Key Points
PTEN is downregulated in Sézary syndrome by different mechanisms, mostly by gene deletions and microRNAs.
PTEN deficiency activates AKT in skin resident but not circulating Sezary cells.
Abstract
Sézary syndrome (SS) is an incurable leukemic variant of cutaneous T-cell lymphoma characterized by recurrent chromosomal alterations, among which, chromosome 10q deletion is very frequent. In this study, we investigated the PTEN status, on locus 10q23, in 44 SS patients; our findings show that PTEN is deleted in 36% of SS cases, whereas PTEN downregulation is observed in almost all of the samples evaluated by quantitative reverse-transcriptase polymerase chain reaction and Western blotting analysis. Neither DNA sequence mutation nor promoter hypermethylation were found at the PTEN locus, but we demonstrate that PTEN level can be also reduced by a group of miRs previously found upregulated and of prognostic relevance in SS; particularly, miR-21, miR-106b, and miR-486 were able to control PTEN abundance either in vitro or in vivo. Finally, because reduced PTEN activates the PI3/AKT-mediated pathway of cell growth and survival, we demonstrate that PTEN deficiency is associated with activated AKT in skin resident but not circulating SS cells, suggesting that the cutaneous milieu may strongly contribute to the SS cell growth. To our knowledge, this is the first study fully exploring the PTEN status in a large cohort of SS patients, unveiling potential elements of clinical utility in this malignancy.
Introduction
Cutaneous T-cell lymphoma (CTCL) represents a malignant expansion of CD4+CD45RO+ memory lymphocytes that show a strong propensity for the skin. The most aggressive entity of CTCLs is Sézary syndrome (SS), a leukemic variant characterized by the presence of malignant lymphocytes named Sezary cells in the skin, lymph nodes, and peripheral blood.1 Despite major therapeutic advances during the last few years,2 SS remains incurable and discovery of new compounds will require a better understanding of the molecular mechanism driving SS cell survival and proliferation, as well as more specific targeting strategies. Recently, studies using both high-density comparative genomic hybridization (CGH) and single nucleotide polymorphism (SNP) array technologies demonstrated that SS is characterized by specific chromosomal gains and deletions. Among these, loss of chromosome 10q, including the PTEN locus at 10q23, appears to be particularly common in these individuals.3-5
PTEN is a nonredundant lipid phosphatase whose main role is to antagonize the PI3K signaling,6 a relevant pathway for cancer development with several members representing potential targets for therapeutics.7 PTEN is one of the most frequently lost or mutated genes in human cancers,8 including T-cell malignancies as T-cell acute lymphoblastic leukemia9 and CTCL, as already demonstrated by loss of heterozygosity (LOH) found within the PTEN locus of mycosis fungoides (MF).10 PTEN plays a critical role in T-cell development; in fact, conditional loss of PTEN in thymocytes leads to T-cell lymphomas.11,12 Conversely, PTEN deletion in mature T-cell subsets induces autoimmunity, increased cytokine releasing, and, in general, a PI3K/AKT hyperactivation through T-cell receptor also in absence of CD28 co-stimulation, thus indicating that PTEN imposes stringent environmental signals for an appropriate T-cell activation.13
In addition to the mutations and genetic alterations, PTEN abundance appears to be finely regulated at the transcriptional/translational level by microRNAs (miRNAs),6 competitive endogenous pseudogene RNAs,14 and promoter hypermethylation,15 and at the posttranslational level by phosphorylation and ubiquitination.15 It has recently been demonstrated that programmed death-1, a gene also found overexpressed in SS,16 regulates the PTEN phosphatase activity through these last regulatory mechanisms, thereby promoting or inhibiting the PI3K/AKT signaling.17
Methods
Patients, tumor, normal cells isolation, and genomic DNA extraction
SS patients were enrolled in this study approved by the Ethical Committee of the Istituto Dermopatico dell’Immacolata, and informed consent was obtained in accordance with the Declaration of Helsinki. Diagnosis of SS was based on the described criteria.20 The major clinical and immunologic characteristics are reported in supplemental Table 1 on the Blood Web site.
Tumor, normal cell isolation, and genomic DNA extraction was performed as described elsewhere.5
Reagents
Hut78 (TIB161), its clonal derivative H9 (HTB 176), and HH (CRL2105) cell lines from American Type Culture Collection (ATCC, Manassas, VA) established from the peripheral blood of CTCL patients were grown in complete RPMI 1640 supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO)
Primers for quantitative RT-PCR (qRT-PCR) and DNA methylation analysis were designed using Primer3 Input 0.4.0 and synthesized by Eurofins MWG Operon. Primer sequences are shown in supplemental Table 2.
DNA methylation analysis
In total, 200 ng of genomic DNA from each sample was treated with a Methylamp DNA modification sample kit (Epigentek, Brooklyn, NY) to convert the unmethylated cytosine to uracil according to the manufacturer’s instructions. CpGenome Universal Methylated DNA was included as a methylated genomic DNA control (Millipore, Billerica, MA). Bisulfite-treated DNA was analyzed by PCR with methylated or unmethylated specific primers designed outside the common region between PTEN and the homolog pseudogene PTENP1.8 PCR products were separated by 2% agarose gel electrophoresis and visualized with ethidium bromide staining.
Genomic RT-PCR analysis
Genomic qRT-PCR was performed on the DNA of 27 SS samples using the GoTaq qPCR Master MIX (Promega) on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems). We used the method described by Hoebeeck et al.21 Briefly, 2 endogenous controls (SDC4, COMT) not affected by copy-number changes were used for normalization of qRT-PCR data; the fold change (FC) values were calculated using an Affymetrix genomic DNA reference diploid sample by the standard curve methods.
PTEN copy-number analysis
In total, 500 ng of genomic DNA was input for the Genome-Wide Human SNP6.0 Array (Affymetrix, Santa Clara, CA), according to the manufacturer’s specifications. CEL files were converted by Genotyping Console Software to perform copy number and LOH analysis. Gains and losses were summarized by the segment reporting tool and displayed in the genotyping console browser. This browser aligns whole-genome to annotated genes and copy-number variations, fluorescence in situ hybridization clones, or customized tracks.
PTEN sequencing
PTEN coding sequence (accession number NM_000314.4) and the 4 regions within 3′UTR containing recognized miR-106b binding sites (2041-2566, 3135-3280, 3261-3419, 3634-3763) were sequenced. Primer pairs for the amplification of each region-of-interest were designed using Ampliseq Designer v.1.2.9 software (Life Technologies) (https://www.ampliseq.com/browse.action). Sequence analysis was conducted using Torrent Suite from Life Technologies. For library preparation, sequencing data analysis, and variants identification, see the supplemental Material and methods.
qRT-PCR
The RNA purification by Trizol was performed according to the manufacturer’s indications (Invitrogen). Mature miRNAs were analyzed using the TaqMan microRNA assay (Applied Biosystems), and Z30 was used as an internal normalized reference.
PTEN and PTENP1 expression analysis was performed with a SYBR Green I dye chemistry and AmpliTaq Gold DNA Polymerase on an ABI PRISM 7000 machine (Applied Biosystems).
Quantitative gene expression analysis was performed by the ΔCT method as described previously.22
Cell-line transfections
Pre-miRNAs (Ambion) and LNA knockdown oligos (Exiqon) were transfected to H9 cells at the final concentration of 100 nM by a Siport transfection reagent (Ambion) following the manufacturer’s instructions.
Western blotting
Cells were lysed in a RIPA buffer supplemented with a phosphatase and protease inhibitor cocktail (Roche Diagnostic, Indianapolis, IN). Proteins were quantified using BCA protein assay (Bio-Rad, Hercules, CA) and resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and probed using standard techniques.23 Membranes were probed with specific primary antibodies for PTEN used 1:500, AKT used 1:2000, phospho-AKT used 1:500 (Cell Signaling), and β-actin diluted 1:1000 (clone AC-40; Sigma-Aldrich) used as an internal standard for loading. Immunodetection was carried out by using appropriate horseradish peroxidase–linked secondary antibodies and enhanced chemiluminescence detection reagents (GE Healthcare, Amersham Biosciences, Little Chalfont, United Kingdom).)
Immunohistochemistry
Paraffin-embedded skin tissues from SS/MF patients were selected from the files of IDI Pathology. Specimens were classified according to the EORTC classification.1 PTEN used 1:100 (clone 6H2.1) and pAKT used 1:15 (clone 14-5) (Dako, Fort Collins, CO) were retrieved in pH9 and high pH buffer (Dako), respectively, by incubation in a bath at 96°C for 30 minutes. Samples were then revealed by the streptavidin-biotin peroxidized labeling method and DAB staining. Sections were counterstained with hematoxylin and mounted with aqueous mounting media.
Statistics
Statistical analyses were carried out with the software GraphPad PRISM 5 (GraphPad Software Inc., La Jolla, CA). Differences were evaluated with 2-tailed Student test and a Pearson correlation test. P ≤ .05 was considered significant.
Results
PTEN is frequently deleted but not mutated in SS
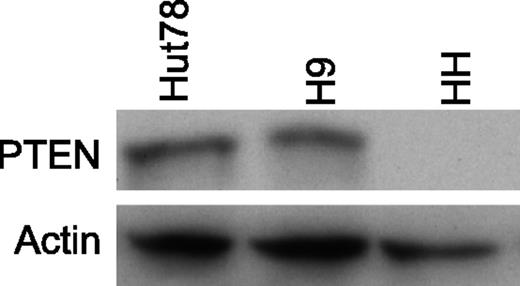
The Affymetrix 10K and SNP6.0 arrays were used to investigate the copy number (CN) status of the PTEN locus in 68 samples derived from 44 SS patients and 3 cell lines (Table 1). The SS group was composed of 27 SS cases available of 28 cases previously investigated from our laboratory with the SNP-10K array and oligonucleotide–comparative genomic hybridization array (aCGH).5 The clinical features of the new enrolled 17 SS are shown in supplemental Table 1. The monoallelic loss of PTEN has been observed in 16 of 44 SS patients (36%); in 3 cases (32, 39, and 53), PTEN was heterozygously deleted in both the T1 and T2 follow-up samples, whereas in 2 cases (61 and 64), the loss became homozygous in the second follow-up, suggesting that this aberration may be correlated with the tumor progression. A biallelic deletion of PTEN was also observed in the HH cell line and confirmed by Western blotting analysis (Figure 1). The SNP mapping arrays also enable unravel uniparental disomy (UPD) events (ie, regions of LOH within a normal diploid CN state); these abnormalities may be explained by a loss of genetic material followed by a re-duplication of the remaining alleles. We found that patient SS28 exhibited UPD encompassing the PTEN locus.
PTEN and PTENP1 genomic status and promoter methylation analysis
| Copy number status of . | PTEN (10q23) and . | PTENP1 (9p13.3) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID . | Diagnosis . | Mapping array . | PTEN locus . | PTEN LOH status . | UPD . | PTEN promoter methylation . | Genomic qRT-PCR . | PTENP1 locus . | PTENP1 LOH status . | PTENP1 UPD . |
| 02 T | SS | 10K | ML | 1 | 0 | ND | ND | WT | 0 | 0 |
| 05 T | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 08 T1 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 08 T2 | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 11 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 15 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 22 T | SS | 10K/aCGH | ML | 1 | 0 | ND | ND | ML | 1 | 0 |
| 23 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | ND | WT | 0 | 0 |
| 23 T2 | SS | SNP 6.0 | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 23 T3 | SS | SNP 6.0 | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 25 T | SS | 10K/aCGH | ML | 0 | 0 | ND | ND | WT | 0 | 0 |
| 27 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 28 T | SS | 10K/aCGH | WT | 1 | 1 | U | 0.61 | WT | 0 | 0 |
| 30 T1 | SS | 10K | WT | 0 | 0 | U | 1.13 | WT | 0 | 0 |
| 30 T2 | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 32 T1 | SS | 10K/aCGH | ML | 1 | 0 | U | ND | WT | 0 | 0 |
| 32 T2 | SS | SNP 6.0 | ML | 1 | 0 | ND | 0.67 | WT | 0 | 0 |
| 33 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 34 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 35 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 36 T | SS | 10K/aCGH | ML | 0 | 0 | U | 0.69 | WT | 0 | 0 |
| 37 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 38 T1 | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 38 T2 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 38 T3 | SS | SNP 6.0 | WT | 0 | 0 | ND | 1,01 | WT | 0 | 0 |
| 39 T1 | SS | 10K/aCGH | ML | 1 | 0 | U | 0.43 | WT | 0 | 0 |
| 39 T2 | SS | SNP 6.0 | ML | 0 | 0 | ND | ND | WT | 0 | 0 |
| 40 T1 | SS | 10K | WT | 0 | 0 | U | 0.86 | WT | 0 | 0 |
| 40 T2 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 40 T3f | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 41 T1 | SS | 10K | WT | 0 | 0 | U | 1.19 | WT | 0 | 0 |
| 41 T2 | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 41 T3 | SS | SNP 6.0 | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 42 T1 | SS | 10K/aCGH | ML | 0 | 0 | ND | ND | WT | 0 | 0 |
| 43 T1 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 43 T2 | SS | 10K/aCGH | WT | 0 | 0 | U | 0.85 | WT | 0 | 0 |
| 43 T3 | SS | SNP 6.0 | WT | 0 | 0 | U | 1.09 | WT | 0 | 0 |
| 44 T1 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 45 T1 | SS | 10K/aCGH | WT | 0 | 0 | U | 1.07 | WT | 0 | 0 |
| 45 T2 | SS | SNP 6.0 | WT | 0 | 0 | ND | 0.72 | WT | 0 | 0 |
| 46 T1 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 48 T1 | SS | 10K/aCGH | ML | 1 | 0 | ND | 0.45 | ML | 1 | 0 |
| 49 T1 | SS | 10K/aCGH | ML | 0 | 0 | ND | ND | WT | 0 | 0 |
| 50 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | ND | WT | 0 | 0 |
| 50 T2 | SS | 10K | WT | 0 | 0 | ND | 1.06 | WT | 0 | 0 |
| 51 T1 | SS | 10K/aCGH | WT | 0 | 0 | U | 1.20 | WT | 0 | 0 |
| 53 T1 | SS | SNP 6.0 | ML | 0 | 0 | U | ND | ML | 0 | 0 |
| 53 T2 | SS | SNP 6.0 | ML | 0 | 0 | ND | 0.49 | ML | 0 | 0 |
| 55 T1 | SS | 10K | ML | 1 | 0 | U | 0.50 | WT | 0 | 0 |
| 57 T1 | SS | SNP 6.0 | ML | 1 | 0 | ND | 0.30 | WT | 0 | 0 |
| 58 T1 | SS | 10K | ML | 0 | 0 | U | 0.44 | WT | 0 | 0 |
| 60 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | ND | WT | 0 | 0 |
| 60 T2 | SS | SNP 6.0 | WT | 0 | 0 | U | 1.20 | WT | 0 | 0 |
| 61 T1 | SS | 10K | ML | 0 | 0 | ND | ND | WT | 0 | 0 |
| 61 T2 | SS | 10K | BL | 0 | 0 | PCR failed | 0.09 | WT | 0 | 0 |
| 62 T1 | SS | 10K | WT | 0 | 0 | U | 1.17 | WT | 0 | 0 |
| 63 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | 1.06 | WT | 0 | 0 |
| 63 T2 | SS | SNP 6.0 | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 64 T1 | SS | SNP 6.0 | ML | 0 | 0 | U | 0.51 | WT | 0 | 0 |
| 64 T2 | SS | SNP 6.0 | BL | 0 | 0 | ND | 0.26 | WT | 0 | 0 |
| 65 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | 1.20 | WT | 0 | 0 |
| 66 T1 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 67 T1 | SS | SNP 6.0 | ML | 0 | 0 | U | 0.47 | WT | 0 | 0 |
| 69 T1 | SS | SNP 6.0 | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 70 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | ND | WT | 0 | 0 |
| Hut78 | Cell line | SNP 6.0 | ML | 1 | 0 | U | ND | ML | 1 | 0 |
| H9 | Cell line | SNP 6.0 | ML | 1 | 0 | U | ND | ML | 1 | 0 |
| HH | Cell line | SNP 6.0 | BL | 0 | 0 | U | ND | WT | 0 | 0 |
| Copy number status of . | PTEN (10q23) and . | PTENP1 (9p13.3) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID . | Diagnosis . | Mapping array . | PTEN locus . | PTEN LOH status . | UPD . | PTEN promoter methylation . | Genomic qRT-PCR . | PTENP1 locus . | PTENP1 LOH status . | PTENP1 UPD . |
| 02 T | SS | 10K | ML | 1 | 0 | ND | ND | WT | 0 | 0 |
| 05 T | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 08 T1 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 08 T2 | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 11 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 15 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 22 T | SS | 10K/aCGH | ML | 1 | 0 | ND | ND | ML | 1 | 0 |
| 23 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | ND | WT | 0 | 0 |
| 23 T2 | SS | SNP 6.0 | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 23 T3 | SS | SNP 6.0 | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 25 T | SS | 10K/aCGH | ML | 0 | 0 | ND | ND | WT | 0 | 0 |
| 27 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 28 T | SS | 10K/aCGH | WT | 1 | 1 | U | 0.61 | WT | 0 | 0 |
| 30 T1 | SS | 10K | WT | 0 | 0 | U | 1.13 | WT | 0 | 0 |
| 30 T2 | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 32 T1 | SS | 10K/aCGH | ML | 1 | 0 | U | ND | WT | 0 | 0 |
| 32 T2 | SS | SNP 6.0 | ML | 1 | 0 | ND | 0.67 | WT | 0 | 0 |
| 33 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 34 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 35 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 36 T | SS | 10K/aCGH | ML | 0 | 0 | U | 0.69 | WT | 0 | 0 |
| 37 T | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 38 T1 | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 38 T2 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 38 T3 | SS | SNP 6.0 | WT | 0 | 0 | ND | 1,01 | WT | 0 | 0 |
| 39 T1 | SS | 10K/aCGH | ML | 1 | 0 | U | 0.43 | WT | 0 | 0 |
| 39 T2 | SS | SNP 6.0 | ML | 0 | 0 | ND | ND | WT | 0 | 0 |
| 40 T1 | SS | 10K | WT | 0 | 0 | U | 0.86 | WT | 0 | 0 |
| 40 T2 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 40 T3f | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 41 T1 | SS | 10K | WT | 0 | 0 | U | 1.19 | WT | 0 | 0 |
| 41 T2 | SS | 10K/aCGH | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 41 T3 | SS | SNP 6.0 | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 42 T1 | SS | 10K/aCGH | ML | 0 | 0 | ND | ND | WT | 0 | 0 |
| 43 T1 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 43 T2 | SS | 10K/aCGH | WT | 0 | 0 | U | 0.85 | WT | 0 | 0 |
| 43 T3 | SS | SNP 6.0 | WT | 0 | 0 | U | 1.09 | WT | 0 | 0 |
| 44 T1 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 45 T1 | SS | 10K/aCGH | WT | 0 | 0 | U | 1.07 | WT | 0 | 0 |
| 45 T2 | SS | SNP 6.0 | WT | 0 | 0 | ND | 0.72 | WT | 0 | 0 |
| 46 T1 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 48 T1 | SS | 10K/aCGH | ML | 1 | 0 | ND | 0.45 | ML | 1 | 0 |
| 49 T1 | SS | 10K/aCGH | ML | 0 | 0 | ND | ND | WT | 0 | 0 |
| 50 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | ND | WT | 0 | 0 |
| 50 T2 | SS | 10K | WT | 0 | 0 | ND | 1.06 | WT | 0 | 0 |
| 51 T1 | SS | 10K/aCGH | WT | 0 | 0 | U | 1.20 | WT | 0 | 0 |
| 53 T1 | SS | SNP 6.0 | ML | 0 | 0 | U | ND | ML | 0 | 0 |
| 53 T2 | SS | SNP 6.0 | ML | 0 | 0 | ND | 0.49 | ML | 0 | 0 |
| 55 T1 | SS | 10K | ML | 1 | 0 | U | 0.50 | WT | 0 | 0 |
| 57 T1 | SS | SNP 6.0 | ML | 1 | 0 | ND | 0.30 | WT | 0 | 0 |
| 58 T1 | SS | 10K | ML | 0 | 0 | U | 0.44 | WT | 0 | 0 |
| 60 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | ND | WT | 0 | 0 |
| 60 T2 | SS | SNP 6.0 | WT | 0 | 0 | U | 1.20 | WT | 0 | 0 |
| 61 T1 | SS | 10K | ML | 0 | 0 | ND | ND | WT | 0 | 0 |
| 61 T2 | SS | 10K | BL | 0 | 0 | PCR failed | 0.09 | WT | 0 | 0 |
| 62 T1 | SS | 10K | WT | 0 | 0 | U | 1.17 | WT | 0 | 0 |
| 63 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | 1.06 | WT | 0 | 0 |
| 63 T2 | SS | SNP 6.0 | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 64 T1 | SS | SNP 6.0 | ML | 0 | 0 | U | 0.51 | WT | 0 | 0 |
| 64 T2 | SS | SNP 6.0 | BL | 0 | 0 | ND | 0.26 | WT | 0 | 0 |
| 65 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | 1.20 | WT | 0 | 0 |
| 66 T1 | SS | 10K | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 67 T1 | SS | SNP 6.0 | ML | 0 | 0 | U | 0.47 | WT | 0 | 0 |
| 69 T1 | SS | SNP 6.0 | WT | 0 | 0 | ND | ND | WT | 0 | 0 |
| 70 T1 | SS | SNP 6.0 | WT | 0 | 0 | U | ND | WT | 0 | 0 |
| Hut78 | Cell line | SNP 6.0 | ML | 1 | 0 | U | ND | ML | 1 | 0 |
| H9 | Cell line | SNP 6.0 | ML | 1 | 0 | U | ND | ML | 1 | 0 |
| HH | Cell line | SNP 6.0 | BL | 0 | 0 | U | ND | WT | 0 | 0 |
0, absent; 1, present; BL, biallelic loss; M, methylated; ML, monoallelic loss; ND, not determined; U, unmethylated; WT, diploid.
Bold denotes SS patients derived from the data set already published in Caprini et al.5
PTEN protein in CTCL cell lines. Western blotting analysis revealing the PTEN level in CTCL cell lines. No PTEN protein is detected in HH cells, in agreement with their PTEN−/− status.
PTEN protein in CTCL cell lines. Western blotting analysis revealing the PTEN level in CTCL cell lines. No PTEN protein is detected in HH cells, in agreement with their PTEN−/− status.
Altogether, we found a hetero/homozygous loss of PTEN in 16 of 44 SS cases (36%) and in all of the cell lines we analyzed, whereas 1 SS patient showed a UPD event.
We validated the SNP arrays performing genomic qRT-PCR on 27 SS samples (see Methods for details). Using the relative standard curve methods, we obtained FC values ranging from 0.72 to 1.2 for SS patients, displaying a normal PTEN diploid status (n = 14); FC values <0.72 were observed in all heterozygous PTEN-deleted samples (n = 11), whereas for SS61 T2 and SS64 T2, showing a homozygous deletion of PTEN, we calculated a FC of 0.09 and 0.26, respectively (Table 1). CTCL cell lines were not validated because endogenous control genes used to normalize genomic qRT-PCR were affected by chromosomal imbalances.
Because PTEN is frequently also inactivated by nucleotide mutation, we decide to perform a target re-sequencing of PTEN coding exons. Twenty-five samples (21 patients plus 4 follow-ups from SS60, SS63, SS50, and SS23) were subjected to deep-sequence analysis, which did not reveal any mutation, indicating that this mechanism does not occur in SS. Also, the sequence of SS28 carrying UPD was wild-type, indicating that this genetic alteration does not affect the gene sequence.
In addition to genetic deletion, PTEN can be transcriptionally silenced by the hypermethylation of the promoter region. Methylation-specific PCR performed on 25 patients and 3 cell lines led us to exclude that PTEN is inactivated by this mechanism in SS (Table 1).
PTEN abundance can be modulated by several miRNAs.24 Recently the PTEN pseudogene, PTENP1 showing an extensive sequence similarity to PTEN, has been described as a modulator of the interaction between PTEN, messenger RNA (mRNA), and miRNAs. By this mechanism, it can protect PTEN from miRNA downregulation and exert a tumor suppressor role.25 The PTENP1 locus at chromosome 9p13.3 is selectively lost in human cancer.25 In contrast to other malignancies, we observed a PTENP1deletion only in 3 of 44 patients (2%) and in the Hut78 and H9 cell lines (Table 1), thus indicating that PTENP1 is not under a strong genomic selective pressure in SS.
PTEN is commonly downregulated in SS
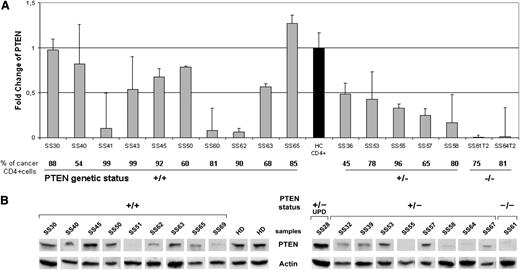
To establish whether the genomic loss of PTEN was also reflected by the loss of PTEN expression, we analyzed the mRNA level in enriched neoplastic CD4+cells of 2 patients with a homozygous deletion (SS61T2 and SS64 T2), 5 patients with a heterozygous deletion (SS36, SS53, SS55, SS57, SS58), and 10 patients with a wild-type PTEN status (SS30, SS40, SS41, SS43, SS45, SS50, SS60, SS62, SS63, SS65) vs sorted CD3+/CD4+ peripheral lymphocytes from 5 healthy donors (HDs). qRT-PCR results showed a significant PTEN downregulation in all SS individuals analyzed (FC mean value, 0.51) vs HDs (P = .038). Moreover, we observed a decreased PTEN expression in SS patients showing a PTEN hetero (+/−) and homozygous (−/−) deletion (FC mean value, 0.24) when compared with those associated to a PTEN wild-type (+/+) status (P = .044), indicating that the PTEN mRNA level is dependent on its genomic status. However, we observed that SS41, SS60, and SS62, with a PTEN+/+ genotype, expressed a very low mRNA level, suggesting that additional PTEN-inactivating mechanism(s) may act at the transcriptional level (Figure 2A).
PTEN levels measured in SS patients. (A) qRT-PCR analysis of PTEN performed in 17 SS patients. The PTEN mRNA level was calculated for each SS patient relatively to a reference pool consisting of 5 healthy controls (black bar). The bottom portion indicates the percentage of clonal TCR vβ+ cells within the CD4+ fraction and the PTEN status represented as wild type (+/+), heterozygous deletion (+/−), and homozygous deletion (−/−). Samples were measured in 2 independent experiments performed in triplicate. (B) Western blotting analysis of PTEN in the purified CD4+ cells of 19 SS individuals grouped by PTEN +/+, +/−, and −/−, and UPD genomic status. Two representative HDs of the 5 analyzed are shown in the figure. Because of the timing of patient recruitment, the gels were run at separate times and the lanes were composed for the figure.
PTEN levels measured in SS patients. (A) qRT-PCR analysis of PTEN performed in 17 SS patients. The PTEN mRNA level was calculated for each SS patient relatively to a reference pool consisting of 5 healthy controls (black bar). The bottom portion indicates the percentage of clonal TCR vβ+ cells within the CD4+ fraction and the PTEN status represented as wild type (+/+), heterozygous deletion (+/−), and homozygous deletion (−/−). Samples were measured in 2 independent experiments performed in triplicate. (B) Western blotting analysis of PTEN in the purified CD4+ cells of 19 SS individuals grouped by PTEN +/+, +/−, and −/−, and UPD genomic status. Two representative HDs of the 5 analyzed are shown in the figure. Because of the timing of patient recruitment, the gels were run at separate times and the lanes were composed for the figure.
Next, we evaluated the PTEN protein level by Western blotting analysis of purified CD4+ neopastic cells from 19 SS patients and 5 HDs. The results revealed that SS samples showed a significantly decreased PTEN protein level of 0.17 ± 0.15 compared with HDs (0.4 ± 0.01) (P = .04). Moreover, in accordance with mRNA data, we found that PTEN abundance is significantly influenced by the gene dosage, as demonstrated by the mean of PTEN densitometric values of 0.2 ± 0.13, observed in PTEN+/+ cases (n = 9) vs 0.09 ± 0.08 values associated with PTEN+/− and PTEN−/− samples (n = 9) (P = .02), whereas a value of 0.59 was associated to SS28 coherently with a functional PTEN diploid status potentially restored by a UPD event. As expected, no PTEN protein was detected in the SS61 T2 sample with homozygous deletion of PTEN (Figure 2B). Overall, these results indicate that mRNA level is consistent with the PTEN protein abundance observed.
PTEN is regulated in vitro by multiple miRNAs and miR-106b correlates with PTEN expression in vivo
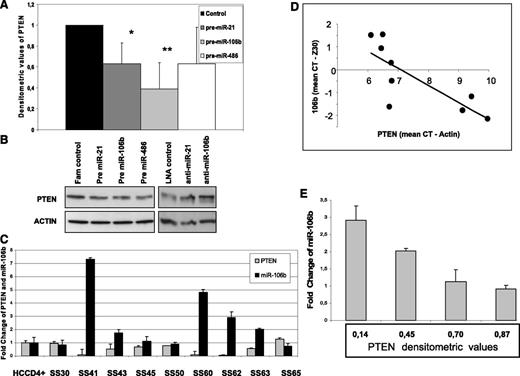
We next asked whether miR-21, miR-106b, miR-214, and miR-486, all correlated with a worst SS prognosis26 and already validated to target PTEN in many cancers,27-30 were also able to regulate PTEN level in our system. Using a loss or gain of function approach, we first overexpressed them in the H9 cell line in which PTEN protein is well detectable (Figure 1). After 48 hours, we revealed by qRT-PCR an overexpression of all miRNAs investigated (supplemental Figure 1); concurrently, we observed a reduction in the PTEN level of 40%, 60%, and 40% after pre–miR-21 (n = 6), pre–miR-106b (n = 6), and pre–miR-486 (n = 3), respectively (Figure 3A-B), whereas pre–miR-214 did not show a consistent downregulation of PTEN (data not shown). qRT-PCR analysis revealed that only pre–miR-106b was able to reduce about 80% of PTEN transcript, whereas all other miRNAs did not influence but rather enhanced it. This latter observation might be explained by PTENP1 mRNA co-expression that, different from PTEN, is targeted by miR-106b but also by miR-214 and miR-486, preserving PTEN from degradation (supplemental Figure 2). In light of these results, we correlated PTENP1 expression with miRNAs levels and PTEN mRNA also in vivo without finding any significant relationship among these genes (data not shown).
PTEN regulations by miRNAs in SS patients. (A) Densitometric values calculated as PTEN/actin showed a PTEN protein reduction in pre–miR-21, pre–miR-106b, and pre–miR-486 transfected H9 cells vs fluorescein-scrambled oligo used as control. Data are given as mean ± SD from 9 independent experiments (*P = .02, **P = .006). (B) Western blotting analysis performed in pre-miRNAs and anti-miRNAs transfected H9 cells showed a reduction or increase in PTEN protein level, respectively. Two of 9 representative electrophoretic patterns are shown. (C-D) A significant inverse relationship between PTEN and miR-106b was observed comparing their mRNA levels measured by qRT-PCR in 9 SS patients with a +/+ PTEN status (R= −0.79, P = .01 for Pearson correlation test). (E) An inverse correlation between PTEN protein levels and miR-106b was also observed in 4 samples simultaneously analyzed by Western blotting and qRT-PCR (R= −0.96, P = .04 for Pearson correlation test).
PTEN regulations by miRNAs in SS patients. (A) Densitometric values calculated as PTEN/actin showed a PTEN protein reduction in pre–miR-21, pre–miR-106b, and pre–miR-486 transfected H9 cells vs fluorescein-scrambled oligo used as control. Data are given as mean ± SD from 9 independent experiments (*P = .02, **P = .006). (B) Western blotting analysis performed in pre-miRNAs and anti-miRNAs transfected H9 cells showed a reduction or increase in PTEN protein level, respectively. Two of 9 representative electrophoretic patterns are shown. (C-D) A significant inverse relationship between PTEN and miR-106b was observed comparing their mRNA levels measured by qRT-PCR in 9 SS patients with a +/+ PTEN status (R= −0.79, P = .01 for Pearson correlation test). (E) An inverse correlation between PTEN protein levels and miR-106b was also observed in 4 samples simultaneously analyzed by Western blotting and qRT-PCR (R= −0.96, P = .04 for Pearson correlation test).
Because H9 express high levels of miR-21 and miR-106b (supplemental Figure 1), we next tested whether knockdown of these miRNAs was able to enhance PTEN expression. Western blotting analysis of anti–miR-21 (n = 3) and anti–miR-106b (n = 3) transfectants revealed an increase in the PTEN level of 1.6 ± 0.9 and 1.8 ± 0.25 FC, respectively, confirming the ability of these miRNAs to modulate in vitro PTEN in the CTCL cell line (Figure 3B). Finally, because miRNAs can affect gene expression via translational inhibition and/or the target mRNA degradation mechanism, we investigated the correlation between miRNAs and PTEN mRNA levels in sorted CD4+ neoplastic cells. To better evaluate the degradation effect of miRNAs on PTEN transcripts, excluding an mRNA reduction because of mono or biallelic deletion of the PTEN gene, we focused our attention on 9 available SS patients associated with a PTEN wild-type status. In agreement with the miR-106b ability to target PTEN mRNA in vitro, we found a significant inverse relationship between miR-106b and PTEN mRNA levels (R = −0.79, P = .01 Pearson correlation test) (Figure 4C-D); 4 samples, also analyzed for protein expression, confirmed this result (R = −0.96, P = .04) (Figure 4E).
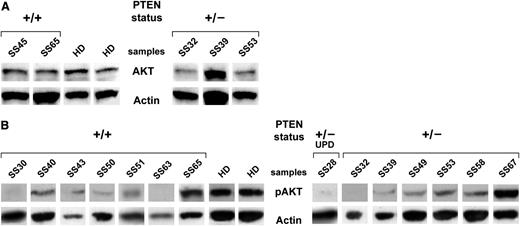
Basal and phospho-AKT levels measured in SS patients. (A) Western blotting analysis for AKT performed in sorted CD4+ lymphocytes from 5 SS patients. Two representative HDs of 5 patients analyzed are shown. (B) Western blotting analysis for phospho-AKT of sorted CD4+ lymphocytes from 14 SS patients. Two representative HDs of 5 patients analyzed are shown. Because of the timing of patient recruitment, the gels were run at separate times and the lanes were composed for the figure.
Basal and phospho-AKT levels measured in SS patients. (A) Western blotting analysis for AKT performed in sorted CD4+ lymphocytes from 5 SS patients. Two representative HDs of 5 patients analyzed are shown. (B) Western blotting analysis for phospho-AKT of sorted CD4+ lymphocytes from 14 SS patients. Two representative HDs of 5 patients analyzed are shown. Because of the timing of patient recruitment, the gels were run at separate times and the lanes were composed for the figure.
Furthermore, in the PTEN target, deep-sequencing approach, the 4 recognized miR-106b binding regions within 3′UTR were included. The results revealed the transition c.*1123 G>A within the miR-106 target site occurring in patient SS23 T1 and T2 samples. The variation appears in 50% of the sequence reads, suggesting it may represent a rare polymorphism whose biological significance, however, could not be further investigated because of the unavailability of the corresponding RNA.
Hence, these data support the notion that, in addition to genomic losses, PTEN is also downregulated in SS by multiple miRNAs both in vitro and in vivo.
Low level of PTEN does not enhance the AKT kinase activity in circulating SS cells
Having demonstrated the low PTEN abundance in SS cells, we next sought to establish whether this deficiency could activate the PI3/AKT pathway. For this purpose, we evaluated by Western blotting the basal AKT amount of CD4+-sorted cells from 5 SS patients and 5 HDs. Our results demonstrated a slight, although not significant, decrease of AKT protein in SS samples when compared with HDs (2.2 ± 0.95 vs 2.61 ± 0.68) (Figure 4A). To further clarify this aspect, we investigated the phospho-Akt level of purified CD4+ cells from 14 SS patients vs 5 HDs using an antibody that recognizes AKT at Ser473. SS cases showed a significantly low amount of phosphorylated/activated AKT vs control (0.12 ± 0.16 vs 0.39 ± 0.12; P = .003), suggesting that the majority of circulating SS cells are quiescent or low-cycling cells (Figure 4B). The only exception is represented by SS65 and SS67 (showing pAKT values of 0.44 and 0.53, respectively) that seem to have an active canonical survival/proliferation pathway mediated by PI3K/AKT.
PTEN and pAKT are detectable in SS skin biopsies
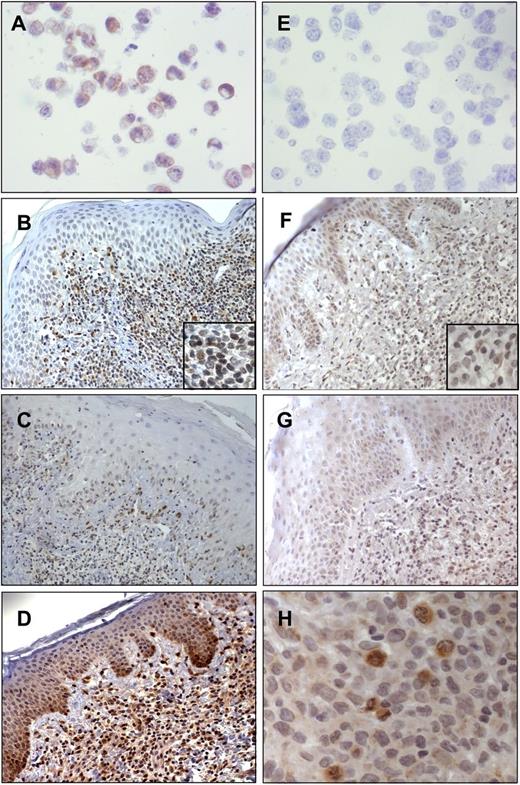
We then evaluated the PTEN expression also in skin-homing SS cells, and using IHC analyses we investigated 24 SS and 10 MF skin biopsies. Paraffin-embedded cell blocks generated from Hut78+/− PTEN and HH−/− PTEN cell lines were used as positive and negative controls, respectively, for PTEN immunoreactivity (Figure 5A,E). In agreement with our biochemical data, we observed a very weak immunostaining, at the cytoplasmic and nuclear levels, in 13 of 24 SS cases (54%) associated with 5% to 30% of malignant lymphocytes (Figure 5B-C), whereas the remaining 11 samples (46%) were consistently negative. Conversely, a moderate nuclear and cytoplasmic PTEN expression was observed in 6 of 10 (60%) of MF coupled with 5% to 30% of neoplastic cells (Figure 5D).
Immunohistochemistry of PTEN and pAKT performed in SS and MF skin biopsies. (A) Hut78 as a positive control for PTEN (original magnification ×40/0.7 numerical aperture); (B-C) PTEN immunoreactivity in SS skin (B, original magnification ×20/0.6 numerical aperture; C, original magnification ×10/0.3 numerical aperture); (D) PTEN immunoreactivity in MF (original magnification ×20/0.6 numerical aperture); (E) HH as a negative control for PTEN (original magnification ×40/0.7 numerical aperture); (F-G) pAKT immunostaining associated with the cytoplasm of keratinocytes and scattered malignant lymphocytes (original magnification ×20/0.6 numerical aperture); and (H) strong pAKT positivity associated with the cytoplasm of SS cells and particularly with actively dividing mitotic cells (original magnification ×40/0.7 numerical aperture). Digital images were acquired with an AxioCam MRc5 camera (Zeiss) and an Axioskop2 microscope (Zeiss).
Immunohistochemistry of PTEN and pAKT performed in SS and MF skin biopsies. (A) Hut78 as a positive control for PTEN (original magnification ×40/0.7 numerical aperture); (B-C) PTEN immunoreactivity in SS skin (B, original magnification ×20/0.6 numerical aperture; C, original magnification ×10/0.3 numerical aperture); (D) PTEN immunoreactivity in MF (original magnification ×20/0.6 numerical aperture); (E) HH as a negative control for PTEN (original magnification ×40/0.7 numerical aperture); (F-G) pAKT immunostaining associated with the cytoplasm of keratinocytes and scattered malignant lymphocytes (original magnification ×20/0.6 numerical aperture); and (H) strong pAKT positivity associated with the cytoplasm of SS cells and particularly with actively dividing mitotic cells (original magnification ×40/0.7 numerical aperture). Digital images were acquired with an AxioCam MRc5 camera (Zeiss) and an Axioskop2 microscope (Zeiss).
Finally, to address the question of whether SS skin resident cells, similarly to circulating neoplastic counterparts, also display an inactive AKT pathway, we analyzed by IHC 21 SS skin as well as psoriasis used as a positive control. In SS cases, pAKT immunoreactivity was detected at the cytoplasmic level, in 21 of 21 (100%) samples, and was associated with 5% to 40% of malignant lymphocytes infiltrating the dermis (Figure 5F-G). Interestingly, we noted that pAKT was strongly expressed in actively duplicating SS cells (Figure 5H). We also found pAKT (Ser473) immunostaining in psoriatic epidermis keratinocytes as already described,31 but we did not detect pAKT staining in healthy skin (supplemental Figure 3).
These findings show a different behavior of SS cells depending on the district localization (Table 2). Overall, these data indicate that, coherently with genomic and expression data, PTEN is downregulated in SS skin resident cells vs MF used as a reference. Furthermore, different from circulating SS cells, we found a moderate percentage of skin-infiltrating neoplastic lymphocytes constantly expressing pAKT in all samples.
Comparison of pAKT expression between circulating and skin resident SS cells concurrently analyzed
| SS patients . | PTEN status . | pAKT amount in circulating SS cells detected by WB* . | pAKT+ SS skin resident cells detected by IHC (%) . |
|---|---|---|---|
| SS32 | ML | 0 | 5% |
| SS39 | ML | 0.01 | 25% |
| SS51 | WT | 0.26 | 40% |
| SS53 | ML | 0.07 | 40% |
| SS67 | ML | 0.5 | 30% |
| HD | WT | 0.39 ± 0.12 | 0† |
| SS patients . | PTEN status . | pAKT amount in circulating SS cells detected by WB* . | pAKT+ SS skin resident cells detected by IHC (%) . |
|---|---|---|---|
| SS32 | ML | 0 | 5% |
| SS39 | ML | 0.01 | 25% |
| SS51 | WT | 0.26 | 40% |
| SS53 | ML | 0.07 | 40% |
| SS67 | ML | 0.5 | 30% |
| HD | WT | 0.39 ± 0.12 | 0† |
Densitometric values calculated as pAKT/actin.
IHC for pAKT in healthy skin is shown in supplemental Figure 3.
Discussion
PTEN is one of the most commonly mutated and deleted tumor suppressors in human cancer. Despite several studies identifying the PTEN locus variously affected by genetic deletion in CTCL,3-5 its involvement in this disease has not been fully investigated. Here, we have performed an extensive study on PTEN status in a large cohort of SS samples to define the level of PTEN deregulation. In line with its tumor-suppressive activity, we found that PTEN is lost in 36% of SS cases examined, indicating that its depletion represents a crucial step in SS pathogenesis that, conversely to MF,10 seems to not be associated with the late phase of disease because PTEN loss frequency is similarly observed in early (37%) and late (33%) samples analyzed before and after 12 months from diagnosis, respectively, thus suggesting that this event occurs during the initial phase of disease to be maintained in tumoral progression (data not shown). The deep sequencing performed on the same SS patient group did not demonstrate any mutations within the PTEN coding region.
A PTEN downregulation, both at the mRNA and the protein level, was observed in almost all of the samples we analyzed. Investigating other possible regulatory mechanisms, we demonstrated that neither PTENP1 pseudogene nor a promoter hypermethylation process are active in SS, whereas we found that miR-21, miR-106b, and miR-486 are able to modulate in vitro PTEN level, and miR-106b also displays a significant inverse relationship with PTEN abundance in vivo, indicating that this miRNA is deeply involved in SS pathobiology.
Experiments of genetic manipulation in mice have demonstrated that subtle reduction of PTEN levels have critical effects for tumorigenesis.19 In view of these findings, we can speculate that PTEN genomic loss, when accompanied by miRNA downregulation, might be more effective at promoting tumorigenesis in SS cells. This hypothesis also seems to be supported by the worst prognosis of SS22, SS30, SS32, SS39, SS43, and SS45 patients in which miR-21, miR-106b, and miR-486 were co-overexpressed26 ; in addition, mouse models overexpressing PTEN targeting miR-17-92 cluster (miR-106b-25 homolog) develop T-cell lymphoma.32
PTEN expression and localization was also evaluated by IHC. Although we were not able to compare IHC and SNP array results because the samples were not concurrently analyzed, we detected a very faint or absent PTEN immunoreactivity in all cases examined, in good agreement with Western blotting results. Similarly to peripheral T-cell lymphomas,33 we observed a nuclear and cytoplasmic localization of PTEN within the SS cells. Nuclear PTEN is found in cells arrested at the G0/G1 phase,34 whereas proliferating cells mainly show cytoplasmic PTEN.35 Thus, our findings suggest that SS cells are not quiescent but they can grow within a cutaneous background. Consistent with this hypothesis, we found AKT activation in SS cells that infiltrate the skin but not in the circulating ones, suggesting that skin microenvironment stimulates SS cells through direct-cell interactions and/or growth factor stimulation. A hypothesis, that PTEN reduction can amplify pathways induced by SS cell adhesion to the cutaneous milieu, thus promoting growth/survival of skin-infiltrating malignant lymphocytes, is supported by various findings: first, in the skin, SS cells interact with Langerhans cells in the intraepidermal Pautrier microabscess; second, SS cells coexpress skin homing receptors as CLA and CCR4 that specifically bind adhesion molecules present on endothelial cells of the skin, such as E-selectin and TARC36 ; and third, SS cells that migrate via chemokines,20,37 induce PI3K pathway, leading to AKT activation.38
The SS genome is characterized by a high chromosomal instability with recurrent gains, losses, or rearrangements.4,5 Genome-wide array analyses have identified genes that might contribute to genetic instability of SS such as TP5339 ; this gene is of particular importance because it is located at chromosome 17p13.3–17p13.1, a region that shows a high frequency of deletion in SS ranging from 71% to 75%.4,5 PTEN controls the P53 stability via MDM2 and interacts with p53, enhancing its acetylation and its DNA binding ability.39,40 Alternatively, p53 positively controls the transcription of PTEN.41 Because PTEN also plays a crucial role in genomic stability,42 we can speculate that reduced PTEN abundance in SS could perturb the positive regulatory relationship between PTEN and p53, contributing to the chromosomal alterations frequently observed in SS.
SS cells arise from central memory T lymphocytes,43 cells able to remember a pathogen persisting for as long as the lifetime of individuals via long-term homeostasis, a condition that implies a great resistance to both spontaneous and FAS-induced apoptosis.44 Recent studies have demonstrated that members of the Forkhead box (FOXO) family of transcription factors are important regulators of quiescence and longevity, playing critical roles in the development and function of memory subsets.45 Akt directly phosphorylates and inactivates members of the FOXO family, detaining them to cytoplasm.46 Interestingly, central memory T lymphocytes express high levels of phosphor-FOXO3a,47 maintaining in this way low levels of proapoptotic and cell cycle arrest FOXO3a target genes like FAS ligand, Bim, and p27, which are downregulated in SS.48-50 Altogether, this evidence strongly suggests that PTEN may enhance the apoptotic resistance of SS cells via FOXO3a, thus contributing with other genetic hits to their malignant expansion.
In conclusion, our data demonstrate that in SS, similarly to other malignancies, genetic deletion of the PTEN locus is highly recurrent. However, other deregulating mechanisms, such as abnormal expression of miRNAs, may influence PTEN expression; particularly, miR-106b shows a significant inverse relationship with PTEN level not only in vitro, but also in vivo. Finally, our results also provide evidence that low PTEN level does not activate AKT in circulating SS cells but, alternatively, it activates AKT in skin-infiltrating SS cells, indicating that skin milieu may strongly contribute to the SS pathogenesis and progression. To our knowledge, this is the first study to fully explore the status of PTEN in SS, shedding light on the mechanisms of its malfunctioning in this malignancy and unveiling information of potential clinical importance.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the medical and the nursing staff of the III Clinical Dermatology Unit of the IDI-IRCCS for continuous and helpful collaboration, Dr Laura Lupini and Marta Russo for scientific expertise and technical assistance to perform sequencing analysis, Dr Emilio Berti for continuous support, and Silvia Truffa for technical support.
This study was supported by the Ministero della Salute and the Associazione Italiana Ricerca sul Cancro (M.G.N.).
Authorship
Contribution: C.C. and M.C.P. performed research and analyzed data; C.L.L., E.P., and A.B. performed biochemical experiments; V.T. performed DNA methylation analysis; B.M. performed immunohistochemistry analysis; F.P. evaluated pathologic specimens; A.F. performed statistical analyses; E.S. performed cytofluorimetric analyses; G.A.L. and M.C. recruited eligible patients for this study and collected clinical information; E.C. and G.R. co-designed the study, co-analyzed the data, and critically revised the paper; and M.G.N. co-designed the study, performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Giandomenico Russo, IDI-IRCCS, Via dei Monti di Creta 104, 00167 Rome Italy; e-mail: g.russo@idi.it.
References
Author notes
C.C. and M.C.P. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal