Key Points
Engineered T-cell receptors can have redundant recognition of alternative protein motifs, resulting in severe cardiac toxicity.
The use of induced pleuripotent stem cells (iPSCs) is a promising approach to identify potential off-target effects of engineered T cells.
Abstract
An obstacle to cancer immunotherapy has been that the affinity of T-cell receptors (TCRs) for antigens expressed in tumors is generally low. We initiated clinical testing of engineered T cells expressing an affinity-enhanced TCR against HLA-A*01–restricted MAGE-A3. Open-label protocols to test the TCRs for patients with myeloma and melanoma were initiated. The first two treated patients developed cardiogenic shock and died within a few days of T-cell infusion, events not predicted by preclinical studies of the high-affinity TCRs. Gross findings at autopsy revealed severe myocardial damage, and histopathological analysis revealed T-cell infiltration. No MAGE-A3 expression was detected in heart autopsy tissues. Robust proliferation of the engineered T cells in vivo was documented in both patients. A beating cardiomyocyte culture generated from induced pluripotent stem cells triggered T-cell killing, which was due to recognition of an unrelated peptide derived from the striated muscle-specific protein titin. These patients demonstrate that TCR-engineered T cells can have serious and not readily predictable off-target and organ-specific toxicities and highlight the need for improved methods to define the specificity of engineered TCRs.
Introduction
T cells can be genetically engineered to express T-cell receptors (TCRs) with a high affinity and specificity for antigens.1-3 The MAGE-A3 antigen has been a leading target for immunotherapeutic approaches to cancer because of its frequent expression in multiple tumor types and restricted expression in normal tissues. However, naturally occurring TCRs to self-antigens are of lower affinity because of thymic selection, a fact that limits the ability of wild-type (WT) (or natural) TCRs to recognize the low antigen–HLA levels typically expressed on tumor cells.4 Therefore, we enhanced the affinity of a TCR specific for the HLA-A*01–restricted MAGE-A3 peptide (EVDPIGHLY). Following extensive molecular, biophysical, and immunologic testing in vitro and in mouse models of a library of TCR candidates, a moderately high-affinity TCR (MAGE-A3a3a) was selected as the lead candidate for clinical evaluation.5 Here we report that the adoptive transfer of autologous T cells engineered to express the MAGE-A3a3a TCR resulted in severe cardiac toxicity that was not due to off-tumor antigen expression or to recognition of epitopes from related cancer-testis antigens. Unexpectedly, toxicity was due to recognition of an epitope derived from an unrelated protein expressed by contracting normal cardiac tissue and one that would not be identified by using typical preclinical screening strategies.
Materials and methods
The affinity-enhanced MAGE-A3a3a TCR has 4 substitutions in the alpha chain of the CDR2 region and retained the WT sequences in the beta chain. The preclinical testing of the TCR has been reported.6
Protocols to test the engineered TCR for patients with the HLA-A*01 allele and whose tumors expressed MAGE-A3 by reverse-transcription polymerase chain reaction (RT-PCR) analysis were initiated at Washington University and the University of Pennsylvania. The protocols were registered at ClinicalTrials.gov (NCT01350401 and NCT01352286).
Manufacturing of the T-cell products
T-cell products were manufactured by using CD25-depleted and monocyte-depleted leukopheresis product. T cells were stimulated with CD3/28 beads and transduced with lentiviral vector as previously described.7 Case 1 T-cell product was in culture for 9 days, during which there was an expansion in total cell number of 7.5 population doublings. Case 2 T-cell product was also in culture for 9 days, during which there was an expansion in total cell number of 5.1 population doublings. Characterization of the infused products is shown in Table 1.
Characterization of infused products
| Criteria . | Case 1* . | Case 2 . |
|---|---|---|
| No. of cells infused | 10.00E+09 | 5.09E+09 |
| % Transduction efficiency | 52.80% | 47.50% |
| No. of transduced cells | 5.28E+09 | 2.42E+09 |
| Viability of sentinel vial | 93.60% | 88.90% |
| % CD3+CD45+ | 98.20% | 99% |
| % CD4 of CD3 | 53.80% | 60.40% |
| % CD8 of CD3 | 45.60% | 39.70% |
| % CD62L+ of CD3 | 97.6% | 94.1% |
| Transduction efficiency via vector copy number | 1.17 copy/cell | 0.8 copy/cell |
| Criteria . | Case 1* . | Case 2 . |
|---|---|---|
| No. of cells infused | 10.00E+09 | 5.09E+09 |
| % Transduction efficiency | 52.80% | 47.50% |
| No. of transduced cells | 5.28E+09 | 2.42E+09 |
| Viability of sentinel vial | 93.60% | 88.90% |
| % CD3+CD45+ | 98.20% | 99% |
| % CD4 of CD3 | 53.80% | 60.40% |
| % CD8 of CD3 | 45.60% | 39.70% |
| % CD62L+ of CD3 | 97.6% | 94.1% |
| Transduction efficiency via vector copy number | 1.17 copy/cell | 0.8 copy/cell |
As per protocol, Case 1 was infused by using a split-dose regimen with 30% and 70% of the dose infused on day +5 and day +6, respectively.
Research sample draws and processing
Research sample processing, freezing, and laboratory analyses were performed in the Translational and Correlative Studies Laboratory at the University of Pennsylvania, which operates under principles of Good Laboratory Practice. Assay performance and data reporting conforms with Minimal Information About T Cell Assays (MIATA) guidelines.8 Samples (peripheral blood, bone marrow) were collected in lavender-top (K2EDTA,) or red-top (no additive) BD vacutainer tubes (Becton Dickinson). Lavender-top tubes were delivered to the laboratory within 2 hours of draw or shipped overnight at room temperature in insulated containers essentially as described9 prior to processing. Samples were processed within 30 minutes of receipt according to established laboratory standard operating procedures. Peripheral blood and marrow mononuclear cells were purified, processed, and stored in liquid nitrogen as described.7 Red-top tubes were processed within 2 hours of draw including coagulation time, and serum was isolated by centrifugation and aliquoted in single-use 100-µL aliquots and stored at −80°C.
qPCR analysis
Whole blood or marrow samples were collected in lavender-top BD vacutainer tubes. Genomic DNA was isolated directly from whole blood and flash-frozen autopsy tissues, and quantitative PCR (qPCR) analysis on genomic DNA samples was performed in bulk by using ABI TaqMan technology and a qualified assay to detect the woodchuck posttranslational regulatory element sequence present in the integrated lentivirus, which contains the MAGE-A3a3a transgene as described.5 ,7 To determine copy number per unit DNA, an 8-point standard curve was generated consisting of 5 to 106 copies of lentivirus plasmid spiked into 100-ng nontransduced control genomic DNA. Each data point (sample, standard curve) was evaluated in triplicate with a positive concentration-time product value in 3/3 replicates with % cell volume less than 0.93% for all quantifiable values. A parallel amplification reaction to control for the quality of interrogated DNA was performed by using 20 ng input genomic DNA from peripheral blood and a primer/probe combination specific for nontranscribed genomic sequence upstream of the CDKN1A gene as described.7 These amplification reactions generated a correction factor to correct for calculated versus actual DNA input. Copies of transgene per microgram of genomic DNA were calculated according to the following formula: copies calculated from standard curve/input DNA (ng) × correction factor × 1000 ng × 0.5 (qualification process–established normalization factor to account for the efficiency of the woodchuck posttranslational regulatory element amplification). Accuracy of this assay was determined by the ability to quantify marking of the infused cell product by qPCR.
Flow cytometry reagents
The following antibodies were used for these studies: anti-CD3 fluorescein isothiocyanate and anti-C8α-PE-Cy7 (both from eBioscience) and anti-Vβ5.1 monoclonal antibody (Beckman Coulter; PN IM2285). To detect the MAGE-A3 TCR α/β heterodimer, a MAGE-A3–specific dextramer reagent (Immudex Corp.) conjugated to allophycocyanin was used (A*0101/EVDPIGHLY/APC; Cat# WA3249-APC).
Multiparameter flow cytometry
Cells were evaluated by flow cytometry after thaw and overnight rest at 2 × 106 cells/mL. Multiparametric immunophenotyping was performed by using approximately 1.0 × 106 total cells/condition and by using fluorescence minus one for dextramer and Vβ5.1 stains. Cells were stained in 100 µL phosphate-buffered saline for 30 minutes on ice using antibody and reagent concentrations recommended by the manufacturer; they were then washed, resuspended in 0.5% paraformaldehyde, and acquired by using an Accuri C6 cytometer equipped with a blue (488 nm) and red (633 nm) laser. Accuri files were exported in flow cytometry standard file format and analyzed by using FlowJo software version 9.5.3 (Treestar). Compensation values were established by using single-antibody stains and BD compensation beads (Becton Dickinson) and were calculated by the software. The gating strategy for T cells was as follows: live cells (forward scatter/side scatter) > CD3+/SSC low.
Soluble factor analysis
Whole blood was collected in red-top BD vacutainer tubes, processed to obtain serum by using established laboratory standard operating procedures, aliquoted for single use, and stored at −80°C. Quantification of soluble cytokine factors was performed by using Luminex bead array technology and kits purchased from Life Technologies (Invitrogen). Assays were performed as per the manufacturer’s protocol, with a 9-point standard curve generated by using a threefold dilution series. The 2 external standard points were evaluated in duplicate and the 5 internal standards in singlicate; all samples were evaluated in duplicate at 1:2 dilution; calculated % cell volume for the duplicate measures was less than 15%. Data were acquired on a FlexMAP-3D system by percent and analyzed by using XPonent 4.0 software and 5-parameter logistic regression analysis. Standard curve quantification ranges were determined by the 80% to 120% (observed/expected value) range. Reported values included those within the standard curve range and those calculated by the logistic regression analysis.
Beating cardiac myocyte iPSC-CM assay
Induced-pluripotent stem-cell (iPSC) –derived cardiomyocyte (iPSC-CM) cells were thawed from liquid nitrogen storage and treated as per the manufacturer’s instructions. Prior to assay, the cells were virally transduced with HLA-A*01 and confirmed positive. iPSC-CM and control target cells were plated at 5 × 104 per well of a 96-well plate and incubated for 24 hours with nontransduced or MAGE-A3a3a–transduced T cells at a ratio of 1:1. Quantification of interferon gamma (IFN-γ) release in cell supernatants was performed by using Luminex bead array technology with reagents purchased from Life Technologies. Samples were processed in triplicate, and assays were performed as per the manufacturer’s instructions.
Results
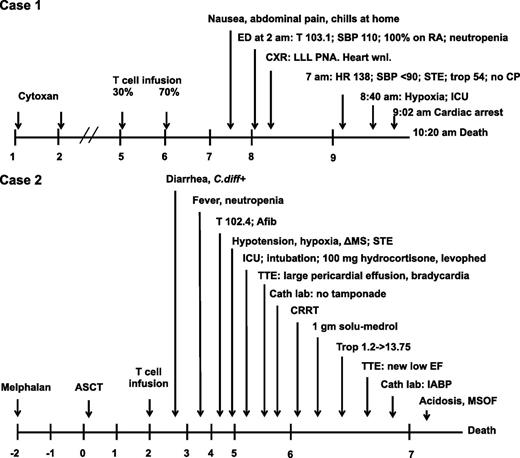
Clinical studies in stage III/IV melanoma and high-risk or relapsed myeloma were initiated. The first 2 patients enrolled on a cohort for patients with the HLA-A*01 allele and whose tumors expressed MAGE-A3 by RT-PCR analysis are reported. The protocol schemas are shown in supplemental Figure 1 (available on the Blood Web site). Both patients developed fever, progressive hypoxia, and hypotension and expired 4 to 5 days after infusion (Figure 1).
Timeline of events after infusion of engineered T cells. ΔMS, altered mental status; Afib, atrial fibrillation; ASCT, autologous stem cell transplant; Cath laboratory, cardiac catheterization laboratory; CP, chest pain; CRRT, continuous renal replacement therapy; CXR, chest x-ray; ED, emergency department; EF, ejection fraction; HR, heart rate; IABP, intra-aortic balloon pump; ICU, intensive care unit (transfer); LLL, left lower lobe; MSOF, multisystem organ failure; PNA, pneumonia; RA, room air; SBP, systolic blood pressure; STE, ST elevations on electrocardiogram; T, temperature; Trop, troponin; TTE, transthoracic echocardiogram; Wnl, within normal limits. 100% on RA, 100% oxygen saturation on room air by pulse oximetry.
Timeline of events after infusion of engineered T cells. ΔMS, altered mental status; Afib, atrial fibrillation; ASCT, autologous stem cell transplant; Cath laboratory, cardiac catheterization laboratory; CP, chest pain; CRRT, continuous renal replacement therapy; CXR, chest x-ray; ED, emergency department; EF, ejection fraction; HR, heart rate; IABP, intra-aortic balloon pump; ICU, intensive care unit (transfer); LLL, left lower lobe; MSOF, multisystem organ failure; PNA, pneumonia; RA, room air; SBP, systolic blood pressure; STE, ST elevations on electrocardiogram; T, temperature; Trop, troponin; TTE, transthoracic echocardiogram; Wnl, within normal limits. 100% on RA, 100% oxygen saturation on room air by pulse oximetry.
Case 1 (UPCC-01611-100)
The patient was a 63-year-old man who was initially diagnosed with a 2.38-mm ulcerated melanoma of the left neck 6 years prior to enrollment. He was diagnosed with stage IIIB (T3bN1aM0) melanoma after undergoing a wide local excision with sentinel lymph node sampling that was positive for melanoma. He received adjuvant IFN-α but recurred at 1 year with mediastinal lymphadenopathy. His tumor tested negative for the BRAF V600E mutation. He was treated first on a protocol with a dendritic cell vaccine, then on a protocol with ipilimumab for 4 years. He had documented disease progression with increasing axillary and mediastinal adenopathy as well as left lung metastases. The 6-cm axillary adenopathy was resected and histologically confirmed as metastatic melanoma. He was enrolled in UPCC-01611. His baseline screening was notable for a normal electrocardiogram (ECG), normal echocardiogram, and pulmonary function tests with a minimal obstructive ventilation defect. On protocol, a dose of cyclophosphamide 60 mg/kg per day was administered for 2 days starting 4 days prior to the cell infusion since this had previously been shown to potentiate the effects of the infused engineered T cells.3 Thus, 5.3 × 109 MAGE-A3a3a TCR cells were administered in the outpatient setting as a split infusion of 30% and 70% total cells on consecutive days (Figure 1). Three days following the first T-cell infusion, he presented to the emergency department with chills, nausea, and abdominal pain. He was found to be neutropenic and febrile to 103.1°F and was treated with broad-spectrum antibiotics (cefepime and vancomycin). The next morning he was found to be hypotensive, tachycardic, and hypoxic. He did not complain of chest pain, but ECG showed diffuse ST elevations, and cardiac enzymes were elevated: troponin I was recorded at 54.4 ng/mL (normal range, 0.00 to 0.24 ng/mL) and creatinine kinase MB at 65 ng/mL (normal range, 0 to 7 ng/mL). Within 30 minutes, he became unresponsive and had a cardiopulmonary arrest; he was initially resuscitated, but he had a second arrest approximately 90 minutes later from which he could not be resuscitated. The patient expired 4 days after the first T-cell infusion. Analysis of 30 cytokines in blood samples collected after T-cell infusion indicated a systemic inflammatory process that was consistent with T-cell activation because IFN-γ, interleukin 5 (IL-5), and IL-8 were elevated approximately 100-fold above baseline (Figure 2).
Peripheral blood cytokines elevated in both patients. Peripheral blood samples were collected in both patients at predetermined time points; specific levels of 30 cytokines were quantified and compared with baseline (preinfusion) samples. Only cytokines that were elevated at least 10-fold over baseline at any time point are shown. Case 2 received granulocyte colony-stimulating factor (G-CSF) as part of standard-of-care therapy following the autologous stem cell transplant. Baseline (preinfusion) serum values of the 9 analytes shown in Case 1 were IL-6, 12.5 pg/mL; IL-15, 30.8 pg/mL; IL-5, 1.82 pg/mL; IFN-γ, 1.68 pg/mL; IL-1Rα, 245 pg/mL; interferon-inducible protein 10 (IP-10), 99.9 pg/mL; IL-2R, 322 pg/mL; monokine induced by gamma interferon (MIG/CXCL9), 30.7 pg/mL; and IL-8, 11.6 pg/mL. Baseline levels of all cytokines analyzed in the serum in Case 2 are detailed in the legend for Figure 4B. FGF, fibroblast growth factor; GM-CSF, granulocyte macrophage CSF; HGF, hepatocyte growth factor; MCP-1, monocyte chemoattractant protein 1; VEGF, vascular endothelial growth factor.
Peripheral blood cytokines elevated in both patients. Peripheral blood samples were collected in both patients at predetermined time points; specific levels of 30 cytokines were quantified and compared with baseline (preinfusion) samples. Only cytokines that were elevated at least 10-fold over baseline at any time point are shown. Case 2 received granulocyte colony-stimulating factor (G-CSF) as part of standard-of-care therapy following the autologous stem cell transplant. Baseline (preinfusion) serum values of the 9 analytes shown in Case 1 were IL-6, 12.5 pg/mL; IL-15, 30.8 pg/mL; IL-5, 1.82 pg/mL; IFN-γ, 1.68 pg/mL; IL-1Rα, 245 pg/mL; interferon-inducible protein 10 (IP-10), 99.9 pg/mL; IL-2R, 322 pg/mL; monokine induced by gamma interferon (MIG/CXCL9), 30.7 pg/mL; and IL-8, 11.6 pg/mL. Baseline levels of all cytokines analyzed in the serum in Case 2 are detailed in the legend for Figure 4B. FGF, fibroblast growth factor; GM-CSF, granulocyte macrophage CSF; HGF, hepatocyte growth factor; MCP-1, monocyte chemoattractant protein 1; VEGF, vascular endothelial growth factor.
The initial autopsy report concluded that the cause of death was a large acute myocardial infarction. The left anterior descending coronary artery showed severe atherosclerotic disease with 95% occlusion of the lumen. There were patchy full-thickness hemorrhages in both ventricles and in the septum. Two scars in the anterior ventricle were noted as indicating prior myocardial infarctions. Of note, there was no indication of prior infarction by history or on the screening ECG and echocardiogram, indicating that the patient had a history of silent myocardial infarctions.
PCR analysis for vector sequences in blood indicated in vivo expansion of the MAGE-A3a3a T cells, which reached more than 100 cells per microliter of blood (Figure 3A). As expected, the total white blood cell count declined because of cyclophosphamide. Analysis of the biodistribution of the engineered T cells from tissues obtained at autopsy was also performed (Figure 3B). The highest levels of engineered T cells were found in spleen, blood, and heart at nearly equivalent levels. Because of the hemorrhagic nature of the myocardial infarction, the T-cell distribution in the heart was initially attributed to infiltration with blood. Lymph node, lung, and liver metastases had similar numbers of engineered T cells, but few were found in brain or testis samples.
Fate of the infused engineered T cells. (A) Expansion of the engineered T cells over time in both patients. Left y-axis shows cell count per microliter of blood (total white blood cell [WBC] count in red; engineered [marked] cells in green). Right y-axis shows the number of copies of DNA used in the engineered T cells as a function of total genomic DNA in peripheral blood mononuclear cells (PBMCs) over time. (B). Distribution of engineered T cells in tissue samples obtained post mortem in both patients. qPCR for vector sequences was performed from total genomic DNA obtained from flash-frozen tissues. Note different scale on y-axis for Case 1 and Case 2.
Fate of the infused engineered T cells. (A) Expansion of the engineered T cells over time in both patients. Left y-axis shows cell count per microliter of blood (total white blood cell [WBC] count in red; engineered [marked] cells in green). Right y-axis shows the number of copies of DNA used in the engineered T cells as a function of total genomic DNA in peripheral blood mononuclear cells (PBMCs) over time. (B). Distribution of engineered T cells in tissue samples obtained post mortem in both patients. qPCR for vector sequences was performed from total genomic DNA obtained from flash-frozen tissues. Note different scale on y-axis for Case 1 and Case 2.
The conclusions from the initial investigation were that myocardial ischemia followed by infarction was a result of underlying coronary disease and a combination of the metabolic demands of neutropenic fever, coupled with anemia, and a cytokine release syndrome. The patient had had a normal ECG and echocardiogram at study entry, which pointed to the need to add enhanced cardiac screening for study entry. As a result, the protocols were amended to include a nuclear cardiac stress test and troponin T for screen and serial ECGs and troponin T to monitor cardiac status after infusion. Information regarding the death was added to the informed consents, and the US Food and Drug Administration and ethics committees approved proceeding with additional patients to receive the MAGE-A3a3a–engineered T cells. These studies were conducted in accordance with the Declaration of Helsinki.
Case 2 (UPCC-01411-100)
The second patient infused with the engineered T cells was enrolled in the myeloma trial (supplemental Figure 1). The patient was a 57-year-old man who was diagnosed with immunoglobulin G kappa plasma cell myeloma 3 years prior to enrollment, when he presented with plasmacytomas of the T4 and T8 vertebral bodies. He was initially treated with radiation therapy, which was complicated by radiation pneumonitis. He completed 1 month of lenalidomide and dexamethasone but did not tolerate further lenalidomide. He was observed for 2 years, and then he was treated with bortezomib and dexamethasone, but his disease progressed. Three months prior to enrollment, he was treated with two cycles of D-PACE (dexamethasone, cisplatin, doxorubicin, cyclophosphamide, and etoposide). He was assessed by a cardiologist because he had a history of hypertrophic obstructive cardiomyopathy that had been diagnosed at age 18 and he had had intermittent atrial fibrillation requiring cardioversion on at least one occasion. His atrial fibrillation was rate-controlled with diltiazem and propranolol, and he was anti-coagulated with aspirin in view of his underlying hematologic malignancy. His nuclear cardiac stress test and baseline troponin T were normal, and his cardiologist concluded that his cardiomyopathy did not pose a contraindication to enrollment in this protocol.
On protocol, the patient underwent stem cell mobilization and harvest and received melphalan at 200 mg/m2 followed by autologous stem cell transplant. A total of 2.4 × 109 MAGE-A3a3a T cells were infused 2 days later (Figure 1). That day, he developed diarrhea, and a stool sample tested positive for Clostridium difficile toxin; he was treated with metronidazole. Two days after the T-cell infusion, the patient became febrile to 102.4°F in the setting of neutropenia. There were no significant changes on ECG other than atrial fibrillation and tachycardia attributed to the fever. He was treated with broad-spectrum antibiotics. The next day he developed hypotension, hypoxia, altered mental status, and ECG changes. He was treated with oxygen, fluids, hydrocortisone, and vasopressors. An echocardiogram showed a large pericardial effusion and suggested pre-tamponade physiology and possibly cardiogenic shock. He was treated with aggressive supportive cardiac and medical intensive care, including high-dose corticosteroids given as hydrocortisone and then 1 g methylprednisolone. Supportive care included placement of an intra-aortic balloon pump for his cardiogenic shock and continuous renal replacement therapy for acute renal failure. In the course of that day, his troponin rose from 1.2 to 13.75 (normal range, 0.0 to 0.03). During the following day, he developed progressive metabolic acidosis and multisystem organ failure due to worsening cardiogenic shock. He expired 5 days after receiving T cells.
Analysis of 30 cytokines in blood over time showed evidence of immune activation, with IL-6 elevated >1000-fold above baseline (Figure 2).
PCR analysis for MAGE-A3a3a TCR sequences in blood revealed robust in vivo expansion of the T cells, which exceeded 400 cells per microliter of blood 3 days after T-cell infusion (Figure 3A). Analysis of engineered T-cell biodistribution by PCR revealed that the highest concentration of engineered T cells were in the blood and pericardial fluid (Figure 3B). Bone marrow, lung, heart, and liver were the tissues with the next highest distribution of engineered cells. Of note, evaluated brain and testis specimens from Case 2 did not contain significant numbers of engineered cells as they did for Case 1. We also calculated the total number of T cells recovered from the total blood volume and from individual organs (supplemental Table 1) and determined that at the time of autopsy, the transduced T cells expanded nearly 200-fold in Case 2 and 17-fold in Case 1.
At autopsy, the most significant finding was cardiac myonecrosis with an unusual lymphoid predominant infiltrate into the myocardium (Figure 4A). In contrast to Case 1, Case 2 had no evidence of coronary artery disease or thrombosis. The histologic findings of myocardial necrosis were most consistent with immunologically mediated damage.10 Immunohistochemistry for CD3 was performed, and it demonstrated that the lymphoid infiltrate was composed of T cells. A similar pattern of CD3+ cell infiltrate was not observed in the other organs and tissues examined, including multiple sections of skeletal muscle. Therefore, CD3+ cells specifically infiltrated the myocardium, resulting in acute cardiac injury and a histological pattern similar to allograft rejection.10,11 Analysis of 30 cytokines in blood and pericardial fluid obtained from Case 2 at autopsy showed evidence of local immune activation in addition to systemic immune activation (Figure 4B).
Analysis of cardiac-specific toxicity. (A) Pathologic and immunohistochemical analysis of myocardium from both patients. Hematoxylin and eosin (H&E) stained sections of myocardium shown at two magnifications for each patient (top panels), demonstrating lymphocytic infiltrate and diffuse myocyte necrosis. Immunohistochemical (IHC) staining with anti-CD3 (lower panels) demonstrates that lymphocytic infiltrates are T cells, shown at two magnifications. (B) Cytokines in the peripheral blood and the pericardial fluid in Case 2 obtained post mortem show evidence of T-cell activation. Thirty cytokines were assayed; each cytokine level shown is normalized to the concentration of that cytokine in a peripheral blood sample obtained at baseline. The patient was given injections of filgrastim (G-CSF) as per standard of care post-ASCT. Baseline levels of cytokines in blood were VEGF, 1.73 pg/mL; IL-1β, 0.5 pg/mL; G-CSF, 10.51 pg/mL; epidermal growth factor (EGF), 23.13 pg/mL; IL-10, 2.54 pg/mL; HGF, 357.83 pg/mL; FGF-basic, 4.45 pg/mL; IFN-α, 70.76 pg/mL; IL-6, 3.42 pg/mL; IL-12, 93.34 pg/mL; regulated upon activation, normal T-cell expressed and secreted (RANTES), 12 717 pg/mL; eotaxin, 76.1 pg/mL; IL-13, 0.53 pg/mL; IL-15, 59.94 pg/mL; IL-17, 0.16 pg/mL; macrophage inflammatory protein 1 alpha (MIP-1α/CCL3), 14.85 pg/mL; GM-CSF, 0.77 pg/mL; MIP-1β, 36.44 pg/mL; MCP-1, 953 pg/mL; IL-5, 0.34 pg/mL; IFN-γ, 2.49 pg/mL; tumor necrosis factor alpha (TNF-α), 1.26 pg/mL; IL-1Rα, 37.66 pg/mL; IL-2, 0.56 pg/mL; IL-7, 15.36 pg/mL; IP-10, 23.14 pg/mL; IL-2R, 205.81 pg/mL; MIg, 10.51 pg/mL; IL-4, 9.91 pg/mL; and IL-8, 10.68 pg/mL.
Analysis of cardiac-specific toxicity. (A) Pathologic and immunohistochemical analysis of myocardium from both patients. Hematoxylin and eosin (H&E) stained sections of myocardium shown at two magnifications for each patient (top panels), demonstrating lymphocytic infiltrate and diffuse myocyte necrosis. Immunohistochemical (IHC) staining with anti-CD3 (lower panels) demonstrates that lymphocytic infiltrates are T cells, shown at two magnifications. (B) Cytokines in the peripheral blood and the pericardial fluid in Case 2 obtained post mortem show evidence of T-cell activation. Thirty cytokines were assayed; each cytokine level shown is normalized to the concentration of that cytokine in a peripheral blood sample obtained at baseline. The patient was given injections of filgrastim (G-CSF) as per standard of care post-ASCT. Baseline levels of cytokines in blood were VEGF, 1.73 pg/mL; IL-1β, 0.5 pg/mL; G-CSF, 10.51 pg/mL; epidermal growth factor (EGF), 23.13 pg/mL; IL-10, 2.54 pg/mL; HGF, 357.83 pg/mL; FGF-basic, 4.45 pg/mL; IFN-α, 70.76 pg/mL; IL-6, 3.42 pg/mL; IL-12, 93.34 pg/mL; regulated upon activation, normal T-cell expressed and secreted (RANTES), 12 717 pg/mL; eotaxin, 76.1 pg/mL; IL-13, 0.53 pg/mL; IL-15, 59.94 pg/mL; IL-17, 0.16 pg/mL; macrophage inflammatory protein 1 alpha (MIP-1α/CCL3), 14.85 pg/mL; GM-CSF, 0.77 pg/mL; MIP-1β, 36.44 pg/mL; MCP-1, 953 pg/mL; IL-5, 0.34 pg/mL; IFN-γ, 2.49 pg/mL; tumor necrosis factor alpha (TNF-α), 1.26 pg/mL; IL-1Rα, 37.66 pg/mL; IL-2, 0.56 pg/mL; IL-7, 15.36 pg/mL; IP-10, 23.14 pg/mL; IL-2R, 205.81 pg/mL; MIg, 10.51 pg/mL; IL-4, 9.91 pg/mL; and IL-8, 10.68 pg/mL.
Additional post hoc investigation
Because of the autopsy findings in Case 2 and the similarities in clinical courses, the cardiac tissue from Case 1 was re-examined. Immunohistochemistry of heart tissue from Case 1 revealed extensive CD3+ T-cell infiltration (Figure 4A). Histopathologically, it was also noted that there was extensive myocyte necrosis and neutrophilic inflammation that was not fully explained by the distribution of atherosclerotic disease in the proximal left anterior descending coronary artery, since the other coronary arteries had only minimal disease. The role of cardiac hypertrophy in Case 2 is unclear. The data suggest that T-cell–mediated acute cardiac injury contributed to acute cardiac failure in both patients.
TCR-engineered T cells could cause injury to normal tissues by any of several mechanisms: (1) mispairing of introduced and endogenous TCR α/β chains resulting in novel specificities, (2) alloreactivity, (3) recognition of an unknown epitope expressed by normal tissue, or (4) expression of the target MAGE-A3 epitope by normal tissue. Mechanisms 1 to 3 would represent examples of off-target toxicity, while mechanism 4 would exemplify on-target off-tumor toxicity.
Mispairing of endogenous TCRs with exogenous TCRs could generate novel specificities and result in a graft-versus-host disease–like syndrome.1 This possibility has been comprehensively studied in mice12 but has not been described in any clinical trials of TCR-transduced T cells to date.3 Although precise quantification of mispaired TCRs in vivo is difficult, the frequency of T cells expressing the Vβ5.1 chain used by the MAGE-A3 TCR can be compared with surface expression of the MAGE-A3–specific α/β heterodimer, which defines the correct α/β TCR pair. By using this method, an increase in the frequency of Vβ5.1 chain relative to the correct α/β TCR pair could indicate mispairing; this was not observed in either patient (Figure 5A).
Elucidation of mechanism of clinical cardiac toxicity. (A) Analysis for expression of the correctly paired MAGE-A3–engineered TCR (MAGE dextramer staining, y-axis) of input (untransduced) T cells, transduced T-cell product, and T cells recovered from PBMCs in both patients at time of death. The x-axis shows staining for specific TCR-β chain (Vβ5.1) used by the engineered MAGE-A3 TCRs. Numbers shown in each quadrant indicate percentages of the gated CD3+ cells. Cell populations that stain only for Vβ5.1 but not dextramer are the sum of (1) endogenous TCR that uses that β chain (ie, population shown in input T cells) and (2) mispaired MAGE-A3–engineered TCR. No expansion of T cells with mispaired TCRs was detected after infusion in the patients. (B) A sample of the T-cell product infused into Case 2, along with fresh MAGE-A3a3a–transduced T cells and untransduced T cells, was tested for IFN-γ production by ELISPOT when cocultured with a large panel of HLA-A1+ cell lines, including one that was MAGE-A3+ (EJM) as a positive control. Bars indicate mean ± standard error of the mean (SEM) of 3 replicates. (C) Activation and cytokine production of MAGE-engineered T cells incubated in vitro with HLA-A*01+, titin-positive, MAGE-A3− beating cardiac myocyte cells derived from iPSC-CM or iCells. The EJM plasmacytoma cell line expresses HLA-A*01 and MAGE-A3 (positive control), and the colo205 cancer cell line expresses HLA-A*01 but not MAGE-A3 or titin (negative control). Controls for the effector cells are nontransduced (ntd) T cells, or no T cells (targets only). Bars indicate mean ± SEM of 3 replicates. *P < .0001 by Student t test.
Elucidation of mechanism of clinical cardiac toxicity. (A) Analysis for expression of the correctly paired MAGE-A3–engineered TCR (MAGE dextramer staining, y-axis) of input (untransduced) T cells, transduced T-cell product, and T cells recovered from PBMCs in both patients at time of death. The x-axis shows staining for specific TCR-β chain (Vβ5.1) used by the engineered MAGE-A3 TCRs. Numbers shown in each quadrant indicate percentages of the gated CD3+ cells. Cell populations that stain only for Vβ5.1 but not dextramer are the sum of (1) endogenous TCR that uses that β chain (ie, population shown in input T cells) and (2) mispaired MAGE-A3–engineered TCR. No expansion of T cells with mispaired TCRs was detected after infusion in the patients. (B) A sample of the T-cell product infused into Case 2, along with fresh MAGE-A3a3a–transduced T cells and untransduced T cells, was tested for IFN-γ production by ELISPOT when cocultured with a large panel of HLA-A1+ cell lines, including one that was MAGE-A3+ (EJM) as a positive control. Bars indicate mean ± standard error of the mean (SEM) of 3 replicates. (C) Activation and cytokine production of MAGE-engineered T cells incubated in vitro with HLA-A*01+, titin-positive, MAGE-A3− beating cardiac myocyte cells derived from iPSC-CM or iCells. The EJM plasmacytoma cell line expresses HLA-A*01 and MAGE-A3 (positive control), and the colo205 cancer cell line expresses HLA-A*01 but not MAGE-A3 or titin (negative control). Controls for the effector cells are nontransduced (ntd) T cells, or no T cells (targets only). Bars indicate mean ± SEM of 3 replicates. *P < .0001 by Student t test.
We next evaluated each patients’ engineered T-cell product against related epitopes from a comprehensive list of other MAGE family members as well as against an alloreactivity panel which covered >95% of known HLA types; no response was detected in either analysis.5 It was also determined that there was no expression of MAGE-A3, or potentially cross-reactive targets MAGE-A6 or MAGE-B18 by RT-PCR in either patients’ heart tissue or in autopsy samples from five other normal hearts.5 Thus, we were unable to establish that alloreactivity or on-target off-tumor reactivity were responsible for the observed toxicity.
To evaluate the potential for off-target off-tumor reactivity being responsible for the observed toxicity, we performed extensive in vitro analyses on the product that was administered to Case 2 and on newly MAGE-A3a3a–transduced T cells. However, neither the patient’s cell product nor fresh MAGE-A3a3a–transduced T cells were activated in response to a large panel of HLA-A1+ cell lines, including cardiac myocytes and endothelial cells (Figure 5B). However, when an HLA-A1+ beating cardiomyocyte culture derived from iPSCs, or iCells, was exposed to MAGE-A3a3a–transduced T cells, the T cells did become activated (Figure 5C), and they killed the cardiomyocytes. Compared with T cells transduced with the WT MAGE-A3 TCR that served as the backbone of the a3a modifications, MAGE-A3a3a–transduced T cells were significantly more sensitive to activation by both iCells and the positive control (HLA-A1+ MAGE-A3+) cell line EJM. Having replicated the clinical findings in vitro, further studies demonstrated that a cross-reactive epitope derived from the human protein titin, presented in the context of HLA-A*01, could be potently recognized by the MAGE-A3 TCR-engineered T cells.5
Titin is a sarcomeric protein expressed in striated muscle,13 and certain mutations in titin are known to cause dilated cardiomyopathy.14 Importantly, standard cultured primary cardiomyocyte cells do not express titin, but actively beating cardiac myocytes derived from iPSCs do express titin. Patient autopsy samples extracted from the heart were confirmed to be highly positive for titin by qRT-PCR, and presentation of titin was confirmed by mass spectrometry of peptides eluted from the surface of cells.5
Discussion
Through an unfortunate series of clinical events, we discovered that T cells engineered to express an affinity-enhanced TCR directed to an epitope of MAGE-A3 are also able to effectively recognize a similar peptide epitope derived from the entirely unrelated protein titin, which is expressed in cardiac tissue. MAGE-A3 has been prioritized by the National Cancer Institute as the top-rated cancer-testis antigen immunotherapy target because of its frequent expression in many types of cancer and lack of expression on adult cells that express HLA class I antigens.15,16 Heretofore, infusions with engineered T cells expressing TCRs with enhanced affinity to another cancer-testis antigen, NY ESO-1, have been safe and have demonstrated clinically evident antitumor responses.17
The observation that the striated muscle-specific protein titin could be targeted by a TCR specific for a cancer-testis antigen is the first clinical example of off-target activity mediated by engineered TCRs. It has been previously reported that affinity-modified TCRs can show lack of specificity, including self-reactivity.18 Titin is the largest gene in the human genome and is not related either structurally or functionally to MAGE-A3 or to other cancer-testis antigens.13 Titin itself is immunologically interesting, because patients with autoimmune diseases such as myasthenia gravis, particularly when associated with thymoma, often have antibodies directed to titin.19 Whether this is a result of antigen spreading in a neuromuscular disease or whether the association with thymoma is mechanistically related to the development of anti-titin antibodies remain unanswered questions. We specifically evaluated but, interestingly, did not observe any T-cell infiltration in skeletal muscle sections in Case 2. Although we cannot fully explain this finding, it appears that skeletal muscle may be immunologically distinct from cardiac muscle; for example, skeletal muscle can be immunologically silent when transduced with adeno-associated viral vectors20 whereas liver cannot.21 Furthermore, cardiac muscle may be particularly susceptible to dysregulated T-cell activation, since mice deficient in the T-cell inhibitory receptor PD-1 develop an enhanced T-cell–mediated myocarditis in some experimental models.22,23
The parental TCR from which the MAGE-A3a3a TCR was derived was originally isolated from a patient with melanoma that expressed the MAGE-A3 antigen.24 It is likely that the low affinity of the parental MAGE-A3–specific TCR, as a consequence of the natural thymic selection process, allowed for expansion of the original T cells in a patient without cardiac toxicity. MAGE-A3 may be a particularly difficult target because another group using an independently derived TCR that was specific for MAGE-A3 peptides presented on HLA-A*02 has reported severe on-target off-tumor toxicity.25 In that study, neurologic toxicity was observed because of the unexpected expression of other members of the MAGE cancer-testis family in the central nervous system. Other instances of on-target off-tumor toxicity have been reported in patients with T cells engineered with a TCR specific for the carcinoembryonic antigen resulting in colitis,26 and TCRs targeting melanoma antigens destroyed normal melanocytes in the skin, ears, and eyes.27
The cardiac toxicity that we observed was severe and, in some respects, resembled graft-versus-host disease limited to a single organ. In Case 2, the process was refractory to immunosuppression with corticosteroids. It should be noted that the toxicity was delayed in onset for several days and was not primarily due to a cytokine storm, as has been reported with T cells expressing chimeric antigen receptors.28,29 We have recently observed that toxicity from chimeric antigen receptor–modified T cells can be aborted with blockade of IL-6 signaling,30 and it is possible that tocilizumab may be applicable to other instances of toxicity from engineered TCR therapy, given that our patients had high levels of IL-6 in the serum and pericardial fluid. However, this would not have been expected to abate the primary off-target cardiac toxicity. Another approach that may be useful would be the incorporation of a conditional suicide gene.31 This approach effectively ablated alloreactive T cells in a clinical trial after human hematopoietic cell transplantation and may prove useful to abrogate on-target or off-target toxicities of TCR-engineered T cells.
The development of engineered, affinity-enhanced TCRs is emerging as a powerful strategy to effectively target tumors and expands the opportunities for engineered TCR-based adoptive T-cell–based therapies.32,33 Although MAGE-A3 remains an attractive target because of its restricted normal tissue expression, the challenge for targeted T-cell therapy remains the identification of suitable epitopes to ensure on-target specificity. Here, we show the need for improved preclinical testing methods to better enable prediction of specificity and illustrate the application of more elaborate cell culture systems which identified the previously unknown cross-reactivity of the MAGE TCR. Finally, this study highlights the potency of engineered T cells, if they are harnessed appropriately, and suggests that reactivity to unknown epitopes expressed on normal essential tissues may represent the gravest safety risk with the use of engineered T cells.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jeffrey Finklestein for sample processing and flow cytometric analyses, Irina Kulikovskaya and Minnal Gupta for quantitative polymerase chain reaction assays, Erica Suppa for Luminex assays, Zhaohui Zheng, Andrea Brennan, and members of the Clinical Cell and Vaccine Production facility for developing methods for clinical scale ex vivo lentiviral transduction and for cell manufacturing, the Human Immunology Core for reagents, and Elizabeth Veloso, Lester Lledo, Joan Gilmore, and Anne Chew for assistance in clinical research support.
Authorship
Contribution: M.V.M. and C.H.J. wrote the manuscript; the protocols were written and designed by G.P.L., E.A.S., A.P.R., and G.B.-S; C.H.J. was the regulatory sponsor with help from M.V.M; Adaptimmune Ltd engineered the MAGE-A3 TCR and was the commercial sponsor. L.E., L.L., A.B., B.M.C., and P.J.C. conducted clinical research; M.K. supervised the laboratory analyses of clinical samples; N.H. and B.K.J. devised experiments uncovering the role of cross-reactivity to titin; B.L.L. supervised cell manufacturing; and all authors discussed and interpreted results and vouch for the data and analyses.
Conflict-of-interest disclosure: N.H., J.H., and J.D. are employees of Immunocore Ltd and G.B.-S., D.P.S., A.B.G., N.J.P., A.D.B., J.E.B., H.K.T.-M., and B.K.J. are employees of Adaptimmune Ltd. The remaining authors declare no competing financial interests.
Correspondence: Carl H. June, 3400 Civic Center Blvd, 8th Floor, Room 08-123, Philadelphia, PA, 19104-5156; email: cjune@exchange.upenn.edu.
References
Author notes
G.P.L., E.A.S., and M.V.M. contributed equally to this study.


![Figure 3. Fate of the infused engineered T cells. (A) Expansion of the engineered T cells over time in both patients. Left y-axis shows cell count per microliter of blood (total white blood cell [WBC] count in red; engineered [marked] cells in green). Right y-axis shows the number of copies of DNA used in the engineered T cells as a function of total genomic DNA in peripheral blood mononuclear cells (PBMCs) over time. (B). Distribution of engineered T cells in tissue samples obtained post mortem in both patients. qPCR for vector sequences was performed from total genomic DNA obtained from flash-frozen tissues. Note different scale on y-axis for Case 1 and Case 2.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/6/10.1182_blood-2013-03-490565/4/m_863f3.jpeg?Expires=1765893141&Signature=r2XXIYVGK2WkxpfvdA3CP3a7IbgDPzxGhxiiyGOzgC-p98bYv~YJXgAy8mdmi0FQiWdPjJJ20KkTAHLRGHUO0UJjvw6PTjhq0QuneGE3cFup9qjNsR5Km8uDD7dq-etTlstyEhthRup5fZ1GvGvwEPGSEBf7-IGN8RaJIhoKmYQuJ5U8JMNinOy~mgw8oUKnpz-6lyzONnnbsJgHY4kvFlUrhLAoC1BlspIrviRT7YdJjTG-wqGX1-H7SNylerUko5lH79EoytaduNkVxCR6grCSTaqJA~ILrna1-jJaBLRylKvnMF46Yqfc7qKrkpV0xIPCKWVfk0XhIEUSqyLJXg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

