analysis to men only. In addition, we assessed the risk of free and total protein S deficiency in individuals with a positive family history.30 Stratified analyses were performed in which the common thrombophilias (ie, blood group non-O, factor V Leiden, and prothrombin G20210A) were compared with normal and low (free) protein S levels. By doing so, one can identify possible interaction or confounding of low free and total protein S levels to the risk of venous thrombosis by the common thrombophilias.
All statistical analyses were performed with SPSS for Windows, version 20.0 (SPSS Inc., Chicago, Ill).
Results
General characteristics
Approximately 50% of controls and patients who were included in the MEGA study were tested for free and total protein S levels. No differences in clinical characteristics between tested participants and nontested participants were observed, indicating that the tested individuals were representative of the whole MEGA group (Table 1). Of note, only 2331 patients were tested for free protein S, whereas 2377 patients were tested for total protein S. Of controls, 2872 were tested for free protein S (n = 1479 partners; n = 1393 RDD controls) and 2940 for total protein S (n = 1481 partners; n = 1459 controls). Some patients or controls were only tested for total protein S and not for free protein S because of laboratory failures. Both total and free protein S levels were reduced in vitamin K antagonist users, in pregnant women, and in women using estrogens (Table 2). Protein S levels were higher in overweight/obese control subjects than in those with normal weight.
Clinical characteristics
| . | Patients in MEGA (n = 4956) . | Patients tested (n = 2377)* . | Controls in MEGA (n = 6297) . | Controls tested (n = 2940) . |
|---|---|---|---|---|
| Male | 2274 (46) | 1083 (46) | 2914 (46) | 1393 (47) |
| Age at enrollment | 49 (13) | 48 (13) | 47 (13) | 48 (12) |
| Vitamin K antagonist use† | 294 (6) | 288 (12) | 34 (0.5) | 30 (1.0) |
| Estrogen use (women) | 1384 (54) | 739 (58) | 934 (29) | 412 (27) |
| Pregnancy/puerperium (women) | 183 (7) | 94 (7) | 87 (3) | 29 (2) |
| Normal weight (BMI 18.5-25 kg/m2) | 1634 (37) | 818 (37) | 2805 (49) | 1354 (48) |
| Overweight (BMI 25-30 kg/m2) | 1834 (42) | 965 (43) | 2112 (37) | 1109 (39) |
| Obesity (BMI ≥ 30 kg/m2) | 907 (21) | 456 (20) | 763 (13) | 365 (13) |
| . | Patients in MEGA (n = 4956) . | Patients tested (n = 2377)* . | Controls in MEGA (n = 6297) . | Controls tested (n = 2940) . |
|---|---|---|---|---|
| Male | 2274 (46) | 1083 (46) | 2914 (46) | 1393 (47) |
| Age at enrollment | 49 (13) | 48 (13) | 47 (13) | 48 (12) |
| Vitamin K antagonist use† | 294 (6) | 288 (12) | 34 (0.5) | 30 (1.0) |
| Estrogen use (women) | 1384 (54) | 739 (58) | 934 (29) | 412 (27) |
| Pregnancy/puerperium (women) | 183 (7) | 94 (7) | 87 (3) | 29 (2) |
| Normal weight (BMI 18.5-25 kg/m2) | 1634 (37) | 818 (37) | 2805 (49) | 1354 (48) |
| Overweight (BMI 25-30 kg/m2) | 1834 (42) | 965 (43) | 2112 (37) | 1109 (39) |
| Obesity (BMI ≥ 30 kg/m2) | 907 (21) | 456 (20) | 763 (13) | 365 (13) |
Continuous variables denoted as mean (standard deviation), categorical data as count (%). Data were missing for some participants in some subgroups.
BMI, body mass index.
There were slightly less patients (n = 2331) and controls (n = 2872) tested on free protein S due to sampling errors.
At time of blood draw.
Association of protein S with venous thrombotic risk factors in control subjects
| . | Free protein S . | Total protein S . | ||||||
|---|---|---|---|---|---|---|---|---|
| Subjects (n) . | Mean level (U/dL) . | Mean difference* (95% CI) . | Mean difference† (95% CI) . | Subjects (n) . | Mean level (U/dL) . | Mean difference* (95% CI) . | Mean difference† (95% CI) . | |
| No vitamin K antagonists | 2858 | 92 | Reference | Reference | 2910 | 102 | Reference | Reference |
| Vitamin K antagonists | 14 | 44 | −50 (−61 to −39) | −52 (−62 to −41) | 30 | 62 | −37 (−48 to −28) | −40 (−49 to −30) |
| Nonestrogen users (women) | 1067 | 88 | Reference | Reference | 1097 | 105 | Reference | Reference |
| Estrogen users (women) | 408 | 79 | −4 (−6 to −1) | −7 (−9 to −5) | 412 | 89 | −9 (−12 to −7) | −11 (−13 to −9) |
| No pregnancy/puerperium (women) | 1473 | 86 | Reference | Reference | 1505 | 101 | Reference | Reference |
| Pregnancy/puerperium (women) | 28 | 70 | −6 (−13 to 2) | −14 (−21 to −6) | 29 | 89 | 1 (−6 to 8) | −7 (−14 to −0.1) |
| Normal weight | 1332 | 87 | Reference | Reference | 1354 | 98 | Reference | Reference |
| Overweight | 1072 | 96 | 6 (4 to 7) | 6 (4 to 7) | 1109 | 105 | 5 (4 to 7) | 5 (3 to 6) |
| Obesity | 358 | 97 | 8 (6 to 11) | 8 (6 to 11) | 365 | 108 | 8 (6 to 10) | 8 (5 to 10) |
| . | Free protein S . | Total protein S . | ||||||
|---|---|---|---|---|---|---|---|---|
| Subjects (n) . | Mean level (U/dL) . | Mean difference* (95% CI) . | Mean difference† (95% CI) . | Subjects (n) . | Mean level (U/dL) . | Mean difference* (95% CI) . | Mean difference† (95% CI) . | |
| No vitamin K antagonists | 2858 | 92 | Reference | Reference | 2910 | 102 | Reference | Reference |
| Vitamin K antagonists | 14 | 44 | −50 (−61 to −39) | −52 (−62 to −41) | 30 | 62 | −37 (−48 to −28) | −40 (−49 to −30) |
| Nonestrogen users (women) | 1067 | 88 | Reference | Reference | 1097 | 105 | Reference | Reference |
| Estrogen users (women) | 408 | 79 | −4 (−6 to −1) | −7 (−9 to −5) | 412 | 89 | −9 (−12 to −7) | −11 (−13 to −9) |
| No pregnancy/puerperium (women) | 1473 | 86 | Reference | Reference | 1505 | 101 | Reference | Reference |
| Pregnancy/puerperium (women) | 28 | 70 | −6 (−13 to 2) | −14 (−21 to −6) | 29 | 89 | 1 (−6 to 8) | −7 (−14 to −0.1) |
| Normal weight | 1332 | 87 | Reference | Reference | 1354 | 98 | Reference | Reference |
| Overweight | 1072 | 96 | 6 (4 to 7) | 6 (4 to 7) | 1109 | 105 | 5 (4 to 7) | 5 (3 to 6) |
| Obesity | 358 | 97 | 8 (6 to 11) | 8 (6 to 11) | 365 | 108 | 8 (6 to 10) | 8 (5 to 10) |
Adjusted for age and gender where applicable.
Adjusted for age, gender (where applicable), and the effects of all other factors in the table.
Risk estimates
Subjects with low free protein S levels (<2.5th percentile, 53 U/dL) or low total protein S levels (<2.5th percentile, <68 U/dL) were not at increased risk of thrombosis when compared with subjects with levels in the 2.5th to 97.5th percentile; OR 0.82 (95% CI, 0.56-1.21) and 0.92 (95% CI, 0.63-1.33), respectively. Estrogen use, pregnancy, or puerperium may transiently decrease protein S levels and have influenced these findings. Therefore, we repeated the analysis after excluding all women who used estrogens, were pregnant, or were in puerperium at the time of venous thrombosis or at the time of blood sampling. This adjustment increased the OR to 1.51 (95% CI, 0.82-2.78) for subjects with low free protein S levels and to 1.34 (95% CI, 0.74-2.44) in subjects with low total protein S levels when compared with subjects with levels in the 2.5th to 97.5th percentile (Table 3). If protein S levels are somehow correlated with lifestyle, patients and partners may have been overmatched. To analyze if this occurred, we compared patients only with RDD controls. Risk estimates for the higher levels of free protein S were somewhat higher than when RDD and partner controls were combined. For the lower (free) protein S levels, however, risk estimates remained close to 1.0.
Risk of venous thrombosis for high or low levels of protein S
| . | . | . | OR (95% CI)* . | ||
|---|---|---|---|---|---|
| . | Patients, n (%) . | Controls, n (%) . | Overall . | Overall minus† . | Overall minus‡ . |
| Total | |||||
| Free protein S antigen | |||||
| >97.5th percentile (>139 U/dL) | 88 (4.3) | 83 (2.9 | 1.55 (1.14-2.11) | 1.81 (1.31-2.50 | 5.82 (3.14-10.78) |
| 97.5th-2.5th percentile (53-139 U/dL) | 1937 (94) | 2705 (95) | Reference | Reference | Reference |
| <2.5th percentile (<53 U/dL) | 44 (2.1) | 70 (2.4) | 0.82 (0.56-1.21) | 1.51 (0.82-2.78) | 1.04 (0.54-2.01) |
| Total protein S antigen | |||||
| >97.5th percentile (>146 U/dL) | 42 (2.0) | 72 (2.5) | 0.82 (0.56-1.21) | 1.00 (0.66-1.53) | 1.02 (0.62-1.68) |
| 97.5th-2.5th percentile (68-146 U/dL) | |||||
| <2.5th percentile (<68 U/dL) | 50 (2.4) | 72 (2.5) | 0.92 (0.63-1.33) | 1.34 (0.74-2.44) | 1.49 (0.71-3.11) |
| Unprovoked‡ | |||||
| Free protein S antigen | |||||
| >97.5th percentile (>139 U/dL) | 33 (6) | 83 (2.9) | 1.74 (1.13-2.67) | 1.72 (1.12-2.65) | 1.86 (1.19-2.89) |
| 97.5th-2.5th percentile (53-139 U/dL) | 504 | 2705 (95) | Reference | Reference | Reference |
| <2.5th percentile (<53 U/dL) | 8 (1.5) | 70 (2.4) | 1.77 (0.84-3.71) | 2.31 (1.06-5.05) | 1.80 (0.80-4.01) |
| Total protein S antigen | |||||
| >97.5th percentile (>146 U/dL) | 16 (2.9) | 72 (2.5) | 1.21 (0.70-2.10) | 1.15 (0.66-2.00) | 1.14 (0.65-2.00) |
| 97.5th-2.5th percentile (68-146 U/dL) | 524 (94) | 2766 (95) | Reference | Reference | Reference |
| <2.5th percentile (<68 U/dL) | 19 (3.4) | 72 (2.5) | 1.19 (0.55-2.57) | 1.59 (0.71-3.58) | 1.75 (0.79-3.85) |
| Provoked§ | |||||
| Free protein S antigen | |||||
| >97.5th percentile (>139 U/dL) | 55 (3.8) | 83 (2.9) | 1.61 (1.12-2.30) | 3.62 (2.20-5.96) | |
| 97.5th-2.5th percentile (53-139 U/dL) | 1390 (94) | 2705 (95) | Reference | Reference | Reference |
| <2.5th percentile (<53 U/dL) | 35 (2.4) | 70 (2.4) | 0.69 (0.45-1.05 | 1.17 (0.53-2.58) | 1.08 (0.49-2.39) |
| Total protein S antigen | |||||
| >97.5th percentile (>146 U/dL) | 25 (1.7) | 72 (2.5) | 0.74 (0.47-1.18) | 0.95 (0.56-1.59) | 1.05 (0.60-1.83) |
| 97.5th-2.5th percentile (68-146 U/dL) | 1431 (96) | 2766 (95) | Reference | Reference | Reference |
| <2.5th percentile (<68 U/dL) | 39 (2.6) | 72 (2.5) | 0.78 (0.52-2.43) | 1.14 (0.54-2.53) | 1.26 (0.55-2.91) |
| . | . | . | OR (95% CI)* . | ||
|---|---|---|---|---|---|
| . | Patients, n (%) . | Controls, n (%) . | Overall . | Overall minus† . | Overall minus‡ . |
| Total | |||||
| Free protein S antigen | |||||
| >97.5th percentile (>139 U/dL) | 88 (4.3) | 83 (2.9 | 1.55 (1.14-2.11) | 1.81 (1.31-2.50 | 5.82 (3.14-10.78) |
| 97.5th-2.5th percentile (53-139 U/dL) | 1937 (94) | 2705 (95) | Reference | Reference | Reference |
| <2.5th percentile (<53 U/dL) | 44 (2.1) | 70 (2.4) | 0.82 (0.56-1.21) | 1.51 (0.82-2.78) | 1.04 (0.54-2.01) |
| Total protein S antigen | |||||
| >97.5th percentile (>146 U/dL) | 42 (2.0) | 72 (2.5) | 0.82 (0.56-1.21) | 1.00 (0.66-1.53) | 1.02 (0.62-1.68) |
| 97.5th-2.5th percentile (68-146 U/dL) | |||||
| <2.5th percentile (<68 U/dL) | 50 (2.4) | 72 (2.5) | 0.92 (0.63-1.33) | 1.34 (0.74-2.44) | 1.49 (0.71-3.11) |
| Unprovoked‡ | |||||
| Free protein S antigen | |||||
| >97.5th percentile (>139 U/dL) | 33 (6) | 83 (2.9) | 1.74 (1.13-2.67) | 1.72 (1.12-2.65) | 1.86 (1.19-2.89) |
| 97.5th-2.5th percentile (53-139 U/dL) | 504 | 2705 (95) | Reference | Reference | Reference |
| <2.5th percentile (<53 U/dL) | 8 (1.5) | 70 (2.4) | 1.77 (0.84-3.71) | 2.31 (1.06-5.05) | 1.80 (0.80-4.01) |
| Total protein S antigen | |||||
| >97.5th percentile (>146 U/dL) | 16 (2.9) | 72 (2.5) | 1.21 (0.70-2.10) | 1.15 (0.66-2.00) | 1.14 (0.65-2.00) |
| 97.5th-2.5th percentile (68-146 U/dL) | 524 (94) | 2766 (95) | Reference | Reference | Reference |
| <2.5th percentile (<68 U/dL) | 19 (3.4) | 72 (2.5) | 1.19 (0.55-2.57) | 1.59 (0.71-3.58) | 1.75 (0.79-3.85) |
| Provoked§ | |||||
| Free protein S antigen | |||||
| >97.5th percentile (>139 U/dL) | 55 (3.8) | 83 (2.9) | 1.61 (1.12-2.30) | 3.62 (2.20-5.96) | |
| 97.5th-2.5th percentile (53-139 U/dL) | 1390 (94) | 2705 (95) | Reference | Reference | Reference |
| <2.5th percentile (<53 U/dL) | 35 (2.4) | 70 (2.4) | 0.69 (0.45-1.05 | 1.17 (0.53-2.58) | 1.08 (0.49-2.39) |
| Total protein S antigen | |||||
| >97.5th percentile (>146 U/dL) | 25 (1.7) | 72 (2.5) | 0.74 (0.47-1.18) | 0.95 (0.56-1.59) | 1.05 (0.60-1.83) |
| 97.5th-2.5th percentile (68-146 U/dL) | 1431 (96) | 2766 (95) | Reference | Reference | Reference |
| <2.5th percentile (<68 U/dL) | 39 (2.6) | 72 (2.5) | 0.78 (0.52-2.43) | 1.14 (0.54-2.53) | 1.26 (0.55-2.91) |
Adjusted for age and gender.
Estrogen users or pregnancy/puerperium.
Minus estrogen users or pregnancy/puerperium; control group confined to random digit dialing subjects only.
Provoked venous thrombosis classified as thrombosis due to trauma, plaster cast use, surgery, pregnancy/puerperium, or immobilization, within 3 months prior to the index date, >4 hours of traveling by air, train, bus, or car in the 8 weeks before the index date, estrogen/progestogen use at time of the index date, and diagnosis of malignancy within 5 years.
When we classified subjects into different percentiles (Table 4), again no increases in risks for venous thrombosis were observed for lower levels of free and total protein S.
Risk of venous thrombosis for strata of protein S levels
| Measurement . | Patients n (%) . | Controls n (%) . | Total OR (95% CI)* . | |
|---|---|---|---|---|
| Overall . | Overall minus† . | |||
| Free protein S antigen | ||||
| 97.5th-75th percentile (105-139 U/dL) | 511 (25.8) | 617 (22.2) | Reference | Reference |
| 50th-75th percentile (91-105 U/dL) | 528 (26.7) | 692 (25.0) | 0.85 (0.72-1.00) | 0.77 (0.64-0.92) |
| 25th-50th percentile (77-91 U/dL) | 507 (25.6) | 738 (26.6) | 0.81 (0.69-0.96) | 0.70 (0.57-0.85) |
| 10-25th percentile (77-66 U/dL) | 291 (14.7) | 449 (16.1) | 0.75 (0.61-0.92) | 0.55 (0.43-0.72) |
| <10th percentile (<66 U/dL) | 144 (7.2) | 279 (10.0) | 0.61 (0.45-0.85) | 0.53 (0.33-0.83) |
| Total protein S antigen | ||||
| 97.5th-75th percentile (115-146 U/dL) | 498 (24.3) | 653 (23.0) | Reference | Reference |
| 50th-75th percentile (101-115 U/dL) | 573 (28.0) | 720 (25.4) | 1.02 (0.86-1.19) | 0.99 (0.82-1.19) |
| 25th-50th percentile (89-101 U/dL) | 500 (24.4) | 723 (25.5) | 0.85 (0.72-1.01) | 0.73 (0.60-0.89) |
| 10-25th percentile (78-89 U/dL) | 296 (14.5) | 470 (16.6) | 0.79 (0.65-0.97) | 0.68 (0.53-0.88) |
| <10th percentile (<78 U/dL) | 180 (8.8) | 272 (9.6) | 0.86 (0.63-1.16) | 0.78 (0.51-1.20) |
| Measurement . | Patients n (%) . | Controls n (%) . | Total OR (95% CI)* . | |
|---|---|---|---|---|
| Overall . | Overall minus† . | |||
| Free protein S antigen | ||||
| 97.5th-75th percentile (105-139 U/dL) | 511 (25.8) | 617 (22.2) | Reference | Reference |
| 50th-75th percentile (91-105 U/dL) | 528 (26.7) | 692 (25.0) | 0.85 (0.72-1.00) | 0.77 (0.64-0.92) |
| 25th-50th percentile (77-91 U/dL) | 507 (25.6) | 738 (26.6) | 0.81 (0.69-0.96) | 0.70 (0.57-0.85) |
| 10-25th percentile (77-66 U/dL) | 291 (14.7) | 449 (16.1) | 0.75 (0.61-0.92) | 0.55 (0.43-0.72) |
| <10th percentile (<66 U/dL) | 144 (7.2) | 279 (10.0) | 0.61 (0.45-0.85) | 0.53 (0.33-0.83) |
| Total protein S antigen | ||||
| 97.5th-75th percentile (115-146 U/dL) | 498 (24.3) | 653 (23.0) | Reference | Reference |
| 50th-75th percentile (101-115 U/dL) | 573 (28.0) | 720 (25.4) | 1.02 (0.86-1.19) | 0.99 (0.82-1.19) |
| 25th-50th percentile (89-101 U/dL) | 500 (24.4) | 723 (25.5) | 0.85 (0.72-1.01) | 0.73 (0.60-0.89) |
| 10-25th percentile (78-89 U/dL) | 296 (14.5) | 470 (16.6) | 0.79 (0.65-0.97) | 0.68 (0.53-0.88) |
| <10th percentile (<78 U/dL) | 180 (8.8) | 272 (9.6) | 0.86 (0.63-1.16) | 0.78 (0.51-1.20) |
Adjusted for age and gender.
Estrogen users or pregnancy/puerperium.
When we restricted the analysis to subjects who did not change hormone use status between thrombosis and blood draw (ie, either consistently used hormones at both time points or did not use hormones at either time point), the ORs were 1.12 (95% CI, 0.69-1.81) and 0.83 (95% CI, 0.48-1.44), respectively. In men, the OR for venous thrombosis in those with low free protein S was 2.15 (95% CI, 0.74-6.29). For total protein S levels, the ORwas 1.57 (95% CI, 0.70-3.49). Interestingly, subjects with free protein S levels in the >97.5th percentile appeared to be at increased risk of venous thrombosis (OR 1.55; 95% CI, 1.14-2.11), which could only partly be attributed to being overweight or to obesity, as further adjustments for being overweight or for obesity attenuated the OR to 1.48 (95% CI, 1.07-2.04) (data not shown).
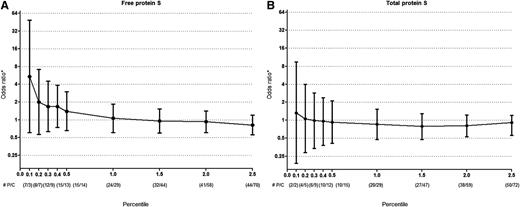
To analyze if the results were sensitive to the cutoff level of free or total protein S, we subsequently compared decreasing cutoff values of free and total protein S levels, respectively, on the risk of venous thrombosis as compared with the reference group (subjects with protein S levels that were between the 2.5th and 97.5th percentile). Although numbers became small, it appeared that a free protein S cut off level of <0.10th (free protein S <33 U/dL) to 0.20th (free protein S <34 U/dL) percentile could identify subjects at high risk of venous thrombosis (Figure 1A). Low, or even extremely low (<0.20th percentile or total protein S <53 U/dL) total protein S levels were not associated with venous thrombosis (Figure 1B). Due to small numbers, the latter analysis could not be performed in subjects that were not pregnant or were not using hormones.
Risk of venous thrombosis for low free and total protein S levels. All ORs are compared with (A) free protein S or (B) total protein S levels between 2.5th-97.5th percentile. Error bars indicate 95% confidence intervals. *Adjusted for age and gender. #P denotes number of patients and C denotes number of controls per percentile.
Risk of venous thrombosis for low free and total protein S levels. All ORs are compared with (A) free protein S or (B) total protein S levels between 2.5th-97.5th percentile. Error bars indicate 95% confidence intervals. *Adjusted for age and gender. #P denotes number of patients and C denotes number of controls per percentile.
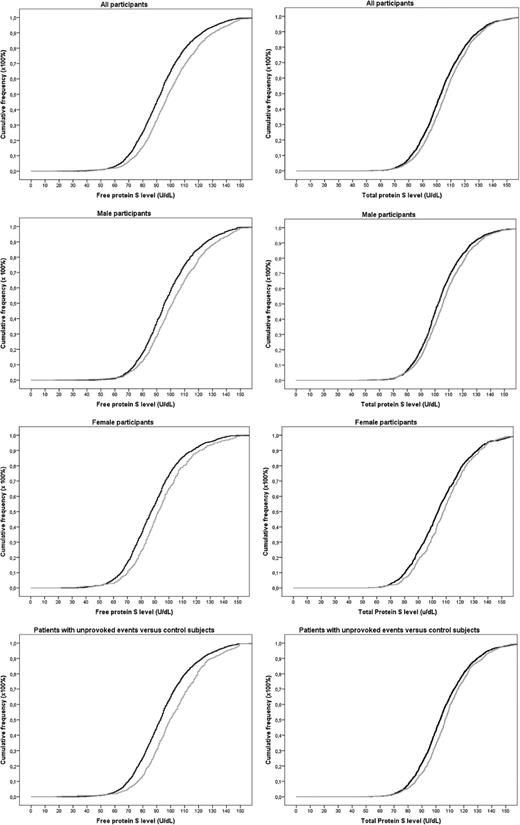
As shown in the cumulative distribution functions, patients had slightly higher free protein S levels (mean 100 U/dL in patients vs 94 U/dL in controls) and total protein S levels (mean 107 U/dL in patients vs 105 U/dL in controls) (Figure 2).
Cumulative distribution functions of free and total protein S levels in patients and controls. Gray lines indicate patients. Black lines indicate controls.
Cumulative distribution functions of free and total protein S levels in patients and controls. Gray lines indicate patients. Black lines indicate controls.
Increased risks for venous thrombosis in protein S deficient subjects are mainly reported from thrombophilic family studies. Therefore, we selected patients with a positive family history and repeated the analysis (Table 5). Again, no effects were apparent. These null findings also could not be explained by confounding of the common thrombophilias or by an interaction between the common thrombophilias and low levels of free and total protein S (Table 6).
Risk of venous thrombosis for high or low levels of protein S, according to family history
| . | . | . | OR (95% CI)* . | |
|---|---|---|---|---|
| . | Patients, n . | Controls, n . | Overall . | Overall minus† . |
| Negative family history | ||||
| Free protein S antigen | ||||
| >97.5th percentile (>139 U/dL) | 936 | 2705 | Reference | Reference |
| <2.5th percentile (<53 U/dL) | 24 | 70 | 0.75 (0.46-1.21) | 1.95 (0.93-4.05) |
| Total protein S antigen | ||||
| 97.5th-2.5th percentile (68-146 U/dL) | 964 | 2766 | Reference | Reference |
| <2.5th percentile (<68 U/dL) | 21 | 72 | 0.65 (0.40-1.07) | 1.41 (0.66-3.00) |
| Positive family history | ||||
| Free protein S antigen | ||||
| 97.5th-2.5th percentile (53-139 U/dL) | 524 | 2705 | Reference | Reference |
| <2.5th percentile (<53 U/dL) | 10 | 70 | 0.86 (0.44-1.70) | 0.92 (0.27-3.08) |
| Total protein S antigen | ||||
| 97.5th-2.5th percentile (68-146 U/dL) | 540 | 2766 | Reference | Reference |
| <2.5th percentile (<68 U/dL) | 12 | 72 | 1.00 (0.53-1.86) | 1.08 (0.37-3.15) |
| . | . | . | OR (95% CI)* . | |
|---|---|---|---|---|
| . | Patients, n . | Controls, n . | Overall . | Overall minus† . |
| Negative family history | ||||
| Free protein S antigen | ||||
| >97.5th percentile (>139 U/dL) | 936 | 2705 | Reference | Reference |
| <2.5th percentile (<53 U/dL) | 24 | 70 | 0.75 (0.46-1.21) | 1.95 (0.93-4.05) |
| Total protein S antigen | ||||
| 97.5th-2.5th percentile (68-146 U/dL) | 964 | 2766 | Reference | Reference |
| <2.5th percentile (<68 U/dL) | 21 | 72 | 0.65 (0.40-1.07) | 1.41 (0.66-3.00) |
| Positive family history | ||||
| Free protein S antigen | ||||
| 97.5th-2.5th percentile (53-139 U/dL) | 524 | 2705 | Reference | Reference |
| <2.5th percentile (<53 U/dL) | 10 | 70 | 0.86 (0.44-1.70) | 0.92 (0.27-3.08) |
| Total protein S antigen | ||||
| 97.5th-2.5th percentile (68-146 U/dL) | 540 | 2766 | Reference | Reference |
| <2.5th percentile (<68 U/dL) | 12 | 72 | 1.00 (0.53-1.86) | 1.08 (0.37-3.15) |
Adjusted for age and gender.
Estrogen users or pregnancy/puerperium.
Risk of venous thrombosis for low levels of protein S, stratified analysis with common thrombophilias
| . | . | Patients n (%) . | Controls n (%) . | OR (95% CI)* . |
|---|---|---|---|---|
| Free protein S antigen | Blood group non-O | |||
| 97.5th-2.5th percentile (53-139 U/dL) | Absent | 556 (28.1) | 1270 (45.9) | Reference |
| <2.5th percentile (<53 U/dL) | Absent | 13 (0.7) | 30 (1.1) | 0.93 (0.48-1.79) |
| 97.5th-2.5th percentile (53-139 U/dL) | Present | 1378 (69.7) | 1429 (51.6) | 2.21 (1.95-2.50) |
| <2.5th percentile (<53 U/dL) | Present | 31 (1.6) | 39 (1.4) | 1.71 (1.05-2.78) |
| Total protein S antigen | Blood group non-O | |||
| 97.5th-2.5th percentile (68-146 U/dL) | Absent | 575 (28.1) | 1300 (45.9) | Reference |
| <2.5th percentile (<68 U/dL) | Absent | 13 (0.6) | 32 (1.1) | 0.86 (0.45-1.65) |
| 97.5th-2.5th percentile (68-146 U/dL) | Present | 1418 (69.4) | 1459 (51.5) | 2.20 (1.95-2.49) |
| <2.5th percentile (<68 U/dL) | Present | 37 (1.8) | 40 (1.4) | 2.00 (1.27-3.18) |
| Free protein S antigen | Factor V Leiden | |||
| 97.5th-2.5th percentile (53-139 U/dL) | Absent | 1633 (82.5) | 2578 (92.9) | Reference |
| <2.5th percentile (<53 U/dL) | Absent | 42 (2.1) | 66 (2.4) | 0.94 (0.63-1.40) |
| 97.5th-2.5th percentile (53-139 U/dL) | Present | 303 (15.3) | 127 (4.6) | 3.79 (3.05-4.71) |
| <2.5th percentile (<53 U/dL) | Present | 2 (0.1) | 4 (0.1) | 0.79 (0.14-4.33) |
| Total protein S antigen | Factor V Leiden | |||
| 97.5th-2.5th percentile (68-146 U/dL) | Absent | 1689 (82.6) | 2630 (92.7) | Reference |
| <2.5th percentile (<68 U/dL) | Absent | 44 (2.2) | 69 (2.4) | 0.94 (0.64-1.39) |
| 97.5th-2.5th percentile (68-146 U/dL) | Present | 307 (15.0) | 136 (4.8) | 3.35 (2.86-4.37) |
| <2.5th percentile (<68 U/dL) | Present | 6 (0.3) | 3 (0.1) | 2.99 (0.75-12.0) |
| Free protein S antigen | Prothrombin G20210A | |||
| 97.5th-2.5th percentile (53-139 U/dL) | Absent | 1843 (93.0) | 2658 (958) | Reference |
| <2.5th percentile (<53 U/dL) | Absent | 42 (2.2) | 69 (2.5) | 0.84 (0.57-1.24) |
| 97.5th-2.5th percentile (53-139 U/dL) | Present | 94 (4.7) | 47 (1.7) | 2.87 (2.01-4.09) |
| <2.5th percentile (<53 U/dL) | Present | 1 (0.1) | 1 (0.0) | 1.46 (0.09-23.4) |
| Total protein S antigen | Prothrombin G20210A | |||
| 97.5th-2.5th percentile (68-146 U/dL) | Absent | 1905 (93.1) | 2718 (95.8) | Reference |
| <2.5th percentile (<68 U/dL) | Absent | 48 (2.3) | 70 (2.5) | 0.93 (0.61-1.35) |
| 97.5th-2.5th percentile (68-146 U/dL) | Present | 92 (4.5) | 48 (1.7) | 2.73 (1.92-3.89) |
| <2.5th percentile (<68 U/dL) | Present | 2 (0.1) | 2 (0.1) | 1.37 (0.19-9.78) |
| . | . | Patients n (%) . | Controls n (%) . | OR (95% CI)* . |
|---|---|---|---|---|
| Free protein S antigen | Blood group non-O | |||
| 97.5th-2.5th percentile (53-139 U/dL) | Absent | 556 (28.1) | 1270 (45.9) | Reference |
| <2.5th percentile (<53 U/dL) | Absent | 13 (0.7) | 30 (1.1) | 0.93 (0.48-1.79) |
| 97.5th-2.5th percentile (53-139 U/dL) | Present | 1378 (69.7) | 1429 (51.6) | 2.21 (1.95-2.50) |
| <2.5th percentile (<53 U/dL) | Present | 31 (1.6) | 39 (1.4) | 1.71 (1.05-2.78) |
| Total protein S antigen | Blood group non-O | |||
| 97.5th-2.5th percentile (68-146 U/dL) | Absent | 575 (28.1) | 1300 (45.9) | Reference |
| <2.5th percentile (<68 U/dL) | Absent | 13 (0.6) | 32 (1.1) | 0.86 (0.45-1.65) |
| 97.5th-2.5th percentile (68-146 U/dL) | Present | 1418 (69.4) | 1459 (51.5) | 2.20 (1.95-2.49) |
| <2.5th percentile (<68 U/dL) | Present | 37 (1.8) | 40 (1.4) | 2.00 (1.27-3.18) |
| Free protein S antigen | Factor V Leiden | |||
| 97.5th-2.5th percentile (53-139 U/dL) | Absent | 1633 (82.5) | 2578 (92.9) | Reference |
| <2.5th percentile (<53 U/dL) | Absent | 42 (2.1) | 66 (2.4) | 0.94 (0.63-1.40) |
| 97.5th-2.5th percentile (53-139 U/dL) | Present | 303 (15.3) | 127 (4.6) | 3.79 (3.05-4.71) |
| <2.5th percentile (<53 U/dL) | Present | 2 (0.1) | 4 (0.1) | 0.79 (0.14-4.33) |
| Total protein S antigen | Factor V Leiden | |||
| 97.5th-2.5th percentile (68-146 U/dL) | Absent | 1689 (82.6) | 2630 (92.7) | Reference |
| <2.5th percentile (<68 U/dL) | Absent | 44 (2.2) | 69 (2.4) | 0.94 (0.64-1.39) |
| 97.5th-2.5th percentile (68-146 U/dL) | Present | 307 (15.0) | 136 (4.8) | 3.35 (2.86-4.37) |
| <2.5th percentile (<68 U/dL) | Present | 6 (0.3) | 3 (0.1) | 2.99 (0.75-12.0) |
| Free protein S antigen | Prothrombin G20210A | |||
| 97.5th-2.5th percentile (53-139 U/dL) | Absent | 1843 (93.0) | 2658 (958) | Reference |
| <2.5th percentile (<53 U/dL) | Absent | 42 (2.2) | 69 (2.5) | 0.84 (0.57-1.24) |
| 97.5th-2.5th percentile (53-139 U/dL) | Present | 94 (4.7) | 47 (1.7) | 2.87 (2.01-4.09) |
| <2.5th percentile (<53 U/dL) | Present | 1 (0.1) | 1 (0.0) | 1.46 (0.09-23.4) |
| Total protein S antigen | Prothrombin G20210A | |||
| 97.5th-2.5th percentile (68-146 U/dL) | Absent | 1905 (93.1) | 2718 (95.8) | Reference |
| <2.5th percentile (<68 U/dL) | Absent | 48 (2.3) | 70 (2.5) | 0.93 (0.61-1.35) |
| 97.5th-2.5th percentile (68-146 U/dL) | Present | 92 (4.5) | 48 (1.7) | 2.73 (1.92-3.89) |
| <2.5th percentile (<68 U/dL) | Present | 2 (0.1) | 2 (0.1) | 1.37 (0.19-9.78) |
Adjusted for age and gender.
Because vitamin K antagonist use was self-reported (and therefore, we might have included subjects with low protein S levels that was due to unreported vitamin K antagonist treatment), we performed an additional sensitivity analysis by examining the distribution of the vitamin K–dependent coagulant factors (ie, factors II, VII, IX, and X) in cases and in controls who did not report vitamin K antagonist usage. All factors were normally distributed, and there were no apparent outliers in the lower range, which suggested that self-reporting was reliable.
Genetic analysis
Copy number variation. First, we tested samples from 2270 consecutive MEGA subjects (1395 patients and 875 controls). An abnormal MLPA pattern was detected in only 1 individual. This individual was a 66-year-old, female patient not using estrogens or vitamin K antagonists at the moment of thrombosis or sample collection. She had an unprovoked first venous thrombosis. Her family history was not available. The patient was heterozygous for a complete deletion of PROS1. Her total protein S was 64 U/dL (<2.5th percentile of the control group) and her free protein S was 22 U/dL (<2.5th percentile of the control group).
The presence of only 1 copy number variation in 2270 samples shows that such variation of PROS1 is at most a rare cause of venous thrombosis. To verify this, we zoomed in on potentially protein S–deficient subjects by selecting DNA from all subjects with low (ie, lower than the 2.5th percentile of the control group) total or low free protein S. Patients who used vitamin K antagonists at the time of the blood draw were also included to guarantee that protein S deficient subjects were not missed. In total, 191 samples were selected (84 patients and 107 controls) in which no new copy number variation of PROS1 was identified.
Sequencing of PROS1
To further investigate the molecular basis of protein S deficiency, we selected the 48 subjects with the lowest levels of free protein S (excluding patients using vitamin K antagonists). All these subjects were below the 1st percentile of the controls (cutoff 46 U/dL); of the 48 selected subjects, 24 were patients and 24 were controls.
Eleven nonsynonymous variations were identified in 10 subjects (5 patients and 5 controls); of these subjects, 1 (subject 25) is a carrier of 2 different variations. Table 7 summarizes the results and gives references for variations previously described. Subjects are numbered according to increasing levels of free protein S. PROS1 coding DNA reference sequence is NM_000313.3 and protein reference sequence is NP_000304.2.
Molecular analysis of PROS1 in subjects with free protein S level below 1st percentile, as determined in MEGA controls
| Subject number* . | Venous thrombosis . | fPS (U/dL) . | tPS (U/dL) . | Sequence change† . | Predicted protein change‡ . | Reference . | Comments . |
|---|---|---|---|---|---|---|---|
| 1 | No | 16 | 123 | — | — | — | |
| 2 | Yes | 18 | 63 | 967delTinsGG | Phe323fs | NOVEL | |
| 3 | Yes | 18 | 68 | — | — | — | — |
| 4 | Yes | 22 | 64 | (?_-133)_(1_?*)del | Complete deletion | 5,10,11 | — |
| 5 | Yes | 30 | 74 | — | — | — | — |
| 6 | Yes | 30 | 58 | 431C>A | Thr144Asn | 40 | Associated with quantitative deficiency of protein S |
| 7 | No | 31 | 54 | — | — | — | — |
| 8 | No | 32 | 50 | — | — | — | — |
| 9 | Yes | 33 | 78 | — | — | — | — |
| 10 | Yes | 33 | 88 | — | — | — | — |
| 11 | No | 34 | 63 | ||||
| 12 | No | 34 | 90 | 370G>A* | — | — | 3′UTR. NOVEL |
| 13 | No | 34 | 66 | — | — | — | — |
| 14 | Yes | 34 | 53 | — | — | — | — |
| 15 | No | 34 | 99 | — | — | — | — |
| 16 | Yes | 36 | 63 | — | — | — | — |
| 17 | Yes | 37 | 66 | 684C>G | Cys228Trp | — | NOVEL |
| 18 | No | 37 | 57 | — | — | — | — |
| 19 | Yes | 38 | 60 | — | — | — | — |
| 20 | No | 39 | 96 | — | — | — | — |
| 21 | Yes | 39 | 81 | — | — | — | — |
| 22 | No | 39 | 65 | — | — | — | — |
| 23 | Yes | 40 | 70 | — | — | — | — |
| 24 | Yes | 40 | 58 | — | — | — | — |
| 25 | No | 40 | 74 | 1301T>C | Val434Ala | rs6803112 | |
| 25 | No | 40 | 74 | 1501T>G | Ser501Ala | 23 | Protein S Heerlen |
| 26 | Yes | 40 | 76 | — | — | — | — |
| 27 | No | 40 | 81 | — | — | — | — |
| 28 | No | 40 | 57 | — | — | — | — |
| 29 | No | 41 | 76 | — | — | — | — |
| 30 | Yes | 41 | 61 | — | — | — | — |
| 31 | Yes | 41 | 53 | — | — | — | — |
| 32 | No | 41 | 81 | 1501T>G | Ser501Ala | 23 | Protein S Heerlen |
| 33 | No | 41 | 82 | — | — | — | — |
| 34 | Yes | 42 | 69 | — | — | — | — |
| 35 | Yes | 42 | 81 | — | — | — | — |
| 36 | No | 42 | 59 | — | — | — | — |
| 37 | Yes | 42 | 43 | — | — | — | — |
| 38 | No | 42 | 96 | — | — | — | — |
| 39 | Yes | 43 | 74 | — | — | — | — |
| 40 | Yes | 43 | 77 | — | — | — | — |
| 41 | No | 43 | 62 | — | — | — | — |
| 42 | No | 43 | 132 | — | — | — | — |
| 43 | No | 43 | 60 | 698G>A | Arg233Lys | 7,41,42 | rs41267007 |
| Probably neutral polymorphism | |||||||
| 44 | Yes | 43 | 60 | 814G>A | Gly272Arg | — | rs41267005 |
| 45 | No | 44 | 61 | 1095T>G | Asn365Lys | — | NOVEL |
| 46 | No | 44 | 63 | — | — | — | — |
| 47 | No | 45 | 88 | — | — | — | — |
| 48 | Yes | 45 | 77 | — | — | — | — |
| Subject number* . | Venous thrombosis . | fPS (U/dL) . | tPS (U/dL) . | Sequence change† . | Predicted protein change‡ . | Reference . | Comments . |
|---|---|---|---|---|---|---|---|
| 1 | No | 16 | 123 | — | — | — | |
| 2 | Yes | 18 | 63 | 967delTinsGG | Phe323fs | NOVEL | |
| 3 | Yes | 18 | 68 | — | — | — | — |
| 4 | Yes | 22 | 64 | (?_-133)_(1_?*)del | Complete deletion | 5,10,11 | — |
| 5 | Yes | 30 | 74 | — | — | — | — |
| 6 | Yes | 30 | 58 | 431C>A | Thr144Asn | 40 | Associated with quantitative deficiency of protein S |
| 7 | No | 31 | 54 | — | — | — | — |
| 8 | No | 32 | 50 | — | — | — | — |
| 9 | Yes | 33 | 78 | — | — | — | — |
| 10 | Yes | 33 | 88 | — | — | — | — |
| 11 | No | 34 | 63 | ||||
| 12 | No | 34 | 90 | 370G>A* | — | — | 3′UTR. NOVEL |
| 13 | No | 34 | 66 | — | — | — | — |
| 14 | Yes | 34 | 53 | — | — | — | — |
| 15 | No | 34 | 99 | — | — | — | — |
| 16 | Yes | 36 | 63 | — | — | — | — |
| 17 | Yes | 37 | 66 | 684C>G | Cys228Trp | — | NOVEL |
| 18 | No | 37 | 57 | — | — | — | — |
| 19 | Yes | 38 | 60 | — | — | — | — |
| 20 | No | 39 | 96 | — | — | — | — |
| 21 | Yes | 39 | 81 | — | — | — | — |
| 22 | No | 39 | 65 | — | — | — | — |
| 23 | Yes | 40 | 70 | — | — | — | — |
| 24 | Yes | 40 | 58 | — | — | — | — |
| 25 | No | 40 | 74 | 1301T>C | Val434Ala | rs6803112 | |
| 25 | No | 40 | 74 | 1501T>G | Ser501Ala | 23 | Protein S Heerlen |
| 26 | Yes | 40 | 76 | — | — | — | — |
| 27 | No | 40 | 81 | — | — | — | — |
| 28 | No | 40 | 57 | — | — | — | — |
| 29 | No | 41 | 76 | — | — | — | — |
| 30 | Yes | 41 | 61 | — | — | — | — |
| 31 | Yes | 41 | 53 | — | — | — | — |
| 32 | No | 41 | 81 | 1501T>G | Ser501Ala | 23 | Protein S Heerlen |
| 33 | No | 41 | 82 | — | — | — | — |
| 34 | Yes | 42 | 69 | — | — | — | — |
| 35 | Yes | 42 | 81 | — | — | — | — |
| 36 | No | 42 | 59 | — | — | — | — |
| 37 | Yes | 42 | 43 | — | — | — | — |
| 38 | No | 42 | 96 | — | — | — | — |
| 39 | Yes | 43 | 74 | — | — | — | — |
| 40 | Yes | 43 | 77 | — | — | — | — |
| 41 | No | 43 | 62 | — | — | — | — |
| 42 | No | 43 | 132 | — | — | — | — |
| 43 | No | 43 | 60 | 698G>A | Arg233Lys | 7,41,42 | rs41267007 |
| Probably neutral polymorphism | |||||||
| 44 | Yes | 43 | 60 | 814G>A | Gly272Arg | — | rs41267005 |
| 45 | No | 44 | 61 | 1095T>G | Asn365Lys | — | NOVEL |
| 46 | No | 44 | 63 | — | — | — | — |
| 47 | No | 45 | 88 | — | — | — | — |
| 48 | Yes | 45 | 77 | — | — | — | — |
Mutations are described accordingly to the Human Genome Variation Society.43
– indicates individuals in whom no mutation was found.
fPS, free protein S; tPS, total protein S; MAF, minor allele frequency.
Numbers are accordingly to increasing levels of free protein S.
PROS1 coding DNA reference sequence is NM_000313.3.
Protein reference sequence is NP_000304.2.
Four variations were novel. Subject 2 (patient) carried an indel mutation (Phe323fs) in the sex hormone-binding globulin (SHBG)-like domain that is predicted to result in a truncated protein; subject 17 (patient) carried a 684C>G (Cys228Trp) substitution in the epidermal growth factor–domain 3; subject 12 (control) carried a nucleotide substitution in the 3′–untranslated region; and subject 45 (control) carried a 1095T>G (Asn365Lys) substitution in the SHBG-like domain.
A complete deletion of PROS1 was detected in subject 4 (patient). Subject 6 is a patient who carried a 431C>A (Thr144Asn) substitution in the EFG-domain 1. Protein S Heerlen (Ser501Ala) was detected in 2control subjects (subjects 25 and 32), and 1 carrier of protein S Heerlen (subject 25) carried a second variation in the SHBG-like domain (13012T>C; Val434Ala). This variation is known as rs6803112 with a reported minor allele frequency (MAF) of 0.097. Subject 43 (control) carried a 698G>A (Arg233Lys) substitution, which is also listed as rs41267007 (MAF 0.002). Finally, control subject 44 carried an 814G>A (Gly272Arg) variation in the epidermal growth factor–domain 4. This variation is listed as rs41267005 (MAF 0.002), but no information regarding protein S levels is available.
Discussion
Conflicting data have been reported regarding venous thrombotic risk in subjects with protein S deficiency within the community.2 In the present study, subjects with low levels of free protein S had an increased risk of unprovoked venous thrombosis (OR 2.31; 95% CI, 1.06-5.05). However, the prevalence of patients with such levels was low (n = 8, or 0.4% of the total patient population), making it impractical to use free protein S levels to identify patients at risk in a clinical setting. The present study confirms the results of a previous population based study (Leiden Thrombophilia Study),20 in which only very low levels of free protein S (ie, <0.10th percentile or <33 U/dL), but not of total protein S, were associated with an increased risk for venous thrombosis (OR 5.44; 95% CI, 0.61-48.78). This increased risk at very low free protein S levels corresponds with findings from family studies of protein S deficiency on venous thrombosis risk (although relative risk estimates in family studies for first venous thrombosis in subjects with very low free protein S levels are even higher than 5),12,31 as well as with findings from family studies that showed the cutoff level of free protein S to identify true inherited protein S deficiency lies far below the commonly used normal range.12,14
Total protein S levels were not able to identify subjects at risk of venous thrombosis, even when these levels were very low. This is in agreement with studies in which free protein S was a better indicator of venous thrombosis risk than total protein S.32 Nevertheless, as shown by its broad CI, this analysis suffers from small sample size (n = 7 of the patient population), indicating that even if very low free protein S levels are associated with venous thrombosis, large numbers of patients with venous thrombosis will need to be tested to identify only a few with a true free protein S deficiency. Such a strategy is unlikely to become clinically useful.
Many patients with venous thrombosis who are on anticoagulant treatment at the time of the blood sampling are identified with a protein S deficiency, which is thought to have clinical consequences.17,33 However, both our study and previous studies show reduced protein S levels in vitamin K antagonist and oral contraceptive users in the absence of a known true (genetic) deficiency.34,35 This reinforces the importance of avoiding the collection of blood for protein S measurements at moments when acquired deficiency can be present.
Molecular analysis of PROS1 in 48 subjects with very low levels of protein S (below the 1st percentile) identified mutations in only 10 subjects. This finding suggests that inherited protein S deficiency is rare in the general population and in patients with thrombosis. Please note, however, that our analysis was limited to copy number variations, exons and their flanking regions, and the 5′- and 3′-UT regions. We cannot exclude that genetic variations in introns or in promoter regions could play a role in protein S deficiency.
Mutations were equally distributed among patients and controls. Three of 6 patients with protein S levels below percentile 0.2 carried mutations that are likely to be detrimental (complete deletion of PROS1, 967delTinsGG, and Thr144Asn). In contrast, mutations in controls were not likely to be detrimental. Protein S Heerlen (known to be associated with type III protein S deficiency) was detected in 2 controls. Another control carried a variation previously described as a neutral polymorphism (Arg233Lys) and a final control carried a variation in the 3′-UT region.
PROS1 deletion was present in only 1 patient, showing that gross copy number variation of PROS1 is rare in the general population.
Some aspects of our study need additional comment. First, we have noted that high levels of free protein S were associated with an increased risk of venous thrombosis. High levels of free protein S have been associated with an increased risk of coronary heart disease in another study.36 Free protein S has been described as a semi-delayed acute phase reactant19,37 and protein S levels were found to be higher in overweight/obese subjects than in subjects with normal weight.25 Obesity is associated with a chronic inflammatory status and increased risk for venous thrombosis.38 It is possible that high levels of free protein S are a consequence of the former. However, adjustment for overweight/obesity only attenuated the risk of venous thrombosis associated with high levels of free protein S, suggesting that other mechanisms could play a role.
Second, our study is a case-control study in which the blood sample was collected after the thrombotic event. Therefore, the possibility that differences in plasma levels of protein S in patients and control subjects were the result of the thrombotic event itself cannot be excluded. However, the blood draw was performed at least 3 months after the thrombotic event, and it is unlikely that the thrombotic event itself caused persistent abnormalities in protein S levels. Furthermore, no differences in protein S levels were observed between patients in whom blood was drawn 3 to 6 months after their thrombotic event compared with those in whom the blood draw took place later (eg, more than 6 to 9, 9 to 12, or >12 months).
Third, free and total protein S were only measured once. This may have led to misclassification of protein S deficiency when protein S levels fluctuate due to transient acquired conditions. However, only use of female hormones and vitamin K antagonists are known determinants of protein S levels. Sensitivity analyses in which oral contraceptive users and pregnant or postpartum women were excluded did not materially affect the risk estimates for total protein S, but did slightly affect those for free protein S. Moreover, subjects who were still on vitamin K antagonists at the time of blood draw were excluded from analyses in which relative risk estimates where determined.
Finally, qualitative analysis of protein S was not performed, as the laboratory analysis of protein S activity levels is known to produce spurious false low levels. Therefore, we may have missed individuals with type II protein S deficiency.39
In summary, low free protein S and low total protein S levels could, at best, rarely identify subjects at risk for venous thrombosis in a population-based study. Only when cutoff levels for free protein S were far below the normal range or when unprovoked venous thrombosis was considered as an outcome event, we found a twofold to fivefold increased risk of venous thrombosis. Protein S testing and subsequent testing on PROS1 mutations, therefore, should not be considered in unselected patients with venous thrombosis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Aat van Wijngaarden for his assistance in PROS1 sequencing, and Rob van Eck for measuring protein S.
This work and the Multiple Environmental and Genetic Assessment of risk factor for venous thrombosis study were supported by grants from The Netherlands Heart Foundation (NHS 2006B160 and NHS 98.113), the Dutch Cancer Foundation (RUL 99/1992), The Netherlands Organisation for Scientific Research (912-09-033-2003), and CAPES (Edital DRI/CGCI/006/2009) (D.D.R.), and by a postdoctoral fellowship of the Netherlands Heart Foundation (2011T12) (W.M.L.).
Authorship
Contribution: M.C.P. performed laboratory analysis, interpreted the data; M.C.P. and D.D.R. drafted the manuscript; D.D.R., I.D.B., and W.M.L. had full access to the database; D.D.R. performed statistical analysis; I.D.B. and C.J.M.D. collected data for the Multiple Environmental and Genetic Assessment of risk factor for venous thrombosis study and contributed to the analysis; A.A.G. standardized the MLPA method; A.A.G., M.C.H.d.V., C.J.M.D., W.M.L., P.H.R., and F.R.R. revised the manuscript; M.C.H.d.V. discussed the results of this study; W.M.L. supervised statistical analysis; P.H.R. conceived and designed the present study and analyzed the data; and F.R.R. designed the MEGA study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pieter H. Reitsma, Einthoven Laboratory for Experimental Vascular Medicine, C7-14g, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, The Netherlands; e-mail: p.h.reitsma@lumc.nl.