A clinical trial is ongoing at Stanford for MCL patients in first remission that interdigitates an autologous CpG-stimulated tumor cell vaccine with autologous peripheral blood stem cell transplant (PSCT) (NCT00490529). In this trial, blood samples collected before and after vaccination and serially post-transplant are assayed for minimal residual disease (MRD) and for T cell repertoire using the LymphoSIGHT™ sequencing method (Faham et al., Blood 2012). We identified a set of T cell clones that appear to be responding to the vaccine, and therefore we investigated whether the number of these clonotypes was correlated with MRD status.
Using universal primer sets, we amplified rearranged IgH variable (V), diversity, and joining (J) gene segments from genomic DNA. Amplified products were sequenced to obtain >1 million reads. The B cell tumor-specific sequence was identified for each patient based on its high frequency in the original tumor biopsy. The presence of the tumor cells was then monitored in serial blood samples with a sensitivity of 1 cell per million leukocytes. The same blood samples were used for amplification, sequencing and analysis of the entire TCRβ repertoire. To facilitate identification of tumor vaccine-induced TCRβ clonotypes, we sequenced the TCRβ repertoire immediately before and after the administration of both the priming vaccination and a booster vaccination. We developed a metric called the vaccine response score (V score). This metric is calculated for each clonotype and reflects the increase in frequency after the initial vaccination AND after the boost. The formula for calculating V score is: V = F1 x F2 x square root [1/ (|F1 – F2| + 1)], where F1 and F2 represent the fold-change of the priming and boost vaccinations, respectively. Clonotypes with a V score >10 were deemed to be vaccination-induced by virtue of these frequency changes.
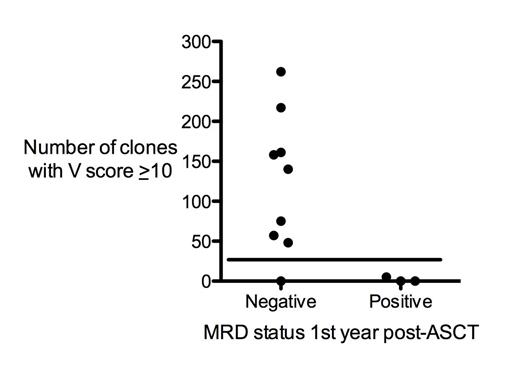
In a series of 12 vaccinated patients, the number of clonotypes with V score ≥ 10 ranged between 0 and 262, with a median of 57. We utilized an antigen-specific analysis to validate that clones with high V scores (≥ 10) were in fact tumor-specific. For this analysis, we incubated peripheral blood mononuclear cells (PBMCs) with the tumor and then sequenced the TCRβ repertoire from cells obtained after culture. Clones that were enriched after culture compared to pre-stimulation PBMCs were deemed to be antigen-specific. These clones that are antigen-specific are highly likely to have a high V score compared to a random frequency-matched set of clones (P two tailed = 1.8 x 10-10), providing further evidence that clones with a high V score are tumor-specific. We then analyzed the relationship between V score and clinical outcome. Patients could be stratified into two groups with “high” (> 25) or “low” (<25) numbers of vaccine-responsive clonotypes. Patients in the high V score group, who had larger numbers of putative tumor-specific T cells, were more likely to have sustained molecular remission during the first-year post-transplant compared with patients in the low V score group (P = 0.018) (Figure 1).
Patients with higher numbers of vaccine-responsive clones (i.e. clones with V score ≥ 10) are more likely to be MRD negative during the first year post-transplant (P=0.018).
Patients with higher numbers of vaccine-responsive clones (i.e. clones with V score ≥ 10) are more likely to be MRD negative during the first year post-transplant (P=0.018).
T cell repertoire analysis identified clonotypes responding to the vaccination, and the presence of these vaccine-specific clonotypes correlates with MRD positivity at the important landmark of one year post-PSCT. Further analysis of additional patients enrolled on the MCL trial is ongoing. This data underscores the prognostic relevance of the sequencing-based V score metric and provides a novel approach for assessment of cancer immunotherapy responses.
Faham:Sequenta: Employment, Equity Ownership, Membership on an entity’s Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal