In this issue of Blood, Cellot et al identify the histone demethylase Jarid1b as a key regulator of renewal and differentiation in hematopoietic stem cells (HSCs).1
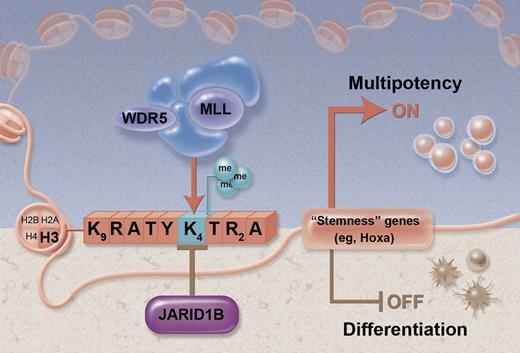
Proposed model for JARID1B activity in HSC regulation. The box of letters depicts the amino-terminal tail of histone H3. By counteracting the trimethylated status of lysine 4 (H3K4), JARID1B represses “stemness” loci and expression of genes associated with multipotency (brown shaded area). Conversely, knockdown of JARID1B shifts the balance toward the methylated status induced by the MLL/WDR5 complex to sustain multipotency (blue shaded area). Figure adapted from Figure 7 in the article by Cellot et al that begins on page 1545. Professional illustration by Alice Y. Chen.
Proposed model for JARID1B activity in HSC regulation. The box of letters depicts the amino-terminal tail of histone H3. By counteracting the trimethylated status of lysine 4 (H3K4), JARID1B represses “stemness” loci and expression of genes associated with multipotency (brown shaded area). Conversely, knockdown of JARID1B shifts the balance toward the methylated status induced by the MLL/WDR5 complex to sustain multipotency (blue shaded area). Figure adapted from Figure 7 in the article by Cellot et al that begins on page 1545. Professional illustration by Alice Y. Chen.
Stem cells are governed by epigenetic mechanisms that can either help maintain the necessary gene expression programs to preserve the stem cell state or ensure the establishment of new cell-specific expression programs during differentiation. One important level of epigenetic control originates from histone modifications such as methylation or acetylation.2 These modifications can be associated with either gene activation or gene repression.2 Histone methylation was originally believed to be a relatively permanent modification but the discovery of enzymes that can demethylate histones, about a decade ago, changed this perception entirely.3 It is now evident that histone methylation is both highly dynamic and reversible.
Since the first histone demethylase, LSD1, was described in 2004, around 30 additional histone demethylating enzymes have been identified, the vast majority belonging to the evolutionary conserved Jumonji C (JmjC) family.3 Functional studies of JmjC proteins have shown that they modulate the expression of specific gene sets associated with fundamental cellular functions such as self-renewal, proliferation, differentiation, and malignant progression.3 Many JmjC demethylases show distinct substrate specificity and have nonoverlapping functions. Therefore, understanding how each one of these enzymes function and how they may coordinately act together to influence gene expression poses a great challenge.
In this context, Cellot et al1 now report on an RNA interference (RNAi) screening strategy to functionally dissect the role of nearly all (23 of 27) known JmjC histone demethylases in HSCs. The Sauvageau laboratory had previously developed RNAi screening paradigms for other gene categories in mouse HSCs.4 In this new screen, Cellot et al1 applied a scoring system based on gene expression profile (the genes preferentially expressed in HSCs got a higher score) and the ability of short hairpin RNAs to alter the in vivo reconstitution kinetics of transplanted HSCs. From this analysis and subsequent validation assays, Cellot et al1 identified Jarid1b as a negative regulator and Jhdm1f as a putative positive regulator of HSC activity. They focused their subsequent work primarily on Jarid1b and could show that Jarid1b knockdown decreased hematopoietic differentiation in vitro while it promoted HSC activity and renewal both in vitro and in vivo. The findings clearly implicate Jarid1b as a novel and crucial mediator of HSC function.
Then Cellot et al1 performed global gene expression profiling to gain insights into the downstream effects of Jarid1b knockdown and to understand how the potential histone modifying effects would affect gene transcription. They found an upregulation of several known genes associated with increased HSC activity, in particular, genes of the 5′ Hoxa cluster.5 Jarid1b has mainly been coupled to demethylation of the H3K4me3 mark that is associated with active gene transcription.3 In agreement with this, Cellot et al1 now have found that several of the upregulated Hoxa genes were potential direct epigenetic targets, as they showed an enrichment of the H3K4me3 mark following Jarid1b knockdown. The proposed working model by Cellot et al,1 based on these findings, is outlined (see figure).
The theory put forward is based on the documented antagonistic actions of the MLL lysine methyltransferases that methylate histone H3K4 to activate transcription of key developmental genes (including the Hox genes), and the Polycomb group (PcG) family enzymes that methylate H3K27 to suppress the same genes.6,7 By counteracting the methylated status of H3K4, Jarid1b would shift the balance toward repression of these genes, and conversely its knockdown would favor gene activation (see figure). This is obviously a highly simplistic model. Many other histone marks and modifying enzymes are most certainly involved and it will take many more years of research to understand the precise function and relationships between these. Nevertheless, the ambitious approach taken here, beginning to dissect out the functional role of individual histone demethylases, is an important first step. Continued efforts in this direction, ideally coupled with gene knockout models, promise to shed new light on these complex processes.8 The RNAi screen perfomed by Cellot et al1 was primarily designed to identify the most prominent and specific targets. Due to stringent inclusion criteria of hits, as well as generic issues related to efficiency and target specificity of RNAi, it is possible that important HSC regulatory functions from other JmjC demethylases could have been missed in the screens.
Given that deregulation of JmjC histone methylases has been linked to cancer development,3 it is noteworthy that mice transplanted with HSCs bearing Jarid1b knockdown showed normal lymphoid and myeloid differentiation and never exhibited any signs of leukemia during long-term follow-up. It would certainly be of great interest to assess whether acquisition of additional genetic alterations providing more proliferative cues (in addition to enhanced stem cell maintenance triggered by Jarid1b knockdown) could lead to malignant transformation. However, it is also possible that the RNAi approach is not sufficient to achieve the maximum effects from the loss of Jarid1b. Future studies using conditional knockout mice for Jarid1b should help determine any potential links with leukemia.
Lessons learned during the last decade about the nonpermanent nature of histone methylation and the activity of histone demethylases emphasize the highly dynamic nature of the epigenetic state. By understanding the key modifiers of this state, such as histone demethylases, new therapeutic options for cancer can be developed. Interfering with the epigenetic state may also provide new opportunities for stem cell expansion. The work by Cellot et al1 indicates that Jarid1b may be one such target for future interventions.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal