Abstract
Recent whole-genome sequencing, whole-exome sequencing, and copy number analysis studies in small sets of patients with chronic lymphocytic leukemia (CLL) at presentation versus progression have demonstrated that clonal evolution has clinical and biologic relevance in CLL. Although the time and cost required to perform whole genome and exome sequencing are improving, challenges still exist in implementing these genomic strategies in real-time clinical practice. Here we performed DNA and RNA sequencing of an extensive panel of all genes known to be recurrently mutated in lymphoid, myeloid, and solid tumor malignancies, at high sequencing depth in patients with CLL at clinical presentation and progression. This allowed us to obtain integrated mutation, copy number, and gene fusion events in CLL and to use this data to obtain new insights into clonal evolution of CLL.
Genomic DNA and total RNA was isolated from 59 CLL samples (including 29 CLL paired patients sampled at the time of initial presentation when no clinical indication for therapy was met and at a later time point of disease progression requiring therapy). The median time between samples was 2.07 years (range 0.08-4.95 years)). Adaptor ligated sequencing libraries were captured by solution hybridization using two custom baitsets targeting 374 cancer-related genes and 24 genes frequently rearranged for DNA-seq, and 272 genes frequently rearranged for RNA-seq. All captured libraries were sequenced to high depth (Illumina HiSeq), averaging >590X for DNA and >20,000,000 total pairs for RNA, to enable the sensitive and specific detection of genomic alterations. All clinical data as well as biomarkers previously validated for clinical use in CLL (IGHV mutational status, CD38 expression, metaphase cytogenetics and FISH) were also obtained for all samples at each time point.
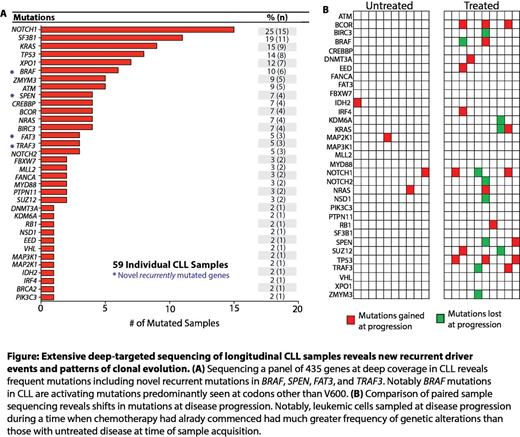
We then performed paired mutational analysis of samples at initial sample acquisition and at disease progression requiring therapy (Figure). Mutational analysis at the time of clonal evolution identified mutations in DNMT3A, EED, IDH2, IRF4, VHL, and RB1 only at the time of disease evolution. Mutations in NOTCH1, KRAS, TP53, NRAS, and BCOR were more common at disease progression than initial presentation. In contrast, mutations in SF3B1 and XPO1 were always retained at disease progression if found at earlier disease states.
These data demonstrate the utility of clinical grade, high throughput targeted sequencing in CLL to identify targetable genomic alterations, discover novel mutations not previously reported, and identify important markers of disease evolution which may represent incipient biomarkers for clinical disease progression. This includes a much more frequent occurrence of targetable mutations in the MAP kinase pathway than previously described in CLL and recurrent loss of function mutations in SPEN and FAT3 in CLL.
Levine:Foundation Medicine, Inc: Consultancy. Heaney:Novartis: Research Funding; Sanofi-Aventis: Consultancy, Research Funding; Onconova: Research Funding; Incyte: Consultancy, Research Funding. Abdel-Wahab:Foundation Medicine, Inc: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal