Abstract
Dipeptidylpeptidase (DPP) 4 has the potential to truncate proteins with a penultimate alanine, proline, or other selective amino acids at the N-terminus. DPP4 truncation of certain chemokines, colony-stimulating factors, and interleukins have recently been linked to regulation of hematopoietic stem/progenitor cells, more mature blood cells, and other cell types. We believe that the potential role of DPP4 in modification of many regulatory proteins, and their subsequent effects on numerous stem/progenitor and other cell-type functions has not been adequately appreciated. This review addresses the potential implications of the modifying effects of DPP4 on a large number of cytokines and other growth-regulating factors with either proven or putative DPP4 truncation sites on hematopoietic cells, and subsequent effects of DPP4-truncated proteins on multiple aspects of steady-state and stressed hematopoiesis, including stem/progenitor cell, and more mature cell, function.
Introduction
The physiology and pathology of organs and tissues depends on normal or disordered regulation of their inherent cell types, respectively. Hematopoietic stem cell (HSC) and hematopoietic progenitor cell (HPC) functions are regulated in paracrine fashion by cell-released cytokines, chemokines, and other growth-modulating factors that induce their effects through specific receptor-mediated intracellular signaling.1-3 Such proteins also regulate other stem and progenitor cell types and influence functions of more mature cells. Although many studies have elucidated activities of these proteins for cell and intracellular effects, little effort has gone into understanding how changes in the biomolecules themselves influence steady-state or stressed cell functions in health and disease, and the roles that enzymes may play in modifying biomolecule activity. This review focuses on dipeptidylpeptidase (DPP) 4, recently shown to influence the functional activity of a number of chemokines, colony-stimulating factors (CSFs), and ILs for effects on HSCs, HPCs, and hematopoiesis.4-6 We believe that DPP4 plays an important and under-recognized role in modifying the activities of many different proteins that influence a multitude of different cell, tissue, and organ responses via protein truncation. Herein we review information on the enzymatic activities of DPP4 and provide a large but not necessarily all-inclusive list of chemokines, other cytokines, and growth modulatory proteins with putative DPP4 truncation sites, focusing on proteins that are known to or may in the future be shown to influence blood cells, and we suggest the means by which DPP4 changes, or might change, the activities of these biomolecules.
DPP4, discovered in 1966, is a 110 Kd member of the prolyoligopeptidase family,7 whose crystal structure has been described in complex with a substrate analog.8 DPP4 functions as a serine protease, selectively cleaving the N-terminal, penultimate proline, or alanine of proteins; other amino acids, such as serine, in the second position, may also be able to be cleaved.9 DPP4 is found as both a type II cell surface protein (CD26) and as a soluble molecule lacking intracellular and transmembrane domains.10 DPP4 is found in serum/plasma, cerebrospinal fluid, synovial fluid, and semen.11,12 The transmembrane form of the protein is ubiquitously expressed in many tissues including bone marrow, venular end of blood vessels, lung, spleen, pancreas, kidney, liver, intestines, and epithelial cells.10,13 It is expressed on embryonic stem cells (ESCs),14 HSCs, and HPCs,4,5 as well as on other more mature blood cells such as memory T cells,15 and may be expressed on other stem and progenitor cell populations.
Although DPP4 may function in a nonenzymatic way, such as described in recent reports in which CD26 is involved in infection of cells by oncoviruses,16 and situations in which natural ligands of DPP4 have been shown to enhance the production of transforming growth factor (TGF)-β on T-cell subsets,17 the enzymatic activity of DPP4 will be the focus of this review because it is important for regulation of steady-state and stressed cellular functions and has been implicated in certain disease states. DPP4 activity is increased in the plasma/serum of people with type 2 diabetes and increased on the surface of T cells in patients with arthritis,18 and increases in DPP4 have been correlated with organ transplant rejection.19-23 Although not much is known about the regulation of DPP4, a heparin sulfate proteoglycan expressed on human and murine stem and progenitor cells, GPC-3 (glypican-3) interacts with tissue factor pathway inhibitor to act as a biological inhibitor of DPP4.24 Understanding preferred targets of DPP4, how DPP4 may alter protein activities for physiological activity and disease pathophysiology,25 as well as regulation of CD26 expression and its enzymatic activity is important and warrants continued investigation.
Effects of DPP4 on HSCs and HPCs
Inhibition of DPP4 enzymatic activity by small peptides such as diprotin A (ILE-PRO-ILE) or VAL-PYR on HPCs expressing CD26 enhances chemotaxis to the chemokine stromal cell–derived factor-1 (SDF-1/CXCL12)4 and homing and engraftment of HSCs.5,26-31 DPP4-truncated CXCL12 was without chemotactic activity but blocked chemotaxis induced by full-length SDF-1.4 Pretreating target populations of human cord blood or mouse bone marrow with diprotin A or using CD26−/− mouse target cells enhanced the CSF activities of granulocyte macrophage (GM)-CSF, granulocyte (G)-CSF, IL-3, and erythropoietin (Epo) for proliferation of HPCs.6 DPP4-truncated CSFs were greatly decreased in CSF activity and blunted the full-length activity of their respective CSF, both in vitro and in vivo.6 At least for GM-CSF, DPP4 truncated GM-CSF bound to the GM-CSF receptor (R) with higher affinity than that of full-length GM-CSF. At a lower amount of truncated to full-length GM-CSF, truncated GM-CSF competitively blocked GM-CSFR binding of full length GM-CSF, and phosphorylation of Jak2 and STAT5. Truncated GM-CSF either prevented hexamer to dodecamer formation of the GM-CSFR complex needed for full GM-CSF activity, or the truncated GM-CSF did not fully signal through the dodecamer GM-CSFR complex.6 Interestingly, the clinical G-CSF preparation (Neupogen) used for mobilization of HSCs and HPCs from bone marrow to blood, has a methionine placed at the N-terminus start site, thus shifting the amino acid sequence so that DPP4 could not truncate the molecule. However, a G-CSF preparation without methionine at the N-terminus, which had a DPP4 truncation site in the correct position, could be enhanced in CSF activity by pretreating target cells with diprotin A to prevent DPP4 truncation of this preparation of G-CSF.6 Interestingly, CD26−/− in mice,32 or diprotin A pretreatment of mice to inhibit DPP4,33 attenuated the HPC-mobilizing capacity of G-CSF, the mechanisms of which have not yet been fully elucidated. This activity likely does not involve direct effects of DPP4 on exogenously administered Neupogen, because, as noted before, this form of G-CSF does not have a truncation site for DPP4.
Taking into consideration effects of DPP4 inhibition on enhanced homing and engraftment of mouse HSC5,6,26-28 and engraftment of human CD34+ cells29-31 in immune-deficient mice, a pilot clinical trial assessed the effects of administration of sitagliptin (a small-molecule inhibitor of DPP4 used to treat type 2 diabetes34,35 ) to patients with high-risk hematologic malignancies receiving single-unit cord blood transplants,36 with encouraging but not yet conclusive results regarding enhanced time to neutrophil engraftment. With the more recent knowledge that DPP4 has negative effects on CSFs6 that nurture immature cell populations in the bone marrow, efforts are underway to modify the dosing schedule of sitagliptin for greater efficacy in enhancing time to engraftment of cord blood. In another transplant setting, diprotin A pretreatment of muscle-derived cells before injection significantly enhanced their engraftment and stimulated sustained proliferation of donor cells in a canine model of Duchenne muscular dystrophy.37 DPP4 inhibition may also be of relevance in transplant settings for the lungs, kidney, and liver.19-23 How these DPP4 phenomena are mechanistically manifested is yet to be determined but may involve blocking DPP4-induced truncation of different proteins.
Effects of DPP4 on leukocyte trafficking and migration
Chemokines play fundamental roles in the immune system for leukocyte trafficking, leukocyte degranulation, and angiogenesis,38 and DPP4 may significantly influence chemokine activity. Similar to DPP4 effects on CXCL12,4-6 DPP4 exerts negative feedback by decreasing the activity of CCL22/MDC. CCL22 attracts monocytes, dendritic cells, activated lymphocytes, and natural killer cells, and reportedly has anti–HIV-1 activity. DPP4-truncated CCL22 fails to desensitize calcium mobilization by full-length CCL22 or thymus- and activation-regulated chemokine in CCR4-transfected cells.39 Truncated CCL22 reduces HUT-78 T-cell chemotactic activity and is 100-fold less potent than full-length CCL22.40 Thus, N-terminal truncation of CCL22 by DPP4 differentially affects its various immunologic functions. Natural NH2-terminally truncated CCL2 (5-76) and CCL8 (6-76) are practically devoid of bioactivity. The truncated forms are potent antagonists of their full-length chemokines for chemotactic potency,41 in a manner similar to that noted for CXCL12-induced chemotaxis of HPCs.4 The CC chemokine, CCL11 (eotaxin), attracts eosinophils to sites of parasitic infection and allergic inflammation. Its chemotactic potency for blood eosinophils and signaling capacity through CCR3 are reduced 30-fold upon truncation by DPP4.42 These examples further highlight a role for DPP4 relevance during inflammation and infection, as well as in steady-state hematopoiesis.
A plethora of proteins with potential DPP4 truncation sites
To determine the extent of proteins with putative DPP4 truncation sites, we initiated a search of amino acid sequences retrieved from the National Center for Biotechnology Information (NCBI) database and from Universal Protein Resource. This search identified a large number of proteins that have a penultimate alanine, proline, or serine in the N-terminus start site. Tables 1 and 2, respectively, list chemokines/other cytokines and additional proteins with potential cell growth regulatory capacities that contain putative penultimate alanine or proline truncation sites for DPP4 activity. Table 3 highlights proteins with a putative serine truncation site. Putative truncation sites are listed for human and mouse proteins, because it is not always the case that both the human and mouse proteins have apparent DPP4 sites. Tables 1 to 3 are not meant to be all inclusive of proteins with DPP4 truncation sites but rather are to demonstrate that there are a wide variety of biologically active molecules that act on many different cell types, tissues, and organ systems that may be affected in significant ways by DPP4, especially because DPP4 is expressed on the cell surface of, and also present within, many cells and it is present in plasma and serum in soluble form.15 Whether or not all proteins listed in Tables 1 to 3 have true DPP4 truncation sites will have to be specifically determined for each of these proteins selectively via mass spectrometry or comparable analysis, and biological assays in vitro and in vivo will be required to determine whether the truncated molecules differ in functional activity from the full-length form of the protein. Classically, DPP4 is considered to cleave specifically at alanine and proline residues in the second, or penultimate, position, with little regard for the first amino acid provided it has a protinated amino group. However, some reports suggest that additional residues in the penultimate position, such as serine, could also be targets of DPP4, but these sites are considered less preferred than the proline or alanine.9 Importantly, certain protein modifications can alter the rate of cleavage. Proteins with proline, hydroxylproline, or N-methylglycine in the third position are thought to prevent cleavage by DPP4.43 This highlights the complexity of potential DPP4 cleavage kinetics, which may differ in vitro and in vivo, and further highlights the necessity of testing the individual DPP4 cleavage potential of each protein of interest.
Chemokines/cytokines with putative penultimate (alanine/proline) truncation site for DPP4
| Chemokines . | Cytokines . | ||||
|---|---|---|---|---|---|
| Peptide . | Species . | N-terminus*† . | Peptide . | Species . | N-terminus*† . |
| CXCL1/GRO-α | M | APIAN… | Epo | H | APPRL... |
| CXCL2/GRO-β/MIP-2α | H | APLAT… | Epo | M | APPRL... |
| CXCL4/PF4 | H | EAEED… | GM-CSF | H | APARS... |
| CXCL5/ENA-78 | M | APSSV... | GM-CSF | M | APTRS... |
| CXCL6/GCP2 | H | GPVSA... | G-CSF | H | ATPLG... |
| CXCL8/IL-8 | H | SAKEL... | G-CSF | M | VPLVT... |
| CXCL9/MIG | H | TPVVR... | IL-3 | H | APMTQ... |
| CXCL10/INP-10 | H | VPLSR... | IL-1 α | H | SAPFS... |
| CXCL10/INP-10 | M | IPLAR... | IL-1 β | H | APVRS... |
| CXCL11/I-TAC | H | IPLAR... | IL-1 α | M | SAPYT... |
| CXCL12/SDF-1 | H | KPVSL... | IL-1 β | M | VPIRQ... |
| CXCL12 (isoform α) | M | KPVSL... | IL-2 | H | APTSS... |
| CXCL12 (isoform β) | M | KPVSL... | IL-2 | M | APTSS... |
| CXCL12(isoform γ) | M | QPDAI... | IL-5 | H | IPTEI... |
| CCL2/MCP-1 | H | QPDAV... | IL-6 | H | VPPGE... |
| CCL2/MCP-1 | M | APYGA... | IL-6 | M | FPTSQ... |
| CCL3/MIP-1α/LD78α | M | APLAA... | IL-8 (6-77) | H | SAEKL... |
| CCL3-L1/LD78β | H | APMGS... | IL-10 | H | SPGQG... |
| CCL4/MIP-1β | H | APMGS... | IL-13 | H | GPVPP... |
| CCL4/MIP-1β | M | APMGS... | IL-17B | H | QPRSP... |
| CCL5/RANTES | H | SPYSS... | IL-17A | M | AAIIP... |
| CCL5/RANTES | M | SPYGS... | IL-17C | M | DPPSW... |
| CCL7/MCP-3 | H | QPVGI... | IL-22 | H | APISS... |
| CCL7/MCP-3 | M | QPDGP... | IL-22 | M | LPVNT... |
| CCL8/MCP-2 | H | QPDSV... | IL-23 α | H | RAVPG... |
| CCL11/Eotaxin | H | GPASV... | IL-23 α | M | VPRSS... |
| CCL11/Eotaxin | M | HPGSI... | IL-27 α | H | FPRPP... |
| CCL12/MCP-5 | M | GPDAV... | IL-27 α | M | FPTDP... |
| CCL16/HCC-4/LEC/LCC-1/LMC | H | QPKVP... | IL-28A | H | VPVAR... |
| CCL19/CKβ11/MIP-3β/ELC/Exodus-3 | M | GANDA... | IL-28B | H | VPVAR... |
| CCL22/MDC/STCP-1 | H | GPYGA... | IL-28A | M | DPVPR... |
| CCL22/MDC/STCP-1 | M | GPYGA... | IL-28B | M | DPVPR... |
| CCL26/Eotaxin-3/MIP-4α | M | HPGSI... | IL-29 | H | GPVPT... |
| CCL27/CTACK/ILC/ESKINE | M | LPLPS... | |||
| Chemokines . | Cytokines . | ||||
|---|---|---|---|---|---|
| Peptide . | Species . | N-terminus*† . | Peptide . | Species . | N-terminus*† . |
| CXCL1/GRO-α | M | APIAN… | Epo | H | APPRL... |
| CXCL2/GRO-β/MIP-2α | H | APLAT… | Epo | M | APPRL... |
| CXCL4/PF4 | H | EAEED… | GM-CSF | H | APARS... |
| CXCL5/ENA-78 | M | APSSV... | GM-CSF | M | APTRS... |
| CXCL6/GCP2 | H | GPVSA... | G-CSF | H | ATPLG... |
| CXCL8/IL-8 | H | SAKEL... | G-CSF | M | VPLVT... |
| CXCL9/MIG | H | TPVVR... | IL-3 | H | APMTQ... |
| CXCL10/INP-10 | H | VPLSR... | IL-1 α | H | SAPFS... |
| CXCL10/INP-10 | M | IPLAR... | IL-1 β | H | APVRS... |
| CXCL11/I-TAC | H | IPLAR... | IL-1 α | M | SAPYT... |
| CXCL12/SDF-1 | H | KPVSL... | IL-1 β | M | VPIRQ... |
| CXCL12 (isoform α) | M | KPVSL... | IL-2 | H | APTSS... |
| CXCL12 (isoform β) | M | KPVSL... | IL-2 | M | APTSS... |
| CXCL12(isoform γ) | M | QPDAI... | IL-5 | H | IPTEI... |
| CCL2/MCP-1 | H | QPDAV... | IL-6 | H | VPPGE... |
| CCL2/MCP-1 | M | APYGA... | IL-6 | M | FPTSQ... |
| CCL3/MIP-1α/LD78α | M | APLAA... | IL-8 (6-77) | H | SAEKL... |
| CCL3-L1/LD78β | H | APMGS... | IL-10 | H | SPGQG... |
| CCL4/MIP-1β | H | APMGS... | IL-13 | H | GPVPP... |
| CCL4/MIP-1β | M | APMGS... | IL-17B | H | QPRSP... |
| CCL5/RANTES | H | SPYSS... | IL-17A | M | AAIIP... |
| CCL5/RANTES | M | SPYGS... | IL-17C | M | DPPSW... |
| CCL7/MCP-3 | H | QPVGI... | IL-22 | H | APISS... |
| CCL7/MCP-3 | M | QPDGP... | IL-22 | M | LPVNT... |
| CCL8/MCP-2 | H | QPDSV... | IL-23 α | H | RAVPG... |
| CCL11/Eotaxin | H | GPASV... | IL-23 α | M | VPRSS... |
| CCL11/Eotaxin | M | HPGSI... | IL-27 α | H | FPRPP... |
| CCL12/MCP-5 | M | GPDAV... | IL-27 α | M | FPTDP... |
| CCL16/HCC-4/LEC/LCC-1/LMC | H | QPKVP... | IL-28A | H | VPVAR... |
| CCL19/CKβ11/MIP-3β/ELC/Exodus-3 | M | GANDA... | IL-28B | H | VPVAR... |
| CCL22/MDC/STCP-1 | H | GPYGA... | IL-28A | M | DPVPR... |
| CCL22/MDC/STCP-1 | M | GPYGA... | IL-28B | M | DPVPR... |
| CCL26/Eotaxin-3/MIP-4α | M | HPGSI... | IL-29 | H | GPVPT... |
| CCL27/CTACK/ILC/ESKINE | M | LPLPS... | |||
CCL, chemokine (C-C motif) ligand; CXCL, chemokine (C-X-C motif) ligand; Epo, erythropoietin; GM-CSF, granulocyte-macrophage colony-stimulating factor; G-GSF, granulocytes colony-stimulating factor; IL, interleukin; Species: M = mouse, H = human.
Sequence retrieved from NCBI database.
Sequence retrieved from UniProt.
Modulatory factors with putative penultimate (alanine/proline) truncation site for DPP4
| Modulatory Factors . | |||||
|---|---|---|---|---|---|
| Peptide . | Species . | N-terminus*† . | Peptide . | Species . | N-terminus*† . |
| Adipsin (propeptide) | H | PPRGR... | IGF-1 isoform 4 | M | GPETL... |
| Adipsin (propeptide) | M | QPRGR... | IGF-1 isoform 5 | M | GPETL... |
| α-1 microglobulin | H | GPVPT... | Inhibin α chain (propeptide) | H | HALGG... |
| α-1 microglobulin | M | DPAST... | Inhibin α chain (propeptide) | M | HAVGG... |
| ANGPTL4 (isoform a) | H | GPVQS... | Inhibin β E chain | H | TPTCE... |
| ANGPTL4 (isoform b) | H | GPVQS... | Inhibin β E chain | M | TPTCE... |
| ANGPTL6 | H | RAGAP... | Lactoferrin | M | KATTV... |
| BDNF (propeptide) | H | APMKE... | Leptin | H | VPIQK... |
| BDNF (propeptide) | M | APMKE... | Leptin | M | VPIQK... |
| bFGF | H | PALPE... | LIF | H | SPLPI... |
| bFGF | M | PALPE... | LIF | M | SPLPI... |
| Bradykinin | H | RPPGF... | LTα | H | LPGVG... |
| Bradykinin | M | RPPGF... | Neurotrophin-3 | H | YAEHK... |
| BMP-4 | H | SPKHH... | Neurotrophin-3 | M | YAEHK... |
| BMP-4 | M | SPKHH... | Notch-3 | H | APPCL... |
| BMP-5 | H | AANKR... | Notch-3 | M | APPCL... |
| BMP-5 | M | AASKR... | NPY | H | YPSKP... |
| c1qTNF5 | H | SPPLD... | NPY | M | YPSKP... |
| c1qTNF5 | M | SPPLD... | Oncostatin M | H | AAIGS... |
| Chromogranin | H | LPVNS... | Osteopontin | H | IPVKQ... |
| Chromogranin | M | LPVNS... | Osteopontin | M | LPVKV... |
| DKK3 | H | APAPT... | PDGFD | M | TPQRA... |
| DNER | H | NPVPA... | Peptide YY | H | YPIKP... |
| Enterostatin | H | APGPR... | Peptide YY | M | YPAKP... |
| Enterostatin | M | APGPR... | Prolactin | H | LPICP... |
| GDF-1 | H | DAEPV... | Prolactin | M | LPICS... |
| GDF-3 | H | AAIPV... | RBP3 | H | GPTHL... |
| GDF-3 | M | AAISV... | RBP3 | M | GPTHL... |
| GDF-5 | H | APLAT... | Somatotropin isoform 1 | H | FPTIP... |
| GDF-5 | M | APLAN... | Somatotropin isoform 2 | H | FPTIP... |
| GDF-6 | H | TAFAS... | Somatotropin isoform 3 | H | FPTIP... |
| GDF-6 | M | TAFAS... | Somatotropin isoform 4 | H | FPTIP... |
| GDF-7 | H | TALAG... | Somatotropin | M | FPAMP... |
| GDF-7 | M | TALAG... | Transferrin | H | VPDKT... |
| GDF-8 | H | GPVDL...§ | Transferrin | M | VPDKT... |
| GDNF (propeptide) | H | SPDKQ... | Trypsinogen | H | APFDD... |
| GDNF (propeptide) | M | SPDKQ... | Vasostatin-1 | H | LPVNS... |
| GHRH | H | YADAI... | Vasostatin-2 | H | LPVNS... |
| GLP-1 (7-36) | H | HAEGT... | VEGF A isoform i | H | APMAE... |
| GLP-2 | H | HADGS... | VEGF A isoform k | H | APMAE... |
| GRP | H | VPLPA... | VEGF A isoform l | H | APMAE... |
| GRP | M | APVST... | VEGF A isoform m | H | APMAE... |
| HCG α | H | APDVQ... | VEGF A isoform n | H | APMAE... |
| HCG α | M | LPDGD... | VEGF A isoform o | H | APMAE... |
| IGF-1 isoform 1 | H | GPETL... | VEGF A isoform p | H | APMAE... |
| IGF-1 isoform 2 | H | GPETL... | VEGF A isoform 1 | M | APTTE... |
| IGF-1 isoform 3 | H | GPETL... | VEGF A isoform 2 | M | APTTE... |
| IGF-1 isoform 4 | H | GPETL... | VEGF A isoform 3 | M | APTTE... |
| IGF-1 isoform 1 | M | GPETL... | Wnt 9b | M | AAYFG... |
| IGF-1 isoform 2 | M | GPETL... | Wnt 10a | H | MPRSA... |
| IGF-1 isoform 3 | M | GPETL... | Wnt 10a | M | VPRSA... |
| Modulatory Factors . | |||||
|---|---|---|---|---|---|
| Peptide . | Species . | N-terminus*† . | Peptide . | Species . | N-terminus*† . |
| Adipsin (propeptide) | H | PPRGR... | IGF-1 isoform 4 | M | GPETL... |
| Adipsin (propeptide) | M | QPRGR... | IGF-1 isoform 5 | M | GPETL... |
| α-1 microglobulin | H | GPVPT... | Inhibin α chain (propeptide) | H | HALGG... |
| α-1 microglobulin | M | DPAST... | Inhibin α chain (propeptide) | M | HAVGG... |
| ANGPTL4 (isoform a) | H | GPVQS... | Inhibin β E chain | H | TPTCE... |
| ANGPTL4 (isoform b) | H | GPVQS... | Inhibin β E chain | M | TPTCE... |
| ANGPTL6 | H | RAGAP... | Lactoferrin | M | KATTV... |
| BDNF (propeptide) | H | APMKE... | Leptin | H | VPIQK... |
| BDNF (propeptide) | M | APMKE... | Leptin | M | VPIQK... |
| bFGF | H | PALPE... | LIF | H | SPLPI... |
| bFGF | M | PALPE... | LIF | M | SPLPI... |
| Bradykinin | H | RPPGF... | LTα | H | LPGVG... |
| Bradykinin | M | RPPGF... | Neurotrophin-3 | H | YAEHK... |
| BMP-4 | H | SPKHH... | Neurotrophin-3 | M | YAEHK... |
| BMP-4 | M | SPKHH... | Notch-3 | H | APPCL... |
| BMP-5 | H | AANKR... | Notch-3 | M | APPCL... |
| BMP-5 | M | AASKR... | NPY | H | YPSKP... |
| c1qTNF5 | H | SPPLD... | NPY | M | YPSKP... |
| c1qTNF5 | M | SPPLD... | Oncostatin M | H | AAIGS... |
| Chromogranin | H | LPVNS... | Osteopontin | H | IPVKQ... |
| Chromogranin | M | LPVNS... | Osteopontin | M | LPVKV... |
| DKK3 | H | APAPT... | PDGFD | M | TPQRA... |
| DNER | H | NPVPA... | Peptide YY | H | YPIKP... |
| Enterostatin | H | APGPR... | Peptide YY | M | YPAKP... |
| Enterostatin | M | APGPR... | Prolactin | H | LPICP... |
| GDF-1 | H | DAEPV... | Prolactin | M | LPICS... |
| GDF-3 | H | AAIPV... | RBP3 | H | GPTHL... |
| GDF-3 | M | AAISV... | RBP3 | M | GPTHL... |
| GDF-5 | H | APLAT... | Somatotropin isoform 1 | H | FPTIP... |
| GDF-5 | M | APLAN... | Somatotropin isoform 2 | H | FPTIP... |
| GDF-6 | H | TAFAS... | Somatotropin isoform 3 | H | FPTIP... |
| GDF-6 | M | TAFAS... | Somatotropin isoform 4 | H | FPTIP... |
| GDF-7 | H | TALAG... | Somatotropin | M | FPAMP... |
| GDF-7 | M | TALAG... | Transferrin | H | VPDKT... |
| GDF-8 | H | GPVDL...§ | Transferrin | M | VPDKT... |
| GDNF (propeptide) | H | SPDKQ... | Trypsinogen | H | APFDD... |
| GDNF (propeptide) | M | SPDKQ... | Vasostatin-1 | H | LPVNS... |
| GHRH | H | YADAI... | Vasostatin-2 | H | LPVNS... |
| GLP-1 (7-36) | H | HAEGT... | VEGF A isoform i | H | APMAE... |
| GLP-2 | H | HADGS... | VEGF A isoform k | H | APMAE... |
| GRP | H | VPLPA... | VEGF A isoform l | H | APMAE... |
| GRP | M | APVST... | VEGF A isoform m | H | APMAE... |
| HCG α | H | APDVQ... | VEGF A isoform n | H | APMAE... |
| HCG α | M | LPDGD... | VEGF A isoform o | H | APMAE... |
| IGF-1 isoform 1 | H | GPETL... | VEGF A isoform p | H | APMAE... |
| IGF-1 isoform 2 | H | GPETL... | VEGF A isoform 1 | M | APTTE... |
| IGF-1 isoform 3 | H | GPETL... | VEGF A isoform 2 | M | APTTE... |
| IGF-1 isoform 4 | H | GPETL... | VEGF A isoform 3 | M | APTTE... |
| IGF-1 isoform 1 | M | GPETL... | Wnt 9b | M | AAYFG... |
| IGF-1 isoform 2 | M | GPETL... | Wnt 10a | H | MPRSA... |
| IGF-1 isoform 3 | M | GPETL... | Wnt 10a | M | VPRSA... |
ANGPTL, angiopoietin-related protein; BDNF, brain-derived neurotrophic factor; bFGF, basis fibroblast growth factor; BMP, bone morphogenetic protein; c1qTNF5, C1q and tumor necrosis factor related protein 5; DKK, Dickkopf-related protein; DNER, Δ and notchlike epidermal growth factor–related receptor; GDF, growth differentiation factor; GDNF, glial cell line–derived neurotrophic factor; GHRH, growth-hormone–releasing hormone; GLP, glucagonlike peptide; GRP, gastrin-releasing peptide; HCG α, glycoprotein hormones α chain; IGF, insulin-like growth factor; LIF, leukemia inhibitory factor; LTα, lymphotoxin-α; Notch, neurogenic locus notch homolog protein; NPY, neuropeptide Y; PDGFD, platelet-derived growth factor D; RBP, retinol-binding protein; VEGF, vascular endothelial growth factor. Species: M = mouse, H = human.
Sequence retrieved from NCBI database.
Sequence retrieved from UniProt.
Sequence only retrieved from NCBI database.
Factors with putative truncation site (serine) for DPP4
| Peptide . | Species . | N-terminus*† . | Peptide . | Species . | N-terminus*† . |
|---|---|---|---|---|---|
| CXCL1 | H | ASVAT... | ANGPT2 | M | YSNFR... |
| CXCL3 | H | ASVVT... | BDNF | H | HSDPA... |
| CXCL7 | H | SSTKG... | BDNF | M | HSDPA... |
| CXCL7 | M | KSDGM... | DLL3 | M | HSFGP... |
| CCL1 | H | KSMQV... | DLL4 | M | GSGIF... |
| CCL1 | M | KSMLT... | DKK3 | M | PSPTV... |
| CCL20 | H | ASNFD... | FZD6 | H | HSLFT... |
| CCL20 | M | ASNYD... | FZD6 | M | HSLFT... |
| IL-3 | M | ASISG... | Glucagon | M | HSQGT... |
| IL-17E | M | VSLRI... | Glucagon | M | HSQGT... |
| IFN-β | H | MSYN... | GM2A | H | SSFSW... |
| LTα | M | LSGVR... | β NGF | H | SSSHP... |
| TGF-β1 | H | LSTCK... | β NGF | M | SSTHP... |
| TGF-β1 | M | LSTCK... | PACAP (1-27) | H | HSDGI... |
| TNF-β | M | LSGVR... | PACAP (1-38) | H | HSDGI... |
| TNF (membrane form) | H | MSTES... | PDGFC | M | ESNLS... |
| TNF (membrane form) | M | MSTES... | PDGFC | H | ESNLS... |
| DEK | H | MSASA... | Wnt 8a | M | ASAWS... |
| DEK | M | MSAAA... | Wnt 8b | H | WSVNN... |
| Peptide . | Species . | N-terminus*† . | Peptide . | Species . | N-terminus*† . |
|---|---|---|---|---|---|
| CXCL1 | H | ASVAT... | ANGPT2 | M | YSNFR... |
| CXCL3 | H | ASVVT... | BDNF | H | HSDPA... |
| CXCL7 | H | SSTKG... | BDNF | M | HSDPA... |
| CXCL7 | M | KSDGM... | DLL3 | M | HSFGP... |
| CCL1 | H | KSMQV... | DLL4 | M | GSGIF... |
| CCL1 | M | KSMLT... | DKK3 | M | PSPTV... |
| CCL20 | H | ASNFD... | FZD6 | H | HSLFT... |
| CCL20 | M | ASNYD... | FZD6 | M | HSLFT... |
| IL-3 | M | ASISG... | Glucagon | M | HSQGT... |
| IL-17E | M | VSLRI... | Glucagon | M | HSQGT... |
| IFN-β | H | MSYN... | GM2A | H | SSFSW... |
| LTα | M | LSGVR... | β NGF | H | SSSHP... |
| TGF-β1 | H | LSTCK... | β NGF | M | SSTHP... |
| TGF-β1 | M | LSTCK... | PACAP (1-27) | H | HSDGI... |
| TNF-β | M | LSGVR... | PACAP (1-38) | H | HSDGI... |
| TNF (membrane form) | H | MSTES... | PDGFC | M | ESNLS... |
| TNF (membrane form) | M | MSTES... | PDGFC | H | ESNLS... |
| DEK | H | MSASA... | Wnt 8a | M | ASAWS... |
| DEK | M | MSAAA... | Wnt 8b | H | WSVNN... |
ANGPTL, angiopoietin-related protein; BDNF, brain-derived neurotrophic factor; CCL, chemokine (C-C motif) ligand; CXCL, chemokine (C-X-C motif) ligand; DKK, Dickkopf-related protein; DLL, Δ-like protein; Epo, erythropoietin; FZD6, frizzled-6; G-GSF, granulocytes colony-stimulating factor; GM2A, ganglioside GM2 activator; GM-CSF, granulocyte-macrophage colony-stimulating factor; IFN, interferon; IL, interleukin; LTα, lymphotoxin-α; NGF, nerve growth factor; PACAP, pituitary adenylate cyclase–activating polypeptide; PDGFD, platelet-derived growth factor D; TGF, transforming growth factor; TNF, tumor necrosis factor.
Sequence retrieved from NCBI database.
Sequence retrieved from UniProt.
Potential effects of DPP4 on ESCs and induced pluripotent stem (iPS) cells
In addition to HSCs, other primitive cells with the capability of both self-renewal and differentiation toward hematopoietic and other cells types, such as ESCs, have the potential to be significantly influenced by DPP4. Several soluble factors have been identified that exert functional effects on ESC self-renewal. Leukemia inhibitory factor (LIF), a member of the IL-6 family, is known to strongly promote self-renewal in mouse ESCs (mESCs).44 LIF has a putative truncation site for DPP4 (Table 2), and CD26 is expressed on mESCs.14 Interestingly, pretreating mESCs with diprotin A enhances the LIF-induced maintenance of mESCs (Ou X, Broxmeyer HE; unpublished observations). Basic fibroblast growth factor (bFGF) is a member of the FGF family. bFGF synergizes with the BMP antagonist noggin to sustain undifferentiated proliferation of human ESCs under feeder-free conditions.45,46 bFGF also stimulates proliferation of all cells of mesodermal origin and many cells of neuroectodermal, ectodermal, and endodermal origin. BMP4 plays well-established roles in embryonic development and lineage determination for mouse and human ESCs, as well as tissue repair. Because bFGF and BMP4 have putative truncation sites for DPP4 (Table 2), efforts are needed to determine possible roles of DPP4 in regulating FGF2 and BMP4 activities, and for determining whether inhibition of DPP4 might better promote human ESC self-renewal or pluripotency.
iPS cells47-50 can be generated via reprogramming of somatic cell types, including immature subsets of cord blood hematopoietic cells,51,52 to an ESC-like state, and human iPS cells require proteins such as FGF2 and BMP4 for their maintenance. Because iPS cells express CD26 (O’Leary HA, Lee MR, Broxmeyer HE; unpublished observations), an investigation of inhibition of DPP4 on iPS cell generation and perhaps differentiation is also warranted.
Models for potential effects of DPP4 truncation of proteins
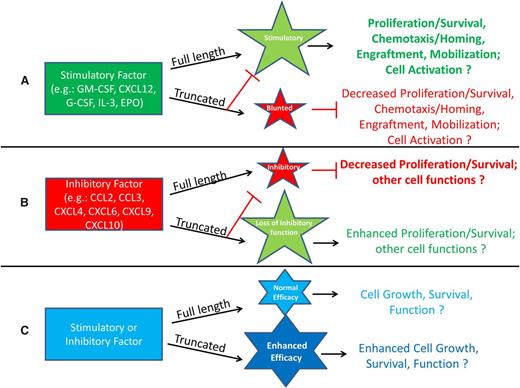
It is clear that we are only just beginning to elucidate the extent of DPP4 effects on hematopoiesis, stem cells, and more mature cell types. This section presents different scenarios of recognized, and also possible, effects of DPP4 truncation of proteins, shown schematically in Figures 1 and 2. Figure 1A demonstrates the possibility that truncation of a positive-acting molecule would lose a large part or all of its activity, and in addition the truncated protein would block or blunt the positive activity of the full-length protein. Examples of this are the truncated forms of SDF-1/CXCL124 and of GM-CSF, G-CSF, IL-3, and Epo.6 These truncated proteins have significantly decreased activity and are able to block the chemotaxis, survival, and CSF activities of the full-length form of their own molecule. Such outcomes may be mediated by those already shown for GM-CSF stimulation of colony formation and subsequent induction of intracellular signaling.6 In this case, DPP4-truncated GM-CSF binds with higher affinity than full length GM-CSF to the GM-CSFR but does not signal well, yet it acts as a competitor (eg, dominant negative form through its blocking of the receptor binding of full-length GM-CSF). In the case of Epo, which has different target receptors and actions, it is not yet clear how DPP4 truncation may modify these different EPO functions.53 Moreover, because other CSFs also have multiple targets, a role for DPP4 truncation of these CSFs and their effects on different target cell populations needs further investigation. Figure 1B presents a situation similar to that in Figure 1A, but here, a protein with suppressive activity loses most, or all, of its inhibitory activity when truncated by DPP4, and the truncated molecule blocks the suppressive activity of the full-length form of the protein, perhaps also through the truncated protein having a higher affinity for its receptor than the full-length form. Evidence for this possibility exists in that inhibition of DPP4 or CD26−/− of target mouse bone marrow cells, or inhibition of DPP4 on cord blood, enhances the suppressive activity of selected members of the chemokine family of molecules, such as CCL2/MCP-1, CCL3/MIP-1α, CXCL4/PF4, CXCL6/GCP-2, CXCL9/MIG, and CXCL10/IP-10, on colony formation of HPC.54 These chemokines have an alanine or proline in the penultimate N-terminus of the start site of these proteins (Table 1). Recently, we found that the DPP4-truncated forms of the chemokines assessed (CCL2, CCL3, CXCL8/IL-8, and CXCL9) lose their suppressive activity and block myelosuppression in vitro and in vivo of the respective full-length forms of their chemokine (Broxmeyer HE; unpublished observations). In both cases (Figure 1A-B), the truncated molecule acts as a competitive inhibitor or dominant negative form of the full-length molecule. This offers a potential means for feedback regulation of the action of their full length molecules. There is also the possibility that truncation by DPP4 may further enhance the stimulatory or inhibitory activity of a molecule beyond that of the full-length version of that molecule (Figure 1C), although examples of this have not yet been shown.
Potential enhancement or inhibition resulting from DPP4 interactions with regulatory proteins. (A) Molecules that are stimulatory in the full-length form may lose activity after DPP4 truncation and act as competitive inhibitors by diminishing the function of the full-length protein through binding to the cognate receptor with higher affinity than that of the full-length molecule. (B) Molecules that are inhibitory in the full-length form may lose their suppressive ability when truncated by DPP4 and by binding to the cognate receptor with higher affinity, allowing for blockage of the full-length molecule’s suppressive activity. (C) Stimulatory or inhibitory molecules may increase their efficacy when truncated.
Potential enhancement or inhibition resulting from DPP4 interactions with regulatory proteins. (A) Molecules that are stimulatory in the full-length form may lose activity after DPP4 truncation and act as competitive inhibitors by diminishing the function of the full-length protein through binding to the cognate receptor with higher affinity than that of the full-length molecule. (B) Molecules that are inhibitory in the full-length form may lose their suppressive ability when truncated by DPP4 and by binding to the cognate receptor with higher affinity, allowing for blockage of the full-length molecule’s suppressive activity. (C) Stimulatory or inhibitory molecules may increase their efficacy when truncated.
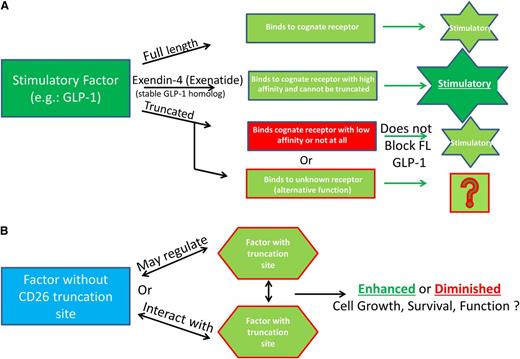
Additional possibilities for functional consequences of DPP4 truncation of regulatory proteins. (A) Truncated molecules do not bind to their receptor and may signal through other receptors. An example of this is full-length GLP-1, which binds to its cognate receptor but is truncated quickly and subsequently is unable to efficiently bind to the GLP-1 receptor and therefore does not inhibit, or alter the function, of the full-length molecule. Exenatide (Byetta) is a synthetic GLP-1 agonist made to mimic the GLP-1 homolog extendin-4, found in the saliva of the Gila monster. Exenatide is used in the treatment of type 2 diabetes because it is stable, binds to the GLP-1 receptor to induce insulin secretion, and is resistant to DPP4 truncation. (B) Factors without truncation sites may regulate factors with truncation sites by altering their expression, or function, and proteins with DPP4 truncation sites may regulate expression of proteins with or without DPP4 truncation sites.
Additional possibilities for functional consequences of DPP4 truncation of regulatory proteins. (A) Truncated molecules do not bind to their receptor and may signal through other receptors. An example of this is full-length GLP-1, which binds to its cognate receptor but is truncated quickly and subsequently is unable to efficiently bind to the GLP-1 receptor and therefore does not inhibit, or alter the function, of the full-length molecule. Exenatide (Byetta) is a synthetic GLP-1 agonist made to mimic the GLP-1 homolog extendin-4, found in the saliva of the Gila monster. Exenatide is used in the treatment of type 2 diabetes because it is stable, binds to the GLP-1 receptor to induce insulin secretion, and is resistant to DPP4 truncation. (B) Factors without truncation sites may regulate factors with truncation sites by altering their expression, or function, and proteins with DPP4 truncation sites may regulate expression of proteins with or without DPP4 truncation sites.
Figure 2A delineates a scenario in which the truncated protein binds to a receptor different from that bound by the full-length protein, a known situation for glucagon-like peptide-1 (GLP-1), a strong antihyperglycemic hormone that induces insulin secretion in a glucose-dependent manner (insulinotropic) and inhibits glucagon secretion. This model could possibly define other molecules, including those active on hematopoietic cells. For GLP-1, this results in the truncated form not inhibiting the full-length form but allows for alterative signaling of the truncated form, potentially through increased binding affinity for another receptor. GLP is rapidly truncated (>98% in ∼3 minutes in vivo), and truncated GLP either binds to the receptor with extremely low affinity (1% that of the full-length form) or not at all.55,56 Truncated GLP-1 does not have the insulinotropic effects seen with full-length GLP-1, but it shows non–GLP-1R-mediated cardioprotective effects.57 This has been shown to be relevant in type 2 diabetes, where DPP4 activity is elevated, and the ability of GLP-1 or other molecules to exert their effects may be diminished. We have evidence that DPP4 activity within bone marrow cells, but not within the serum, is increased after the stress of high- and low-dose nonlethal radiation,6 and this may occur during other situations that stress the hematopoietic system. In such cases, proteins that are N-terminally modified or have greater affinity for the receptors of interest would be of clinical use, as is the case for GLP-1 and extendin-4, and perhaps for Neupogen, which has a methione at the N-terminus that shifted the amino acid sequences away from that of a DPP4-truncatable site. Extendin-4 is a homologous protein of GLP-1, found in 1992 and isolated from the saliva of a Gila monster, that has agonist activity with GLP-1.58,59 Extendin-4 was used as the basis for development of exenatide (Byetta), a synthetic drug that is unable to be truncated by DPP4, has higher affinity for the GLP-1 receptor than that of full-length GLP-1, and is currently used clinically in patients with type 2 diabetes with the same glucose regulation and insulinotropic effects as GLP-1, but without potential for DPP4 inhibition.60
Other models exist such that factors without truncation sites may regulate factors with truncation sites by altering their expression, or function, and proteins with DPP4 truncation sites may regulate expression of proteins with or without truncation sites (Figure 2B). Several proteins without putative DPP4 truncation sites regulate proteins with DPP4 sites and therefore have the potential to alter multiple cell types and processes as well as potentially increasing or decreasing cellular activity/expression of DPP4/CD26. TGF-β1 does not have a DPP4 truncation site but regulates IGF-1 in mesenchymal stromal cells and osteoblasts, resulting in suppression of osteoblast differentiation.61 TGF-β1 increases expression of VEGF.62,63 TGF-β1 and IL-13 have been shown in the context of inflammation to act synergistically to increase the expression of CCL11.64 Of note, IGF-1, VEGF, IL-13, and CCL11 have putative DPP4 truncation sites (Tables 1 and 2), suggesting that a multitude of interactions between factors with and without DPP4 truncation sites as well as DPP4 truncation of these factors may be relevant in physiological models and chronic conditions such as inflammation. Also to be considered are effects of DPP4-truncated proteins on the production/release of other proteins (eg, IL-1, IL-6, IL-8, CSFs, etc), as well as the potential synergistic or suppressive effects that may be inherent when combinations of proteins are working in collaboration to elicit biological effects. As some examples of this, regulation of HSCs and hematopoiesis relies on interactions with cells of the bone marrow microenvironment, which produce cytokines, chemokines, and other growth-modulating factors, some of which have putative DPP4 truncation sites or are regulated by proteins that have such truncation sites. Parathyroid hormone (PTH) and osteopontin (OPN) regulate Notch and Jagged levels, and subsequently the number of osteoblasts in the niche, resulting in alterations in HSC numbers.65-67 In addition to the role that osteoblasts play in the maintenance of HSCs, they also play a critical role in B lymphopoiesis via CXCL12, IL-7, stem cell factor (SCF), IL-6, and IL-3, and for erythropoiesis via HIF pathway modulation of EPO.68,69 With new insight into the proteins that can potentially be cleaved by DPP4 (Tables 1,Table 2-3), this raises interesting speculation about what we really know, or do not know, about niche regulation and how full length vs truncated cytokines may regulate both numbers and lineage distribution of the supportive and hematopoietic cells in the niche. For example, PTH, which does not have a truncation site, is a negative regulator of DPP4 and allows for stem-cell mobilization from the bone marrow to sites of ischemic injury.70 Proteins such as CXCL12, OPN, EPO, IL-6, and IL-3, which are important in niche regulation potentially via PTH, have putative DPP4 truncation sites (Tables 1,Table 2-3), whereas others (Notch-1, Jagged-1, IL-7) do not. Thus, some of the factors regulating the niche may be acting via their truncated, not full-length, forms. For example, OPN is a bone matrix protein expressed by both osteoblasts and osteoclasts and is contextual in its regulation of HSCs, with publications suggesting both negative and positive regulation of HSCs.66,67 OPN is N-terminally cleaved by thrombin, which results in its chemoattractant properties,71 suggesting that it may also be regulated by N-terminal cleavage by DPP4, leading to alterations in function. Recent studies, investigating stem cell fate in the vascular niche have elucidated the importance of SCF and pleiotropin, which do not have DPP4 truncation sites. When SCF was deleted from endothelial cells or leptin receptor–expressing perivascular stromal cells, HSC numbers were diminished, and after myelosuppression, bone marrow HSC were decreased and hematopoietic regeneration blunted in PTH (−/−) mice.72,73 Of interest here is that leptin has a putative DPP4 truncation site (Table 2). Endothelial cells are important for hematopoiesis.74,75 Pleiotropin induces transdifferentiation of monocytes into functional endothelial cells and is itself regulated by platelet-derived growth factor (PDGF) and nerve growth factor (NGF),76 both of which have a putative DPP4 truncation site (Tables 2 and 3). In addition, endothelial progenitor cells (EPCs) are mobilized from the marrow to sites of ischemia, tumors, and so on, via G-CSF, GM-CSF, FGF, OPN, CCL5, CCL2, and CXCL1277,78 (all of which contain putative DPP4 truncation sites). EPCs act directly to form vessels or have an indirect function by secreting angiogenic factors. CXCL12 (DPP4 truncation site)-induced chemotaxis of EPCs can be enhanced with IL-3 (DPP4 truncation site), and EPC function can be modulated by angiopoietin-2, VEGF, CCL11, and CXCL1 (which have putative DPP4 truncation sites), which can lead to neovascularization, angiogenesis, and repair from ischemic injury,79,80 processes that involve blood cells.
Conclusions
A large, but not yet all inclusive, number of biologically active proteins with putative truncation sites for DPP4 has been identified, presenting many unanswered questions regarding how this ubiquitous enzyme may modulate many different hematopoietic and other cell functions through its effects on different cytokines, chemokines, and other proteins.81 In addition, it is important that the protein sequences in the databases, with putative DPP4 truncation sites, be checked frequently to ensure that the sequences have not been updated. For example, the sequence for TGF-β originally showed a DPP4 truncation site; however, the sequence has recently been modified and no longer shows a DPP4 site. Ultimately, biochemical and biological (in vitro and in vivo) studies are needed to verify for each protein whether the putative DPP4 truncation sites are true truncation sites, especially with regard to differing alanine, proline, serine, or other potential DPP4 truncation sites at the N-terminus of each molecule. If so, it is important to determine whether the truncated form is or is not changed in activity from that of its full-length form and whether the truncated form influences the activity of the full-length form, and if so, what that influence is. Such efforts could be extremely rewarding in terms of increased understanding of the in vitro and in vivo regulation of many stem, progenitor, and more mature hematopoietic and other types of cells. This information could have therapeutic relevance.
Acknowledgments
H.A.O. is supported by a postdoctoral stipend to H.E.B. from the National Institutes of Health (T32 DK07519). The laboratory work of these investigators is supported by Public Health Service Grants from the National Institutes of Health: R01 HL056416, R01 HL67384, R01 HL112669, P01 DK090948 (H.E.B.)
Authorship
Contribution: All authors were involved in the concepts, writing, and editing of this study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hal E. Broxmeyer, Department of Microbiology and Immunology, Indiana University School of Medicine, 950 West Walnut St, R2-302, Indianapolis, IN 46202-5181; e-mail: hbroxmey@iupui.edu.
References
Author notes
X.O. and H.A.O. contributed equally to this study.