Key Points
Using a mouse genetic mouse model of Ph+ B-lineage ALL, endogenous antiapoptotic MCL-1 is required for leukemia survival.
In BCR-ABL+ B-lineage ALL human and mouse cells, combining TKIs with small-molecule inhibitors of BCL-2 can potentiate sensitivity to cell death.
Abstract
The response of Philadelphia chromosome (Ph+) acute lymphoblastic leukemia (ALL) to treatment by BCR-ABL tyrosine kinase inhibitors (TKIs) has been disappointing, often resulting in short remissions typified by rapid outgrowth of drug-resistant clones. Therefore, new treatments are needed to improve outcomes for Ph+ ALL patients. In a mouse model of Ph+ B-lineage ALL, MCL-1 expression is dysregulated by the BCR-ABL oncofusion protein, and TKI treatment results in loss of MCL-1 expression prior to the induction of apoptosis, suggesting that MCL-1 may be an essential prosurvival molecule. To test this hypothesis, we developed a mouse model in which conditional allele(s) of Mcl-1 can be deleted either during leukemia transformation or later after the establishment of leukemia. We report that endogenous MCL-1’s antiapoptotic activity promotes survival during BCR-ABL transformation and in established BCR-ABL+ leukemia. This requirement for MCL-1 can be overcome by overexpression of other antiapoptotic molecules. We further demonstrate that strategies to inhibit MCL-1 expression potentiate the proapoptotic action of BCL-2 inhibitors in both mouse and human BCR-ABL+ leukemia cell lines. Thus, strategies focused on antagonizing MCL-1 function and expression would be predicted to be effective therapeutic strategies.
Introduction
Therapeutic strategies cure more than 80% of children with acute lymphoblastic leukemia (ALL), but some patients require intensive treatment and develop complications due to side effects.1 Furthermore, for adults the ALL survival rate remains below 40%, in part due to the dominant role of the BCR-ABL oncogene in adult ALL cases.1,2 In Philadelphia chromosome (Ph+) leukemia, the characteristic t(9;22) chromosomal translocation fuses a breakpoint cluster region (BCR) derived from human chromosome 22 to a portion of the c-ABL proto-oncogene from chromosome 9, leading to the formation of the BCR-ABL fusion oncoproteins p210BCR-ABL (p210) and p185BCR-ABL (p185) that are typically detected in chronic myelogenous leukemia (CML) and Ph+ ALL cells, respectively.3,4 Both p185 and p210 encode constitutively active tyrosine kinases essential for cell transformation.5
The treatment of BCR-ABL–expressing CML has been revolutionized by tyrosine kinase inhibitors (TKIs) of BCR-ABL like imatinib (IM), which induces and maintains remission in the majority of CML cases without serious side effects when administered as a single agent.6 However, ∼5% of CML patients per year develop resistance typically due to secondary mutations in the BCR-ABL oncogene.7 In contrast, the response of Ph+ B-lineage ALL (B-ALL) to treatment by TKIs results in short remissions and is typified by rapid outgrowth of drug-resistant clones.8 As a result, both pediatric and adult Ph+ ALL patients require maximally intensive combination chemotherapy and stem cell transplants, but even these measures are often unable to achieve long-term survival.1
Apoptosis is necessary to regulate the proper development and maintenance of tissue homeostasis in metazoans by eliminating damaged or obsolete cells. However, apoptotic dysregulation can lead to a variety of human pathologies including cancer. Intrinsic apoptosis is triggered when signals activate and/or induce BCL-2 homology domain-3 (BH3)-only molecules, promoting BAX and BAK activation facilitating the release of cytochrome c from the mitochondria that initiates apoptosome assembly and drives downstream caspase activation.9,10 Genetic loss of both Bax and Bak prevents the induction of intrinsic apoptosis.9 Antiapoptotic family members such as BCL-2, BCL-XL, BCL-w, BFL-1, and MCL-1 antagonize this process and as a result are often overexpressed to prevent cell death in malignant cells.11
A mouse model of human Ph+ B-ALL has been established by transducing Arf-deficient (Arf−/−) mouse bone marrow (BM) cells with either the p210 or p185 fusion proteins generating rapidly growing BCR-ABL–expressing pre–B lineage cells that lose interleukin-7 (IL-7) dependence.12 Cytokine signaling by IL-7 can overcome the effects of TKI treatment on the leukemic cells, whereas inhibition by a Janus kinase inhibitor blocked IL-7’s protective effects.12 Furthermore, leukemic cells derived from common γ-chain–deficient BM were more sensitive in vivo than wild-type leukemic cells to IM treatment.13 Thus, cytokine signaling can antagonize the effects of TKI treatment.
To identify whether Mcl-1 is a therapeutic target for treating Ph+ B-ALL, a mouse model of Ph+ B-ALL was developed in which Mcl-1 could be genetically ablated either during transformation or later in BCR-ABL–transformed cells. We demonstrate that endogenous MCL-1 expression is required both during initiation and maintenance of BCR-ABL+ leukemia. Furthermore, we demonstrate that TKIs can potentiate the killing induced by a selective BCL-2 antagonistic small molecule (navitoclax) in mouse and human BCR-ABL+ leukemic cell lines.
Materials and methods
Mice
Mcl-1–conditional, Bim-conditional, Bax-conditional Bak−/−, Arf−/−, and Rosa-ERCreT2 mice have been described previously.14-17 For in vivo deletion, mice transplanted with p185+ Rosa-ERCreT2 leukemia received 5 doses of tamoxifen (TAM) (1 µg per dose; Sigma-Aldrich, St. Louis, MO) emulsified in sunflower oil vehicle (Sigma-Aldrich) by gavage. All mice were bred in accordance with St. Jude Children’s Research Hospital (SJCRH) animal care and use committee.
Plasmids, expression constructs, and generation of mutants
Mcl-1GRRL changes glycine-243 to arginine and arginine-244 to leucine in mouse Mcl-1 by site-directed mutagenesis. Mutagenic polymerase chain reaction (PCR) primer sequences are available by request. Human BCL-XL and BCL-2 were from Dr D. Green (SJCRH, Memphis, TN). BCR-ABL (p185) plasmid was from Dr O. Witte (University of California Los Angeles, Los Angeles, CA). Stable cells were generated by retroviral transduction and puromycin selection (2 µg/mL final; Sigma-Aldrich).
Ecotropic retroviral production and cell transduction
Retroviruses were produced as previously described.18
BM transplantation
BM was harvested and transduced for 3 hours with retrovirus and 8 µg/mL polybrene in the absence of cytokines and was washed, and 1 million cells were injected into lethally irradiated (1100 rad) C57BL/6 recipients (The Jackson Laboratory, Bar Harbor, ME) by tail-vein intravenous route. Peripheral blood was monitored for leukemia, and recipients were observed for morbidity. Each transplant was performed on at least 2 initial BM infections. For secondary transplants, leukemic BM was mixed 1:1 with control BM and transplanted into lethally irradiated recipients (at least 10 recipients per initial leukemia).
Immunohistochemistry
Specimens were fixed in 10% formalin, paraffin embedded, and sectioned 4 µm thick. Sections were stained with hematoxylin and eosin and deparaffinized, and immunohistochemistry for Pax5 detection was performed using goat anti-human Pax5 (SC1974; Santa Cruz Biotechnology, Dallas, TX,). Goat polymer (GHP 516; BioCare Medical, Concord, CA) with deaminobenzidine detection system (TA125 HDX; ThermoShandon, Kalamazoo, MI) was used for visualization. Representative images from leukemic mice are presented.
Cells and cell culture
Mouse p185+Arf−/− B-ALL and human Ph+ cell lines (TOM1,19 OP-1,20 and BV17321 ) were grown in RPMI (Life Technologies, Carlsbad, CA) with 20% fetal bovine serum, 55 µM 2-mercaptoethanol, 2 mM glutamine, penicillin, and streptomycin. When indicated, recombinant IL-7 (20 ng/mL; Peprotech, Rocky Hill, NJ) was added to the culture media. IM (Novartis, Basal, Switzerland), dasatinib (DAS) (Bristol-Myers Squibb, New York City, NY), and navitoclax (ABT-263; Selleckchem, Houston, TX) solubilized in dimethylsulfoxide were added at indicated concentrations.
Western blotting, coimmunoprecipitation, and antibodies
Protein expression from cell lysates and immunoprecipitation was performed as previously described.22 Each immunoblot is representative of at least 3 separate experiments performed independently. The antibodies used were the following: anti–MCL-1 (Rockland Immunochemical, Gilbertsville, PA), anti-BIM (BD Biosciences, Franklin Lakes, NJ), anti–BCL-XL (BD Biosciences), anti–BCL-2 (Clone 6C8), phosphospecific and total STAT5 antisera (Dr J. Ihle, SJCRH), anti-phosphospecific and total ABL antisera (Cell Signaling, Beverly, MA), anti-Cre (EMD Chemical, Billerica, MA), anti–BCL-w (Cell Signaling), anti–BFL-1 (Pierce Biotechnology, Rockford, IL), anti-PARP (Cell Signaling), and antiactin mouse monoclonal (Millipore, Billerica, MA). Secondary antisera were anti-rabbit or anti-mouse horseradish peroxidase conjugated (Jackson ImmunoResearch, West Grove, PA).
Cell death experiments
Cell viability was determined by staining with annexin V–fluorescein isothiocyanate and propidium iodide (PI) (BD Biosciences) and flow cytometry. For experiments on mouse cell lines, each experiment was performed on at least 2 independently generated cells from in vitro–transduced mouse BM. Three separate experiments were performed, each in triplicate.
Analyses of Cre and Mcl-1 5′ LoxP site sequencing
PCR-amplified products of Cre complementary DNA (cDNA) (4 cell lines, 15 clones analyzed) or 5′ LoxP sites (3 lines, 12 clones) from isolated control (Mcl-1wt) or Mcl-1f/f p185-transformed leukemic cell lines were cloned and sequenced.
Results
BCR-ABL maintains MCL-1 expression in pre–B cells
Treatment of p185 BCR-ABL–expressing (hereafter referred to as p185+) Arf−/− cells with TKIs results in cell-cycle arrest and apoptosis induction that can be attenuated by exogenous cytokines including IL-7.12,13 To determine how inhibition of BCR-ABL signaling affects the expression of members of the BCL-2 family, p185+Arf−/− cells were treated with TKIs (Figure 1A-B). Exposure to either TKI, which blocked STAT5 and BCR-ABL phosphorylation, decreased MCL-1 expression but did not alter BCL-2 or BCL-XL expression (Figure 1A-B). The loss of MCL-1 was alleviated by adding exogenous IL-7 to the cultures, suggesting that signaling can bypass drug-dependent inhibition of BCR-ABL signaling and maintain MCL-1 expression (Figure 1A-B). Although other cytokines could render leukemic cells resistant to TKIs, IL-7 was the most potent in preventing TKI-induced cell-cycle arrest and apoptosis (H. Singh and R. K. Guy, SJCRH, written communication).
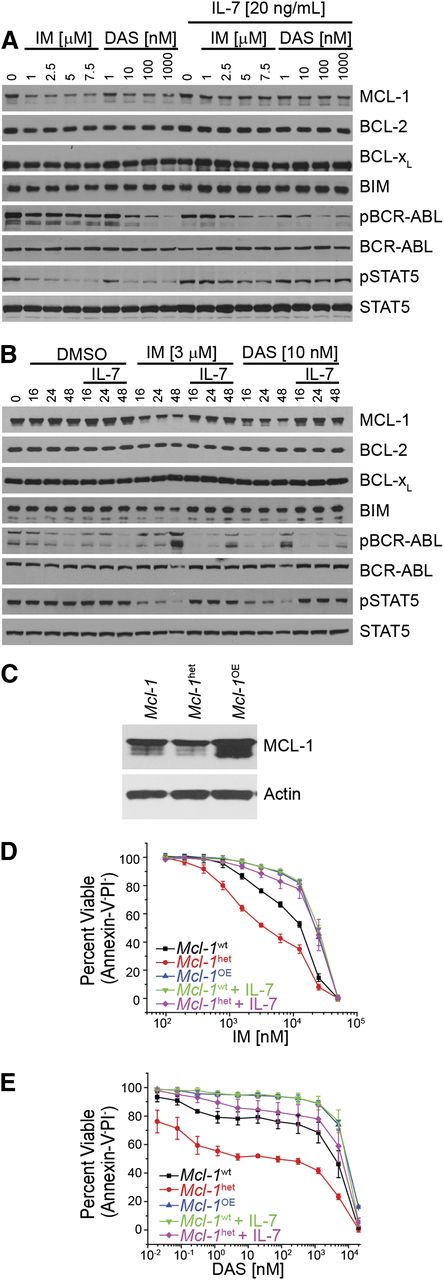
MCL-1 expression dictates response mediated by inhibition of BCR-ABL. (A-B) In vitro–generated BCR-ABL (p185)–expressing Arf−/− cells were treated with IM or DAS in the presence or absence or IL-7 (20 ng/mL) at varying concentrations (24 hours) (A) or with the indicated IM or DAS concentrations for different amounts of time (hours) (B). Cell lysates were resolved and immunoblotted for indicated targets. Anti-ABL antibody was used to detect the BCR-ABL fusion protein, and pBCR-ABL and pSTAT5 represent phosphorylated proteins. (C) Immunoblot of p185-expressing cells that were derived from wild-type Mcl-1 (Mcl-1wt), haploinsufficient (Mcl-1het, 1 floxed Mcl-1 allele deleted using p185-IRES-Cre retrovirus), or overexpressing ectopic Mcl-1 in addition to endogenous (Mcl-1OE) on an Arf−/− genetic background. Cell lysates were resolved and immunoblotted for MCL-1 and actin (loading control). (D-E) Cells described in (C) or p185-expressing Arf−/− pre–B cells were treated for 24 hours at indicated doses with IM (D) or DAS (E) after which cell death was determined. Cells were cultured in the presence of IL-7 (20 ng/mL) where indicated. Annexin-V and PI double-negative cells were scored as viable. Data points represent the average of 3 independent experiments (n = 3), and the error bars denote the standard error of the mean (SEM).
MCL-1 expression dictates response mediated by inhibition of BCR-ABL. (A-B) In vitro–generated BCR-ABL (p185)–expressing Arf−/− cells were treated with IM or DAS in the presence or absence or IL-7 (20 ng/mL) at varying concentrations (24 hours) (A) or with the indicated IM or DAS concentrations for different amounts of time (hours) (B). Cell lysates were resolved and immunoblotted for indicated targets. Anti-ABL antibody was used to detect the BCR-ABL fusion protein, and pBCR-ABL and pSTAT5 represent phosphorylated proteins. (C) Immunoblot of p185-expressing cells that were derived from wild-type Mcl-1 (Mcl-1wt), haploinsufficient (Mcl-1het, 1 floxed Mcl-1 allele deleted using p185-IRES-Cre retrovirus), or overexpressing ectopic Mcl-1 in addition to endogenous (Mcl-1OE) on an Arf−/− genetic background. Cell lysates were resolved and immunoblotted for MCL-1 and actin (loading control). (D-E) Cells described in (C) or p185-expressing Arf−/− pre–B cells were treated for 24 hours at indicated doses with IM (D) or DAS (E) after which cell death was determined. Cells were cultured in the presence of IL-7 (20 ng/mL) where indicated. Annexin-V and PI double-negative cells were scored as viable. Data points represent the average of 3 independent experiments (n = 3), and the error bars denote the standard error of the mean (SEM).
To assess whether MCL-1 expression correlates with the response of p185+Arf−/− cells to TKI inhibition, we generated p185+Arf−/− cells from either wild-type (Mcl-1wt) or Mcl-1 heterozygous BM. Loss of 1 Mcl-1 allele decreased MCL-1 protein levels by ∼50% (Figure 1C). We transduced and selected for p185+Arf−/− cells that constitutively overexpress MCL-1 protein (Figure 1C). When p185+Arf−/− cells expressing different levels of MCL-1 were treated with TKIs, MCL-1 expression level correlated with the sensitivity of the p185+Arf−/− cells to TKI treatment (Figure 1D-E). Consistent with its ability to induce MCL-1, IL-7 treatment reduced the cell-killing activity of both IM and DAS similar to the effects of MCL-1 overexpression and could enhance protection of Mcl-1 heterozygous cell lines (Figure 1D-E). Therefore, MCL-1 expression levels respond to growth factor and BCR-ABL signaling and dictate TKI sensitivity.
Requirement for MCL-1 during in vitro leukemia initiation
To determine whether MCL-1 is an important survival molecule during the transformation of pre–B cells with BCR-ABL, a genetic approach was used to delete 1 or both Mcl-1 genomic alleles by transducing BM cells from Mcl-1f/f, Mcl-1f/wt, or Mcl-1wt mice with a p185-IRES-Cre vector (Figure 2A). Simultaneous expression of p185 with Cre efficiently drove outgrowth of Mcl-1wt or Mcl-1f/wt p185-transformed cells; however, the outgrowth of p185+ cells from Mcl-1f/f BM was delayed (Figure 2A). Genomic analysis of the p185-transformed cells from the cultures confirmed that the Mcl-1 locus was deleted from p185+Mcl-1f/wt BM cultures, but there was no evidence of Mcl-1 deletion from p185+Mcl-1f/f BM cultures, indicating selection against Mcl-1 loss from cells during transformation (Figure 2C).
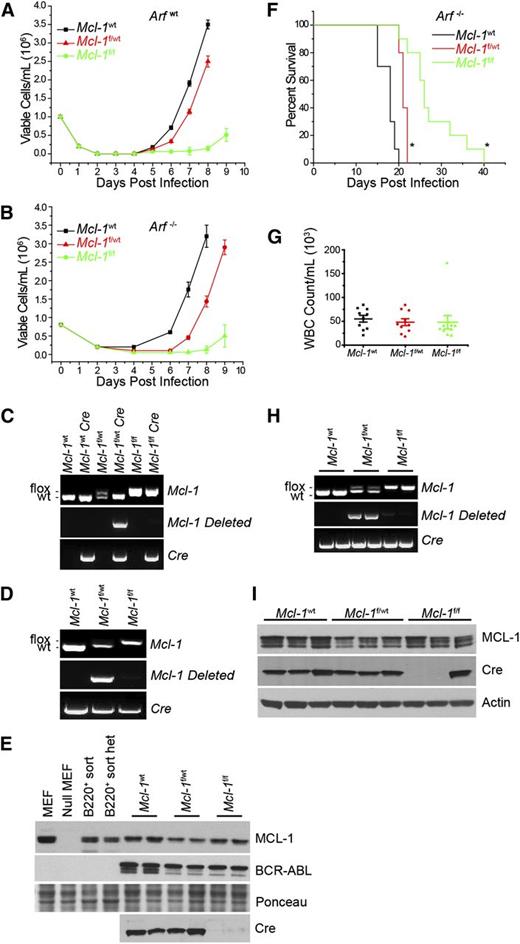
Requirement for MCL-1 during in vitro and in vivo BCR-ABL leukemia initiation. (A-B) BM from wild-type Mcl-1 (Mcl-1wt), Mcl-1f/wt, or Mcl-lf/f mice on Arfwt (A) or Arf−/− (B) genetic backgrounds was transduced with p185-IRES-Cre retrovirus to transform and delete the floxed allele. Viable cell concentration was assessed every 24 hours. Each time point represents an average of 3 independent experiments (n = 3), and the error bars denote SEM. Data represent 10 separate cultures from 2 independent mice. (C-D) Representative Mcl-1, Mcl-1–deleted, and Cre genomic genotyping preformed post outgrowth from Arfwt (C) and Arf−/− (D) genetic backgrounds. (E) Immunoblot analysis from cells that grew out in culture from the Arf−/− experiment detailed in (B) represent 2 independent experiments. Lysates were blotted for MCL-1, BCR-ABL (as detected by anti-ABL antibody), Cre, and Ponceau stain (loading control). (F) Arf−/− BM from Mcl-1wt (n = 10 mice), Mcl-1f/wt (n = 10 mice), or Mcl-1f/f (n = 10 mice) were transduced with p185-IRES-Cre retrovirus and immediately transplanted into lethally irradiated (1100 rad) C57BL/6 recipients. Data represent 10 individual leukemia recipients. BM from 2 independent donor mice was used. Recipient mice where monitored daily and euthanized when moribund. Asterisk (*) denotes P < .001 by log-rank test when compared with Mcl-1wt. (G) Whole blood cell (WBC) counts were taken from each mouse at the time of being euthanized. Each point represents 1 mouse, the horizontal line indicates the averages, and the error bars represent SEM. (H) Representative Mcl-1, deleted Mcl-1, and Cre genotyping of BM cells isolated from 2 representative moribund mice. (I) Representative immunoblots of BM from 3 individual moribund mice were blotted for MCL-1, Cre, and actin (loading control).
Requirement for MCL-1 during in vitro and in vivo BCR-ABL leukemia initiation. (A-B) BM from wild-type Mcl-1 (Mcl-1wt), Mcl-1f/wt, or Mcl-lf/f mice on Arfwt (A) or Arf−/− (B) genetic backgrounds was transduced with p185-IRES-Cre retrovirus to transform and delete the floxed allele. Viable cell concentration was assessed every 24 hours. Each time point represents an average of 3 independent experiments (n = 3), and the error bars denote SEM. Data represent 10 separate cultures from 2 independent mice. (C-D) Representative Mcl-1, Mcl-1–deleted, and Cre genomic genotyping preformed post outgrowth from Arfwt (C) and Arf−/− (D) genetic backgrounds. (E) Immunoblot analysis from cells that grew out in culture from the Arf−/− experiment detailed in (B) represent 2 independent experiments. Lysates were blotted for MCL-1, BCR-ABL (as detected by anti-ABL antibody), Cre, and Ponceau stain (loading control). (F) Arf−/− BM from Mcl-1wt (n = 10 mice), Mcl-1f/wt (n = 10 mice), or Mcl-1f/f (n = 10 mice) were transduced with p185-IRES-Cre retrovirus and immediately transplanted into lethally irradiated (1100 rad) C57BL/6 recipients. Data represent 10 individual leukemia recipients. BM from 2 independent donor mice was used. Recipient mice where monitored daily and euthanized when moribund. Asterisk (*) denotes P < .001 by log-rank test when compared with Mcl-1wt. (G) Whole blood cell (WBC) counts were taken from each mouse at the time of being euthanized. Each point represents 1 mouse, the horizontal line indicates the averages, and the error bars represent SEM. (H) Representative Mcl-1, deleted Mcl-1, and Cre genotyping of BM cells isolated from 2 representative moribund mice. (I) Representative immunoblots of BM from 3 individual moribund mice were blotted for MCL-1, Cre, and actin (loading control).
In clinically aggressive Ph+ B-ALL, the genetic deletion of the INK4-ARF locus occurs frequently.23 Furthermore, it has been shown that in the murine model of BCR-ABL B-ALL, Arf inactivation renders leukemic cells resistant to TKI treatment, enhances their survival in vivo, and facilitates the emergence of BCR-ABL kinase mutant, TKI-resistant clones.12 To test the requirement for MCL-1 in the in vitro establishment of BCR-ABL–dependent cells, BM cells from Mcl-1f/fArf−/−, Mcl-1f/wtArf−/−, or Mcl-1wtArf−/− mice were transduced with a p185-IRES-Cre vector. Transduction with p185 and Cre-recombinase drove outgrowth of Mcl-1wt or Mcl-1f/wt p185-transformed cells on an Arf−/− background (Figure 2B). In contrast, delayed outgrowth of p185+ cells from Mcl-1f/fArf−/− BM was observed (Figure 2B), similar to that observed in Arfwt cultures (Figure 2A). Genomic analysis of the p185+ cells from the cultures confirmed that deletion of the Mcl-1 locus was detectable from p185+Mcl-1f/wtArf−/− BM cultures, but there was no Mcl-1 deletion from p185+Mcl-1f/fArf−/− BM cultures, demonstrating that a strong selection against Mcl-1 loss also occurs even in the absence of Arf (Figure 2D). Analysis of protein expression confirmed that p185-IRES-Cre transduction in Mcl-1f/wtArf−/− BM causes a 50% reduction in MCL-1 protein levels; whereas, MCL-1 protein expression remained unchanged in Mcl-1f/fArf−/− BM cells indicating a lack of Mcl-1 deletion (Figure 2E). The p185-IRES-Cre–transduced Mcl-1f/fArf−/− cells in the cultures failed to express Cre protein despite possessing the Cre cDNA, implying that these cells must have mutated Cre to avoid deleting Mcl-1 (Figure 2D-E). Sequencing of Cre cDNA amplified from p185-IRES-Cre–transduced Mcl-1f/fArf−/− cells indicated that ∼40% of clones analyzed exhibited nonsense mutations in Cre and that ∼40% of clones possessed missense mutations with some possessing both types (supplemental Figure 1A; see the Blood Web site). In contrast, sequencing of control (p185-IRES-Cre–transduced Mcl-1wtArf−/−) leukemic cells did not reveal any Cre mutations illustrating the pressure against Mcl-1 deletion in the p185-IRES-Cre–transduced Mcl-1f/fArf−/− cells (data not shown). These data indicate that in Arfwt or Arf−/− genetic backgrounds, MCL-1 expression is required for BCR-ABL–dependent pre–B-cell transformation in vitro.
Requirement for MCL-1 during in vivo leukemia initiation
To test whether Mcl-1 is required for leukemia initiation in vivo, BM from Mcl-1f/fArf−/−, Mcl-1f/wtArf−/−, or Mcl-1wtArf−/−mice was transduced with a p185-IRES-Cre vector and transplanted into lethally irradiated (1100 rad) C57BL/6 recipient mice. Recipients receiving p185-IRES-Cre–transduced Mcl-1wtArf−/− BM cells rapidly succumbed to leukemia (Figure 2F). The loss of even a single allele of Mcl-1 (p185-IRES-Cre–transduced Mcl-1f/wtArf−/− BM) significantly delayed the onset of the lethal leukemia (Figure 2F). Transplanting p185-IRES-Cre–transduced Mcl-1f/fArf−/− BM cells, in which both Mcl-1 alleles should be deleted, delayed leukemia onset even more than loss of a single Mcl-1 allele; however, eventually all the recipients developed fatal leukemia (Figure 2F).
Although the Mcl-1 background affected the time of leukemia latency, analysis of the moribund mice did not reveal any pathological differences in the leukemia as measured by leukocyte counts, liver mass, spleen size, and histopathology (Figure 2G; supplemental Figure 2). Genomic analysis indicated that the Mcl-1 locus was deleted from cells recovered from moribund mice transplanted with p185-IRES-Cre–transduced Mcl-1f/wtArf−/− BM cells. In contrast, there was no evidence of Mcl-1 deletion in the leukemic blasts isolated from mice transplanted with p185-IRES-Cre–transduced Mcl-1f/fArf−/− BM cells, indicating in vivo selection against Mcl-1 deletion during BCR-ABL–driven leukemogenesis (Figure 2H). Protein analysis confirmed that leukemia from mice transplanted with p185-IRES-Cre–transduced Mcl-1f/wtArf−/− BM expressed less MCL-1 protein than Mcl-1wt leukemic cells, consistent with deletion of the conditional Mcl-1 allele (Figure 2I). However, MCL-1 protein was still detectable in the leukemic cells from mice transplanted with p185-IRES-Cre–transduced Mcl-1f/fArf−/− BM indicating a lack of deletion. This was due, at least in part, to the failure to express Cre protein due to nonsense mutations in Cre (Figure 2I; supplemental Figure 1B). Additionally, in other leukemic samples we also detected missense mutations that resulted in mutant Cre protein (supplemental Figure 1C). Thus, when Cre was coexpressed during the transformation with p185, there was a strong selection against Mcl-1 deletion during in vitro and in vivo leukemogenesis.
Requirement for antiapoptotic function in initiation of leukemia in vitro
To assess whether MCL-1 loss induces the death of the cells, BM was isolated from Mcl-1f/fBaxf/fBak−/− mice for p185-IRES-Cre transduction, and the outgrowth of leukemic cells from the cultures followed (Figure 3A). Control Mcl-1wtBaxf/fBak−/− BM cells rapidly expanded from culture consistent with a loss of both proapoptotic effectors BAX and BAK (Figure 3A). Similarly, leukemia from the Mcl-1f/fBaxf/fBak−/−–transduced BM expanded from the cultures but exhibited a delay when compared with Mcl-1wtBaxf/fBak−/− or Mcl-1wt cultures (Figure 3A). In contrast, the outgrowth from Mcl-1f/f cultures was dramatically delayed (Figure 3A). Expression analyses indicated that unlike the p185+ cells that grew from the Mcl-1f/f cultures, which fail to delete MCL-1 or express Cre protein, the p185+ cells from Mcl-1f/fBaxf/fBak−/− cultures deleted MCL-1, BAX, and BAK and expressed the Cre-recombinase (Figure 3B,D). Thus, removing the intrinsic death pathway, by Bax and Bak ablation, rescued the outgrowth of Mcl-1–deleted p185+ cells in vitro implying that a consequence of MCL-1 loss is apoptosis induction.
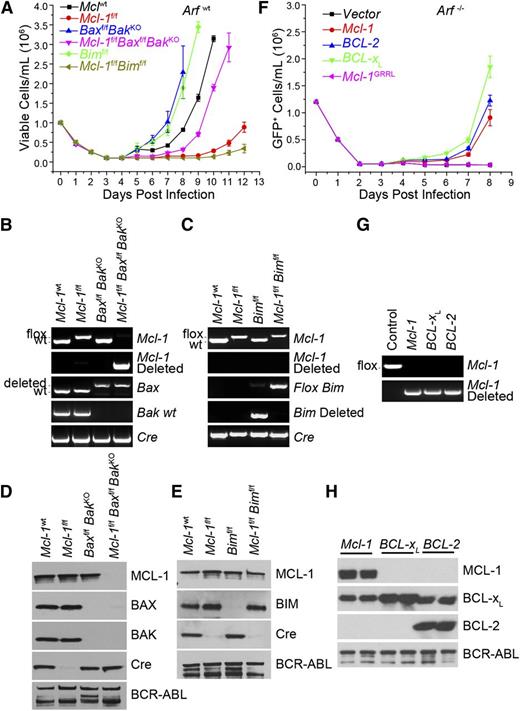
Requirement for antiapoptotic function in initiation of BCR-ABL leukemia. (A) BM from Mcl-1wt, Mcl-lf/f, Baxf/fBaxKO, Baxf/fBaxKOMcl-1f/f, Bimf/f, and Mcl-1f/fBimf/f mice (on an Arfwt background) was transduced with p185-IRES-Cre retrovirus to transform and delete floxed alleles. Viable cell concentration was determined every 24 hours. Each time point represents the average of 3 independent experiments (n = 3) each composed of 2 separate initiations, and the error bars denote the SEM. (B) Representative Mcl-1, Mcl-1–deleted, Bax, Bak, and Cre genomic genotyping post outgrowth from experiment detailed in panel A. (C) Representative Mcl-1, Mcl-1–deleted, Bim, Bim-deleted, and Cre genomic genotyping shown from post-outgrowth cells detailed in panel A. (D) Representative immunoblot of the cells that grew out in culture from the experiment detailed in panel A. Lysates were blotted for MCL-1, BAX, BAK, Cre, and BCR-ABL (as detected by anti-ABL antibody) (loading control). (E) Representative immunoblot analysis of the cells that grew out in culture from the experiment detailed in panel A. Lysates were blotted for MCL-1, BIM, Cre, and BCR-ABL (loading control). (F) Mcl-1f/fArf−/− pre–B cells were cotransduced with Mcl-1, BCL-2, BCL-XL, or Mcl-1GRRL constructs and p185-IRES-Cre retrovirus. Cell viability was determined every 24 hours. Each time point represents 3 independent experiments (n = 3) composed of 2 separate initiations, and the error bars denote the SEM. (G) Representative Mcl-1 and Mcl-1–deleted genotyping from Mcl-1f/fArf−/− pre–B cells cotransduced with indicated construct and p185-IRES-Cre retrovirus post outgrowth. (H) Lysates from Mcl-1f/fArf−/− pre–B cells cotransduced with indicated constructs and p185-IRES-Cre retrovirus post outgrowth were western blotted for MCL-1, BCL-XL, BCL-2 (human-specific antibody), and BCR-ABL (loading control). Results indicate 2 independent cultures.
Requirement for antiapoptotic function in initiation of BCR-ABL leukemia. (A) BM from Mcl-1wt, Mcl-lf/f, Baxf/fBaxKO, Baxf/fBaxKOMcl-1f/f, Bimf/f, and Mcl-1f/fBimf/f mice (on an Arfwt background) was transduced with p185-IRES-Cre retrovirus to transform and delete floxed alleles. Viable cell concentration was determined every 24 hours. Each time point represents the average of 3 independent experiments (n = 3) each composed of 2 separate initiations, and the error bars denote the SEM. (B) Representative Mcl-1, Mcl-1–deleted, Bax, Bak, and Cre genomic genotyping post outgrowth from experiment detailed in panel A. (C) Representative Mcl-1, Mcl-1–deleted, Bim, Bim-deleted, and Cre genomic genotyping shown from post-outgrowth cells detailed in panel A. (D) Representative immunoblot of the cells that grew out in culture from the experiment detailed in panel A. Lysates were blotted for MCL-1, BAX, BAK, Cre, and BCR-ABL (as detected by anti-ABL antibody) (loading control). (E) Representative immunoblot analysis of the cells that grew out in culture from the experiment detailed in panel A. Lysates were blotted for MCL-1, BIM, Cre, and BCR-ABL (loading control). (F) Mcl-1f/fArf−/− pre–B cells were cotransduced with Mcl-1, BCL-2, BCL-XL, or Mcl-1GRRL constructs and p185-IRES-Cre retrovirus. Cell viability was determined every 24 hours. Each time point represents 3 independent experiments (n = 3) composed of 2 separate initiations, and the error bars denote the SEM. (G) Representative Mcl-1 and Mcl-1–deleted genotyping from Mcl-1f/fArf−/− pre–B cells cotransduced with indicated construct and p185-IRES-Cre retrovirus post outgrowth. (H) Lysates from Mcl-1f/fArf−/− pre–B cells cotransduced with indicated constructs and p185-IRES-Cre retrovirus post outgrowth were western blotted for MCL-1, BCL-XL, BCL-2 (human-specific antibody), and BCR-ABL (loading control). Results indicate 2 independent cultures.
Proapoptotic BIM participates in the induction of cell death in BCR-ABL+ leukemia treated with TKIs.24-26 To test whether Bim loss could allow Mcl-1–deleted p185+ cells to survive, Mcl-1f/fBimf/f BM was transduced with p185-IRES-Cre and cultured. Surprisingly, although loss of Bim alone hastened the outgrowth of cultures when compared with controls, codeletion of Mcl-1 and Bim delayed the outgrowth even more than Mcl-1 deletion alone (Figure 3A). Furthermore, the cells that grew out of the Mcl-1f/fBimf/f cultures failed to delete the genomic DNA for either Mcl-1 or Bim and maintained expression of both proteins (Figure 3C,E). Similar to Mcl-1f/f cultures, the Mcl-1f/fBimf/f cells transduced with p185-IRES-Cre failed to express Cre-recombinase, again due to mutations in Cre (Figure 3E; supplemental Figure 1D). Thus, the loss of BIM expression was insufficient to allow the outgrowth of p185+Mcl-1–deleted cells, indicating that additional proapoptotic molecules must induce death upon Mcl-1 deletion.
We mutated MCL-1’s BH3-domain binding groove by mutating glycine-243 to arginine and arginine-244 to leucine (referred to as MCL-1GRRL). To validate that this mutant abolishes MCL-1’s antiapoptotic function, MCL-1GRRL was stably expressed to similar levels as MCL-1wt in simian virus 40–transformed Mcl-1f/f Rosa-ERCreT2 murine embryonic fibroblasts (MEFs) that, when treated with TAM, induced deletion of the endogenous Mcl-1 (supplemental Figure 3A). Mcl-1 deletion rendered MEFs susceptible to staurosporine-induced death, and ectopic expression of MCL-1wt rendered them resistant (supplemental Figure 3B). In contrast, stable expression of the MCL-1GRRL mutant could not protect the MEFs from apoptosis indicating that the mutant is nonfunctional in antagonizing death (supplemental Figure 3B). MCL-1wt co-immunoprecipitated with proapoptotic BIM; however, no interaction was detectable between BIM and MCL-1GRRL protein (supplemental Figure 3C). These data demonstrate that the MCL-1GRRL lacks the ability to sequester proapoptotic modulators, such as BIM, and therefore cannot prevent apoptosis.
To test whether MCL-1’s antiapoptotic function is essential for leukemia in vitro initiation, Mcl-1f/fArf−/− pre–B cells were cotransduced with IRES-GFP vector expressing either vector alone, MCL-1wt, or MCL-1GRRL along with p185-IRES-Cre to simultaneously transform and delete endogenous Mcl-1. Expression of Mcl-1wt but not empty vector efficiently promoted the outgrowth of p185-IRES-Cre–transduced Mcl-1f/fArf−/− cells from the cultures (Figure 3F). In contrast, the expression of MCL-1GRRL was insufficient to promote the outgrowth of cells from the cultures indicating that MCL-1’s antiapoptotic function is required for promoting leukemic initiation in vitro (Figure 3F).
Because MCL-1’s ability to inhibit cell death appears to be required for leukemic cell survival, we tested whether overexpression of other antiapoptotic BCL-2 family members might suffice to protect the BM cells during in vitro leukemic initiation. Mcl-1f/fArf−/− BM was transduced with both p185-IRES-Cre and MIG-vector expressing Mcl-1wt, human BCL-2, and human BCL-XL, then cultured in the absence of growth factors, and monitored daily for the outgrowth of GFP+ cells. Expression of Mcl-1wt but not empty vector promoted the outgrowth of p185-IRES-Cre–transduced Mcl-1f/fArf−/− cells from the cultures (Figure 3F). Additionally, expression of either BCL-2 or BCL-XL supported the outgrowth of p185-IRES-Cre–transduced Mcl-1f/fArf−/− cells indicating that ectopic expression of antiapoptotic molecules rescued the survival of the cells lacking Mcl-1 (Figure 3F). Mcl-1 deletion was readily detectable by PCR from p185-IRES-Cre–transduced Mcl-1f/fArf−/− cells expressing MCL-1, BCL-2, or BCL-XL indicating that deletion in the cells ectopically expressing antiapoptotic molecules was tolerated (Figure 3G). Likewise, analyses of the cultures after cotransduction demonstrated deletion of MCL-1 and corresponding expression of ectopic MCL-1, BCL-2, or BCL-XL (Figure 3H). These data indicated that concomitant expression of exogenous antiapoptotic BCL-2 family members rescued the death induced by deletion of endogenous Mcl-1 during p185-IRES-Cre transformation.
In vitro requirement for MCL-1 in maintenance of leukemia survival
BCR-ABL transformation results in the activation of many cellular signaling pathways that promote the proliferation and survival of leukemic cells.27,28 To identify whether MCL-1 is also required for the continual survival of BCR-ABL–transformed cells, we established IL-7–dependent immortalized but not transformed Mcl-1f/fArf−/− pre–B cells generated from Mcl-1f/fArf−/− mouse BM. These cells were transduced with p185-IRES-Luciferase to promote cytokine and stroma-independent growth (referred to hereafter as p185+Mcl-1f/fArf−/− B-ALL cells).12 To test whether MCL-1 is essential for promoting the survival of fully transformed cells, either vector or Mcl-1wt cDNA was stably expressed in the cells. The p185+Mcl-1f/fArf−/− B-ALL cells stably expressing either MSCV-vector or Mcl-1 were then transduced with Cre-IRES-GFP (MIG-Cre), and the growth of GFP-expressing cells was followed. In vector-expressing cells, there was no outgrowth of GFP+ cells indicating that Cre expression prevented the expansion of the p185+Mcl-1f/fArf−/− B-ALL cells (Figure 4A). Cre expression induced death in the cultures, but because the dead cells are GFP–, they could not be differentiated from non-MIG-Cre–transduced cells (data not shown). Even when IL-7 was added to the cultures, Cre expression prevented the outgrowth of p185+Mcl-1f/fArf−/− B-ALL cells indicating that exogenous growth factor additionally was insufficient to overcome Mcl-1 deletion (Figure 4A). In contrast, expression of MCL-1wt in the p185+Mcl-1f/fArf−/− B-ALL cells promoted GFP+ Cre-expressing p185+Mcl-1f/fArf−/− B-ALL cell expansion demonstrating that ectopic MCL-1wt expression could rescue the apoptosis induced by Mcl-1 deletion (Figure 4A-B).
Requirement for antiapoptotic function in leukemia maintenance. (A) In vitro–generated Mcl-1f/fArf−/− p185+ cells stably expressing either vector or ectopic Mcl-1 were transduced with Cre-IRES-GFP to delete the endogenous Mcl-1. The total number of GFP+ cells was measured every 24 hours. IL-7 was used at a final concentration of 20 ng/mL. Each point represents the average of 3 independent experiments (n = 3), and the error bars denote the SEM. (B) Immunoblot analysis of p185+Mcl-1f/fArf−/− pre–B cells before and after Cre-IRES-GFP transduction was performed. Lysates were western blotted for MCL-1, Cre, and actin (loading control). (C) Mcl-1f/fArf−/− p185+ pre–B cells stably expressing indicated constructs were transduced with Cre-IRES-GFP to delete endogenous Mcl-1. The total number of GFP+ cells was measured every 24 hours. Each point represents the average of 3 independent experiments (n = 3), and the error bars denote the SEM. (D) Immunoblot analysis shown for Mcl-1f/fArf−/− p185+ pre–B cells before and after Cre-IRES-GFP transduction for MCL-1, BCL-2 (human-specific antibody), BCL-XL, Cre, and Actin (loading control).
Requirement for antiapoptotic function in leukemia maintenance. (A) In vitro–generated Mcl-1f/fArf−/− p185+ cells stably expressing either vector or ectopic Mcl-1 were transduced with Cre-IRES-GFP to delete the endogenous Mcl-1. The total number of GFP+ cells was measured every 24 hours. IL-7 was used at a final concentration of 20 ng/mL. Each point represents the average of 3 independent experiments (n = 3), and the error bars denote the SEM. (B) Immunoblot analysis of p185+Mcl-1f/fArf−/− pre–B cells before and after Cre-IRES-GFP transduction was performed. Lysates were western blotted for MCL-1, Cre, and actin (loading control). (C) Mcl-1f/fArf−/− p185+ pre–B cells stably expressing indicated constructs were transduced with Cre-IRES-GFP to delete endogenous Mcl-1. The total number of GFP+ cells was measured every 24 hours. Each point represents the average of 3 independent experiments (n = 3), and the error bars denote the SEM. (D) Immunoblot analysis shown for Mcl-1f/fArf−/− p185+ pre–B cells before and after Cre-IRES-GFP transduction for MCL-1, BCL-2 (human-specific antibody), BCL-XL, Cre, and Actin (loading control).
In vitro requirement for MCL-1’s antiapoptotic function in leukemia maintenance
To test whether MCL-1’s antiapoptotic function is required for the survival of already existing transformed leukemic cells, Mcl-1wt and Mcl-1GRRL mutant cDNA or vector were stably expressed in p185+Mcl-1f/fArf−/− B-ALL cells. The cells were then transduced in parallel with MIG-Cre, and the growth of GFP-expressing (Cre+) cells was followed. MCL-1wt expression promoted the efficient outgrowth of GFP+ cells from the cultures, but no GFP+ cells were expanded from cultures expressing empty vector (Figure 4C). Similarly, expression of the nonfunctional MCL-1GRRL mutant could not support the outgrowth of Mcl-1–deleted cells (Figure 4C). These data indicate that BCR-ABL+Arf−/− B-ALL cells required MCL-1’s antiapoptotic function.
Protein analyses demonstrated that endogenous MCL-1, BCL-2, and BCL-XL are expressed in the unmanipulated BCR-ABL–transformed B-ALL cells, but the loss of MCL-1 was not tolerated in these cells indicating that the endogenous amounts of BCL-2 and BCL-XL are insufficient to maintain survival (Figure 4D). To address whether overexpression of other antiapoptotic BCL-2 family members could overcome Mcl-1 deletion in the BCR-ABL–transformed B-ALL cells, BCL-2 or BCL-XL were stably expressed in the p185+Mcl-1f/fArf−/− B-ALL cells and transduced with MIG-Cre, and the outgrowth of GFP-expressing cells was followed. Ectopic expression of human BCL-2 or BCL-XL promoted the survival and outgrowth of GFP+ cells from the cultures (Figure 4C). Furthermore, protein analysis of the GFP+ cells after MIG-Cre transduction confirmed that these cells lacked MCL-1 expression when they expressed exogenous antiapoptotic cDNAs (Figure 4D). Therefore, these data indicate that even transformed BCR-ABL+ B-ALL cells require endogenous MCL-1 to promote their survival and growth, but that the requirement for MCL-1 can be supplanted by overexpression of other antiapoptotic BCL-2 family members.
In vivo requirement for antiapoptotic MCL-1 function in leukemia maintenance
We bred Mcl-1 conditional mice to mice bearing a TAM-activatable Cre (Rosa-ERCreT2) to delete Mcl-1 in leukemic cells in vivo by TAM administration. The CreERT2 fusion protein is expressed regardless of the TAM treatment but only localizes to the nucleus when TAM is present. BM from Mcl-1f/f Rosa-ERCreT2 or Mcl-1wt Rosa-ERCreT2 (control) mice were infected with the p185-IRES-GFP virus and transplanted into lethally irradiated recipients. After ∼30 days, recipient mice exhibiting signs of leukemia were euthanized, and leukemic BM (>50% GFP+ cells) was mixed 1:1 with CD45.1+ wild-type BM and intravenously transplanted into lethally irradiated recipients. Five days after the secondary transplant, recipients either received 5 doses of TAM (1 µg per dose per day) or vehicle and were monitored for signs of leukemia progression and euthanized when moribund. Both TAM- and vehicle-treated mice harboring control leukemic cells rapidly developed a fatal leukemia indicating that TAM-induced Cre activation did not alter leukemia progression (Figure 5A). Strikingly, although the p185-transformed, Mcl-1f/f Rosa-ERCreT2 vehicle-treated group all succumbed to leukemia (median survival 20 days), the progression of leukemia in the TAM-treated group was significantly delayed (median survival 46 days) with ∼40% TAM-treated mice remaining leukemia free for >200 days since leukemia transplant (Figure 5A).
Requirement for MCL-1 during in vivo BCR-ABL leukemia maintenance. (A) Mcl-1f/f Rosa-ERCreT2 (Arfwt) or Mcl-1wt Rosa-ERCreT2 BM was transduced with p185-IRES-GFP and transplanted into lethally irradiated (1100 rad) C57BL/6 recipients that were monitored for leukemia initiation. After leukemia initiation, BM from the leukemic mice (2 independent leukemia donors per genotype) was harvested and mixed 1:1 with control CD45.1 congenic BM and transplanted into secondary, lethally irradiated (1100 rad) C57BL/6 recipients (∼10 secondary recipients each for 2 separate leukemic donors). After 5 days, the secondary recipients were treated with 5 daily doses of TAM (1 µg/d) to activate Cre (n = 13 mice for Mcl-1f/f Rosa-ERCreT2 and n = 10 for Mcl-1wt Rosa-ERCreT2 donors) or control vehicle (n = 9 mice for Mcl-1f/f Rosa-ERCreT2 and n = 9 for Mcl-1wt Rosa-ERCreT2 donors) by gavage. Mice were monitored daily and euthanized when moribund. Asterisk (*) denotes P < .001 by log-rank test. (B) White blood cell (WBC) counts were taken from moribund, secondary recipients described in panel A at the time of being euthanized. Bars indicate the averages (n = 6 mice for TAM-treated Mcl-1f/f Rosa-ERCreT2 donors, n = 9 for TAM-treated Mcl-1wt Rosa-ERCreT2 donors, n = 9 mice for vehicle-treated Mcl-1f/f Rosa-ERCreT2 donors, and n = 9 for vehicle-treated Mcl-1wt Rosa-ERCreT2 donors), and error bars indicate SEM. (C) Representative immunoblot shown of GFP+ BM isolated from TAM-treated, moribund mice that received p185+Mcl-1f/f Rosa-ERCreT2 leukemic BM (each lane represents a recipient mouse). Lysates were blotted for MCL-1, BCR-ABL (as detected by anti-ABL antibody), and ERCreT2 protein as a loading control (note: ERCreT2 fusion protein is present in all cells but is only activated by TAM). (D) Representative Mcl-1, Mcl-1–deleted, and Cre genotyping of GFP+ BM isolated from TAM-treated, moribund mice that received p185+Mcl-1f/f Rosa-ERCreT2 leukemic BM (each lane represents a recipient mouse). (E) Histopathological examination of moribund vehicle-treated or TAM-treated mice that received p185+Mcl-1f/f Rosa-ERCreT2 leukemic BM. Images of indicated tissues (×50 magnification) are representative of 9 vehicle-treated mice and 4 moribund, TAM-treated mice analyzed. Hematoxylin and eosin (H&E) and PAX5 immunohistochemistry are morphologically and immunophenotypically indicative of B-lineage leukemia.
Requirement for MCL-1 during in vivo BCR-ABL leukemia maintenance. (A) Mcl-1f/f Rosa-ERCreT2 (Arfwt) or Mcl-1wt Rosa-ERCreT2 BM was transduced with p185-IRES-GFP and transplanted into lethally irradiated (1100 rad) C57BL/6 recipients that were monitored for leukemia initiation. After leukemia initiation, BM from the leukemic mice (2 independent leukemia donors per genotype) was harvested and mixed 1:1 with control CD45.1 congenic BM and transplanted into secondary, lethally irradiated (1100 rad) C57BL/6 recipients (∼10 secondary recipients each for 2 separate leukemic donors). After 5 days, the secondary recipients were treated with 5 daily doses of TAM (1 µg/d) to activate Cre (n = 13 mice for Mcl-1f/f Rosa-ERCreT2 and n = 10 for Mcl-1wt Rosa-ERCreT2 donors) or control vehicle (n = 9 mice for Mcl-1f/f Rosa-ERCreT2 and n = 9 for Mcl-1wt Rosa-ERCreT2 donors) by gavage. Mice were monitored daily and euthanized when moribund. Asterisk (*) denotes P < .001 by log-rank test. (B) White blood cell (WBC) counts were taken from moribund, secondary recipients described in panel A at the time of being euthanized. Bars indicate the averages (n = 6 mice for TAM-treated Mcl-1f/f Rosa-ERCreT2 donors, n = 9 for TAM-treated Mcl-1wt Rosa-ERCreT2 donors, n = 9 mice for vehicle-treated Mcl-1f/f Rosa-ERCreT2 donors, and n = 9 for vehicle-treated Mcl-1wt Rosa-ERCreT2 donors), and error bars indicate SEM. (C) Representative immunoblot shown of GFP+ BM isolated from TAM-treated, moribund mice that received p185+Mcl-1f/f Rosa-ERCreT2 leukemic BM (each lane represents a recipient mouse). Lysates were blotted for MCL-1, BCR-ABL (as detected by anti-ABL antibody), and ERCreT2 protein as a loading control (note: ERCreT2 fusion protein is present in all cells but is only activated by TAM). (D) Representative Mcl-1, Mcl-1–deleted, and Cre genotyping of GFP+ BM isolated from TAM-treated, moribund mice that received p185+Mcl-1f/f Rosa-ERCreT2 leukemic BM (each lane represents a recipient mouse). (E) Histopathological examination of moribund vehicle-treated or TAM-treated mice that received p185+Mcl-1f/f Rosa-ERCreT2 leukemic BM. Images of indicated tissues (×50 magnification) are representative of 9 vehicle-treated mice and 4 moribund, TAM-treated mice analyzed. Hematoxylin and eosin (H&E) and PAX5 immunohistochemistry are morphologically and immunophenotypically indicative of B-lineage leukemia.
At the time of being euthanized, moribund mice from both the TAM-treated and control groups exhibited GFP+ cells in their peripheral blood and organs consistent with a p185+ leukemia (Figure 5B; supplemental Figure 4). The resulting leukemic (GFP+) cells from the TAM-treated moribund mice still expressed MCL-1 despite TAM treatment (Figure 5C). To address how the leukemic cells had escaped deletion, the 5′ LoxP sites of the Mcl-1 conditional allele were amplified from leukemic cells isolated from moribund, TAM-treated recipients bearing p185-transformed Mcl-1f/f Rosa-ERCreT2 leukemic BM. Sequencing revealed deletions and mutations in the 5′ loxP Mcl-1 site in 80% of the surviving leukemic cells (supplemental Figure 5A). Intriguingly, in some moribund TAM-treated mice, we detected the deletion of 1 Mcl-1 genomic allele but also found mutations in the 5′ loxP site of the other (nondeleted) allele (Figure 5D; supplemental Figure 5B). Histopathologically, the leukemia arising in the moribund mice was pathologically similar whether the mice received TAM or vehicle treatment and uniformly expressed Pax5, a marker of B-lineage leukemia (Figure 5E; supplemental Figure 3). These data indicate that Mcl-1 deletion in vivo delayed leukemia progression and that those TAM-treated recipients that succumbed to disease exhibited selection against Mcl-1 deletion in the leukemic cells.
Potentiation of TKI activity by small-molecule BCL-2 inhibitors in mouse BCR-ABL leukemia
The small-molecule inhibitor navitoclax (ABT-263) specifically inhibits the antiapoptotic function of BCL-2 and BCL-XL and exhibits efficacy in BCL-2–dependent cancers with limited toxicity.29-31 However, a common feature mediating tumor cell resistance to navitoclax and its progenitor compound (ABT-737) is the expression of other antiapoptotic proteins, like MCL-1, which are not targeted by these compounds.32-34 As MCL-1 expression often correlates with tumor cell resistance to the cell-killing activity of navitoclax, we tested the death sensitivity of in vitro–generated p185+Mcl-1f/fArf−/− B-ALL cell lines in which endogenous Mcl-1 was deleted by Cre and replaced by other antiapoptotic molecules. As expected, the BCR-ABL B-ALL cells expressing exogenous BCL-2 or BCL-XL, but lacking endogenous Mcl-1 expression, were more sensitive to navitoclax than p185+Mcl-1wtArf−/− B-ALL cells (Figure 6A). Furthermore, p185+Mcl-1wtArf−/− B-ALL cells overexpressing MCL-1 were even more resistant to navitoclax than p185+Mcl-1wtArf−/− B-ALL cells indicating that MCL-1’s expression level correlated with navitoclax sensitivity (Figure 6A).
Potentiation of mouse BCR-ABL+leukemia to combining TKI and small-molecule BCL-2 inhibitors. (A) In vitro–generated Mcl-1–deleted Arf−/− p185+ cells stably expressing the indicated constructs were treated with indicated doses of navitoclax (ABT-263) for 24 hours after which the percent viable cells was determined (PI-negative cells were scored as viable). Each point represents the average of 3 independent experiments, and the error bars denote the SEM. p185+Arf−/− cells serve as a control. (B) Arf-null p185+ cells were treated with navitoclax for 24 hours at indicated doses. Lysates were immunoprecipitated with anti-BIM antibody, and immune complexes were resolved and western blotted for MCL-1, BCL-2 (human specific antibody), BCL-XL, BIM, PARP, and actin (loading control). Endogenous BIM serves as the control for equal immunoprecipitation. (C-D) p185+Arf−/− cells were treated with IM (C) or DAS (D) for 24 hours at indicated doses. Lysates were immunoprecipitated with anti-BIM antibody, and immune complexes were resolved and western blotted for MCL-1, BCL-2, BCL-XL, BIM, PARP, and actin (loading control). Endogenous BIM served as the control for equal immunoprecipitation. (E-F) p185+Arf−/− cells were treated with indicated doses of navitoclax and/or IM (E) or DAS (F) for 24 hours after which the percent viable cells was determined. Annexin-V/PI–negative cells were scored as viable. IL-7 was added to a final concentration of 20 ng/mL. Bars represent the average of 3 independent experiments done in triplicate, and the error bars denote the SEM. Asterisk (*) indicates P < .001 by 2-way analysis of variance with a Bon Ferroni posttest between treatments linked with horizontal line.
Potentiation of mouse BCR-ABL+leukemia to combining TKI and small-molecule BCL-2 inhibitors. (A) In vitro–generated Mcl-1–deleted Arf−/− p185+ cells stably expressing the indicated constructs were treated with indicated doses of navitoclax (ABT-263) for 24 hours after which the percent viable cells was determined (PI-negative cells were scored as viable). Each point represents the average of 3 independent experiments, and the error bars denote the SEM. p185+Arf−/− cells serve as a control. (B) Arf-null p185+ cells were treated with navitoclax for 24 hours at indicated doses. Lysates were immunoprecipitated with anti-BIM antibody, and immune complexes were resolved and western blotted for MCL-1, BCL-2 (human specific antibody), BCL-XL, BIM, PARP, and actin (loading control). Endogenous BIM serves as the control for equal immunoprecipitation. (C-D) p185+Arf−/− cells were treated with IM (C) or DAS (D) for 24 hours at indicated doses. Lysates were immunoprecipitated with anti-BIM antibody, and immune complexes were resolved and western blotted for MCL-1, BCL-2, BCL-XL, BIM, PARP, and actin (loading control). Endogenous BIM served as the control for equal immunoprecipitation. (E-F) p185+Arf−/− cells were treated with indicated doses of navitoclax and/or IM (E) or DAS (F) for 24 hours after which the percent viable cells was determined. Annexin-V/PI–negative cells were scored as viable. IL-7 was added to a final concentration of 20 ng/mL. Bars represent the average of 3 independent experiments done in triplicate, and the error bars denote the SEM. Asterisk (*) indicates P < .001 by 2-way analysis of variance with a Bon Ferroni posttest between treatments linked with horizontal line.
Navitoclax has been proposed to promote apoptosis by displacing proapoptotic BH3-only family members from antiapoptotic molecules, thus facilitating proapoptotic effector (BAX and BAK) activation and cell death.35,36 To test this hypothesis, in vitro–generated p185+Arf−/−Mcl-1wt cells (referred to as BCR-ABL B-ALL cells) were treated with navitoclax and immunoprecipitated with anti-BIM antibodies to measure the BH3-only occupancy of antiapoptotic BCL-2 family members, as BIM interacts with BCL-2, BCL-XL, and MCL-1.37,38 As the navitoclax dose increased, the amount of BCL-2 and BCL-XL immunoprecipitating with BIM decreased, and there was a corresponding increase in MCL-1 protein associated with BIM (Figure 6B). These data indicated that navitoclax displaced BIM from BCL-2 and BCL-XL and suggest that the liberated BIM could then interact with MCL-1 (Figure 6B). BIM is a relevant proapoptotic molecule as Bim-deficient p185-transformed leukemic cells, derived from Bim-conditional BM transduced with p185-IRES-Cre retrovirus, are partially resistant to TKI and navitoclax treatment consistent with previously published reports (supplemental Figure 6).24-26,35 These data suggest that treatment with navitoclax may be less effective at killing BCR-ABL B-ALL cells because the leukemic cells may have sufficient MCL-1 to “buffer” the release of proapoptotic molecules, such as BIM, from BCL-2 and BCL-XL.
Treatment of BCR-ABL B-ALL cells with TKIs decreased the amount of MCL-1 protein expressed and induced cell death but did not modulate BCL-2, BCL-XL, or BIM expression (Figure 1A-B and Figure 6C-D). However, TKI treatment did not disrupt the interaction between BCL-2, BCL-XL, or MCL-1 with proapoptotic BIM as detected by co-immunoprecipitation (Figure 6C-D). Because TKIs decreased MCL-1 protein expression, we hypothesized that we could potentiate the efficacy of navitoclax on BCR-ABL B-ALL cells by combining navitoclax with TKI treatment to decrease MCL-1 expression in the cells. We treated BCR-ABL B-ALL cells with low doses of IM (Figure 6E) or DAS (Figure 6F) that each induce ∼10% apoptosis and coadministered increasing doses of navitoclax. As the navitoclax was increased, we observed potentiation of the TKIs indicating a synergistic effect of combining navitoclax and TKI treatment (Figure 6E-F). Growth factor signaling by IL-7 can partially overcome the death induced by TKI treatment; therefore, we tested whether IL-7 could overcome the potentiation of combining TKI treatment and navitoclax. Although exogenous IL-7 treatment reduced the effectiveness of TKI treatment and reduced the killing observed, a statistically significant effect of combining TKIs with navitoclax was observed (Figure 6E-F). These data demonstrate that combining TKIs and navitoclax can enhance activity against BCR-ABL B-ALL cells and overcome the resistant effects of growth factor signaling.
Potentiation of TKI activity by small-molecule BCL-2 inhibitors in human BCR-ABL leukemia cell lines
To test whether similar potentiation of cell death can be observed in human Ph+ leukemia cell lines, we tested whether a similar relationship between BCR-ABL signaling and MCL-1 expression could be observed. In OP-120 and TOM119 (Ph+ B-ALL lines) and BV17321 (Ph+ CML blast crisis line), we observed that treatment with suboptimal doses of either TKI decreased MCL-1 expression but did not alter the expression of other antiapoptotic BCL-2 family members (ie, BCL-2, BCL-XL, or BFL-1) and only modestly decreased BCL-w in OP-1 cells (Figure 7). Therefore, among antiapoptotic BCL-2 family members, only MCL-1 expression declines robustly in response to TKI treatment. We hypothesized that we may be able to potentiate the efficacy of navitoclax on the Ph+ cell line by combining with TKI treatment to decrease MCL-1 expression. Thus, we treated the human Ph+ cell lines with doses of TKIs that each induced ∼10% apoptosis and coadministered increasing navitoclax doses (Figure 7). As navitoclax was increased in the cultures, we observed a potentiation of the TKIs to induce apoptosis (Figure 7). These data indicate that like our observations in mouse BCR-ABL cell lines, combining TKIs and navitoclax in human Ph+ cell lines can also lead to enhanced activity. Although MCL-1 expression is most overtly decreased in response to TKI treatment, it is possible that combining TKI and navitoclax may affect targets other than MCL-1 leading to cell death. These data suggest that combining TKIs and navitoclax may be effective in treating human Ph+ leukemia.
Potentiation of Ph+human cell lines to combining TKI and small-molecule BCL-2 inhibitors. Human Ph+ cell lines: OP-1 Ph+ B-ALL (A) TOM1 Ph+ B-ALL (B), and BV173 Ph+ CML blast crisis (C) cell lines were treated with navitoclax, IM, or DAS at indicated doses for 24 hours after which cell lysates were analyzed for expression of MCL-1, BCL-2, BCL-w, BFL-1, and BCL-XL. For potentiation experiments, the percentage of viable cells was determined by annexin-V and PI negativity. Bars represent the average of 3 independent experiments, each done in triplicate, for each cell type, and the error bars denote the SEM. Asterisk (*) indicates P < .001 by 2-way analysis of variance with a Bon Ferroni posttest between treatments linked with horizontal line.
Potentiation of Ph+human cell lines to combining TKI and small-molecule BCL-2 inhibitors. Human Ph+ cell lines: OP-1 Ph+ B-ALL (A) TOM1 Ph+ B-ALL (B), and BV173 Ph+ CML blast crisis (C) cell lines were treated with navitoclax, IM, or DAS at indicated doses for 24 hours after which cell lysates were analyzed for expression of MCL-1, BCL-2, BCL-w, BFL-1, and BCL-XL. For potentiation experiments, the percentage of viable cells was determined by annexin-V and PI negativity. Bars represent the average of 3 independent experiments, each done in triplicate, for each cell type, and the error bars denote the SEM. Asterisk (*) indicates P < .001 by 2-way analysis of variance with a Bon Ferroni posttest between treatments linked with horizontal line.
Discussion
It has been reported that genetic deletion of Bcl2l1 (encoding the BCL-X antiapoptotic protein) in a mouse model of BCR-ABL B-ALL did not adversely affect leukemogenesis and paradoxically resulted in the increased proliferation of the BCL-X–deficient B-ALL cells.39 In contrast, our experiments reveal that MCL-1 is the essential endogenous promoter of survival both during the initial transformation of B-lineage progenitors with BCR-ABL and also in p185-transformed leukemic cells. The requirement for MCL-1 is critical as both in vitro and in vivo leukemic cells exhibit strong selection pressure against Mcl-1 deletion as mediated by mutagenesis of Cre or mutation of 1 of the loxP sites flanking the Mcl-1 locus. The dependence on MCL-1 is striking as both B-lineage progenitors and leukemic cells simultaneously express other antiapoptotic molecules; however, the endogenous expression of these molecules is insufficient to allow survival of the cells without MCL-1.
One potential rationale for the absolute dependence on MCL-1 is that it possesses unique functions that cannot be compensated for by other antiapoptotic BCL-2 family members.18 However, although MCL-1 may have additional roles beyond preventing cell death, our data demonstrate that either loss of both proapoptotic effectors (BAX and BAK) or ectopic expression of human BCL-2 or human BCL-XL can rescue the survival and growth of Mcl-1–deleted leukemic cells. Therefore, the primary function of MCL-1 in this mouse model of Ph+ B-ALL is to block apoptosis.
Our results are consistent with a model in which the “pool” of antiapoptotic MCL-1 in cells is the critical regulator of cellular survival. Signaling, either through either cytokine receptors or by the BCR-ABL oncofusion, promotes MCL-1 expression to counter proapoptotic molecules in the cells. When signaling diminishes, either due to a lack of cytokines or after TKI treatment, MCL-1 levels decrease becoming insufficient to counter the proapoptotic molecules. Supporting this model, treatment of primary chronic lymphocytic leukemia with TKIs also reduces MCL-1 expression.40 In contrast, when exogenous BCL-2 and BCL-XL are overexpressed in the cells, it expands the “pool” of antiapoptotic modulators allowing the cells to counter proapoptotic expression despite a loss of MCL-1 function.
A dominant role for MCL-1 in promoting leukemic cell survival is also seen in mouse models of acute myelogenous leukemia (AML).41,42 Strikingly, whether the AML cells were driven by c-Myc, MLL-ENL, MLL-AF9, Mixl1, AML1-ETO9a, or Hox19, the resultant leukemia were similarly dependent on MCL-1.41,42 Similar data exist for human CML cells driven by the BCR-ABL oncofusion.43 Furthermore, in these models, MCL-1’s primary function is to prevent apoptosis as ectopic expression of other antiapoptotic family members promoted leukemia survival in these models.41,42 Therefore, strategies to inhibit MCL-1’s antiapoptotic function would be predicted to be therapeutically beneficial. However, MCL-1 is an important modulator of the survival of a number of normal cell lineages including, but not limited to, hematopoietic lineages.14,44,45 Thus, it may be difficult to inhibit MCL-1 in malignant cells without inducing toxicity in normal cells. An alternative strategy may be to modulate MCL-1 expression levels to reduce the size of the antiapoptotic “pool” in cancer cells to foster lethality of malignant cells without inducing toxicity to normal cell lineages.
MCL-1 is a transcriptional target of cytokine and growth factor signaling.14,44,46 Additionally, MCL-1 protein is short lived being a proteasome substrate.47,48 MCL-1 degradation is also regulated, as posttranslational modifications play a critical role in regulating its elimination.49,50 MCL-1 can also undergo ubiquitinylation-independent protease degradation.22 MCL-1’s labile nature raises the possibility that it may be possible to simply perturb its expression. Indeed, combining inhibitors of cellular signaling to potentiate the death induced by TKIs has already shown promise in CML models,51-53 and in some cases, these combined therapies decreased MCL-1 expression.54 Identification of these pathways downstream of BCR-ABL signaling may identify further ways by which MCL-1 expression levels can be modulated in leukemic cells.
When used alone, navitoclax is relatively ineffective at treating BCR-ABL B-ALL cell lines in vitro, presumably as it displaces proapoptotic molecules (ie, BIM and others) from BCL-2 and BCL-XL, but the liberated proapoptotic molecules can still be sequestered by MCL-1.32,33 However, when TKIs are combined with navitoclax, cell death is potentiated as there is less MCL-1 available to sequester proapoptotic molecules. Although we cannot rule out the possibility that combining these inhibitors may have effects beyond merely modulating MCL-1 function, MCL-1 is the only antiapoptotic molecule whose expression is significantly altered by TKI treatment. Our synergy experiments are similar to those in BCR-ABL models of CML or gastrointestinal stromal cell tumor cells in which combinatorial treatment with TKIs and BH3 mimetics like navitoclax has been shown to be synergistic.26,55,56
MCL-1 is a critical prosurvival molecule required to promote the survival of B-cell progenitor populations during BCR-ABL transformation as well as for the continued survival of the BCR-ABL B-ALL cells. These findings highlight the potential for targeting MCL-1 protein expression and function in treating this BCR-ABL B-ALL. We submit that although specific MCL-1 inhibitors may be effective at treating BCR-ABL leukemia, approaches that would alter the expression levels of MCL-1 should also be effective at sensitizing this leukemia to other treatment modalities including BCL-2 inhibitors such as navitoclax.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the input of the St. Jude Biochemistry Department, members of the Opferman laboratory, and helpful discussions with Drs J. Ihle, T. Gruber, C. Mullighan, and C. Sherr.
This work was supported by grants from the National Institutes of Health National Heart, Lung, and Blood Institute (R01HL102175) (J.T.O.) and the American Cancer Society (119130-RSG-10-255-01-LIB) (J.T.O.), a Cancer Center Support Grant P30CA021765, and ALSAC.
Authorship
Contribution: R.T.W. and J.T.O. conceived the study and designed the experiments; B.K. and J.M. performed the experiments, analyzed data, and prepared figures; B.K., J.M., R.M.P., and H.S. generated reagents and discussed findings; J.E.R. analyzed pathology; and J.T.O. supervised the project and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joseph T. Opferman, St. Jude Children’s Research Hospital, MS 340, Room D4007C, 262 Danny Thomas Place, Memphis, TN 38105-3678; e-mail: Joseph.Opferman@stjude.org.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal